Introduction
Colon cancer is one of the leading causes of
cancer-associated mortality worldwide, with >800,000 recorded
cases in 2018, for which radical surgery is the standard treatment
(1). The prognosis of patients with
colon cancer remains poor primarily due to recurrence (2). Upregulation of the expression of
oncogenes is an important contributor to the etiology of cancer,
and can promote abnormal proliferation and cell cycle progression
(3). Hence, it is imperative to
develop novel and reliable prognostic markers for patients with
colon adenocarcinoma.
The eukaryotic initiation factor 3 (EIF3) is a
complex translation initiation factor composed of 13 subunits
(EIF3A to EIF3M) that is involved in mRNA modulation (4). The EIF3 complex is required for key
steps in the initiation of protein synthesis (5). Dysregulated EIF3 subunits have been
implicated in neurodegenerative disorders (such as Parkinson's
disease), infection and tumorigenesis (6). Previous studies have demonstrated that
EIF3 subunits regulate the AMP-activated protein kinase α (AMPKα),
AKT/PI3K/mTOR and stress-activated kinase/JNK signaling pathways
and the BCL-2 family of proteins, and play an important role in the
development and growth of colon neoplasms (7,8).
EIF3M encodes a protein of 42.5 kDa that is necessary for
maintaining the structural integrity and translation initiation
function of EIF3, and is also crucial for mouse embryonic
development (9). EIF3M is
upregulated in colon cancer and involved in the regulation of
tumorigenesis-related genes, including migration inhibitory factor
(MIF) and metallothionein 2 (MT2) (10,11).
Silencing EIF3M expression leads to apoptosis of the HCT-116 colon
cancer cell line (11). A previous
study demonstrated that zinc family member 1 (ZIC1) was upregulated
in liposarcoma, and knockdown of ZIC1 in liposarcoma cell lines was
associated with the degradation of EIF3M (12). Hence, EIF3M may be a pro-survival
downstream target of ZIC1. These studies suggest that EIF3M
expression is essential for carcinogenesis and could be used to
develop a novel therapy for various cancer types.
Due to no studies reporting its prognostic role in
the colon carcinoma, the present research investigated EIF3M
expression in colon cancer by using a variety of laboratory
techniques in conjunction with the Oncomine database, and its
clinicopathological and prognostic value in patients with colon
adenocarcinoma was explored.
Materials and methods
Tissue samples
This study was approved by the Kunshan First
People's Hospital Ethics Committee (Kunshan, China) and written
informed consent was obtained from all the patients. The clinical
and pathological data of 82 patients with colon adenocarcinoma
(ratio male:female, 0.78:1) who had not received any radiotherapy
or chemotherapy before surgery were reviewed. All cases were
diagnosed with adenocarcinoma of the colon and underwent radical
surgery at Kunshan First People's Hospital between January 2010 and
December 2012. Patients were diagnosed with Dukes' stage B or C
disease, and received 8 courses of XELOX regimen (oxaliplatin
combined with capecitabine; 130 mg/m2 oxaliplation IV on
the first day and 2,000 mg/m2/day capecitabine for two
weeks) (13). The mean age of the
patients was 55.69±12.54 years, and the follow-up duration ranged
from 3–60 months. The serum of 20 patients with colon
adenocarcinoma patients at Dukes' stage B or C before surgery and
80 healthy controls was collected to perform ELISAs. Additionally,
20 pairs of fresh-frozen colon tumors and matched normal tissues
(>5.0 cm from tumor tissues) obtained from patients with colon
adenocarcinoma were collected for total protein and mRNA
extraction. The levels of CEA, CA19-9 and CA12-5 were investigated
by ELISA in the laboratory department of Kunshan First People's
Hospital (Kunshan, China) when patients were hospitalized.
Immunohistochemistry (IHC) and
evaluation of immunohistochemical staining
Tissues were fixed in 10% formalin at 20°C for 8 h
and then embedded in paraffin blocks. 5-µm paraffin-embedded
sections were used for EIF3M immunohistochemical staining with an
SP Rabbit and Mouse HRP kit (cat. no. CW2069M, CoWin Biosciences).
Endogenous peroxydase enzymes blocking buffer was used at 20°C for
10 min. And then, normal goat serum was also used for blocking at
20°C for 10 min. These two blocking reagents were constituent parts
of this kit. The primary antibody, EIF3M rabbit polyclonal antibody
(cat. no. bs-9033R, BIOSS), was diluted at 1:100 in
phosphate-buffered saline (PBS). PBS without primary antibodies was
used as a negative control. The SP Rabbit and Mouse HRP kit (cat.
no. CW2069M; CoWin Biosciences) was used to conduct a secondary
incubation at 20°C for 10 min, according to the manufacturer's
protocol. Two pathologists independently evaluated the
immunoreactivity scores (IRS) for EIF3M expression through a
semi-quantitative assessment system. Slides were photographed using
an inverted light microscope (magnification, ×400; Nikon
Corporation). The IRS values were a combination of a score for the
staining intensity and a score of the percentage of cells. The
staining intensity was defined as: 0, no staining; 1, mild
staining; 2, moderate staining; and 3, strong staining. The scores
for the percentage of cells were defined as: ‘0–100%’ = (0, 0%; 1,
1–10%; 2, 11–20%; 3, 21–30%; 4, 31–40%; 5, 41–50%; 6, 51–60%; 7,
61–70%; 8, 71–80%; 9, 81–90%; and 10, 91–100%.). The total IRS was
calculated by multiplying the staining intensity score by the
staining percentage score, and ranged from 0 to 30. Any
disagreement was resolved by discussion. EIF3M expression was
considered positive only when IRS >10.
Western blotting
A total of 20 pairs of fresh-frozen specimens were
used for western blotting. Total protein of each tissue was
extracted using RIPA lysis buffer (Beyotime Institute of
Biotechnology) containing protease inhibitor cocktail (Pierce;
Thermo Fisher Scientific, Inc.). The supernatants were collected
and their protein concentration was measured using a BCA protein
assay kit (Pierce; Thermo Fisher Scientific, Inc.), 20 µg of each
sample was loaded per lane on 8–16% gels (Beyotime Institute of
Biotechnology) and proteins were separated by SDS-PAGE (EMD
Millipore). Proteins were transferred to PVDF membranes (Beyotime
Institute of Biotechnology). Membranes were blocked in 5% non-fat
dry milk with TBST for 1 h at room temperature. EIF3M rabbit
polyclonal antibody (1:500; BIOSS) and β-actin mouse monoclonal
antibody (cat. no. AF0003; 1:1,000; Beyotime Institute of
Biotechnology) were used to incubate membranes at 4°C overnight.
Secondary antibodies (1:1,000; anti-rabbit, cat. no. A0208; and
anti-mouse, cat. no. A0216; both Beyotime Institute of
Biotechnology) conjugated with horseradish peroxidase were used at
37°C for 2 h, and then detected using an Enhanced Chemiluminescence
Detection system (Beyotime Institute of Biotechnology). The
relative densities were quantified with a digital imaging analyzer,
ImageJ version 1.4.1 (National Institutes of Health). EIF3M
expression was normalized to β-actin.
RT-qPCR
TRIzol reagent (Thermo Fisher Scientific, Inc.) was
used to isolate the total RNA from each tissue, and 2 µg RNA was
reverse transcribed using the SuperScript II RNase-Reverse
Transcriptase system (Invitrogen; Thermo Fisher Scientific, Inc.)
at 50°C for 15 min, and then 85°C for 2 min. qPCR was performed
using an iQ5 real-time PCR detection system (Bio-Rad Laboratories,
Inc.) with the SYBR Premix Ex Taq™ kit (Takara Bio, Inc.). The PCR
cycling conditions were as follows: 94°C for 4 min, followed by 40
cycles of 95°C for 1 min, 60°C for 1 min and 72°C for 1 min. PCR
primers were designed as follows: EIF3M forward,
5′-ATGTAACAGGCCAAGTGAATC-3′ and reverse,
5′-CACAGGTGTATTGTACGAGCAT-3′ (198 bp); and β-actin forward,
5′-GGGAAATCGTGCGTGACATTAAGG-3′ and reverse,
5′-CAGGAAGGAAGGCTGGAAGAGTG-3′ (185 bp). The relative expression of
EIF3M was expressed using the 2−ΔΔCq method, where
ΔΔCq=(CtTumor-eIF3m-CtTumor-β-actin)-(CtNormal-eIF3m-CtNormal-β-actin)
(14).
ELISA
The EIF3M ELISA kit (cat. no. S00143, Shanghai
Yuanye Biotechnology Co., Ltd) was used to detect the levels of
EIF3M in serum of 20 patients with colon adenocarcinoma patients
and 80 healthy controls. Following the manufacturer's instructions,
the OD value for each sample was detected using a microplate reader
(wavelength, 450 nm), and the level of EIF3M was calculated using a
standard curve drawn using Excel 2016 (Microsoft Corporation).
Oncomine database analysis
The following 6 datasets were selected from the
Oncomine database (https://www.oncomine.org/): Skrzypczak Colorectal (69
samples); Skrzypczak Colorectal 2 (15 samples); Graudens Colon (30
samples); Ki Colon (91 samples); TCGA Colorectal (123 samples) and
Hong Colorectal (82 samples) (15–18).
These datasets compare expression of EIF3M in colon or colorectal
carcinoma with expression in normal tissues.
Statistical analysis
SPSS 20.0 (IBM Corp.) and GraphPad 6.0 (GraphPad
Software, Inc.) were used for statistical analyses, and P<0.05
was considered to indicate a statistically significant difference.
Pearson's χ2 test was used to analyze the association of
EIF3M expression with clinicopathological characteristics.
Continuous variables, expressed as the mean ± SD, were analyzed
using a Student's t-test; a paired t-test was used for comparing
tumors with adjacent normal tissues, and an unpaired t-test was
used for comparing the serum of patients with colon adenocarcinoma
with that of the healthy controls. In addition, Cox univariate and
multivariate regression analysis and Kaplan-Meier curves with log-
rank test were also used to analyze overall survival (OS) and
disease-free survival (DFS).
Results
Expression of EIF3M in colon
adenocarcinoma
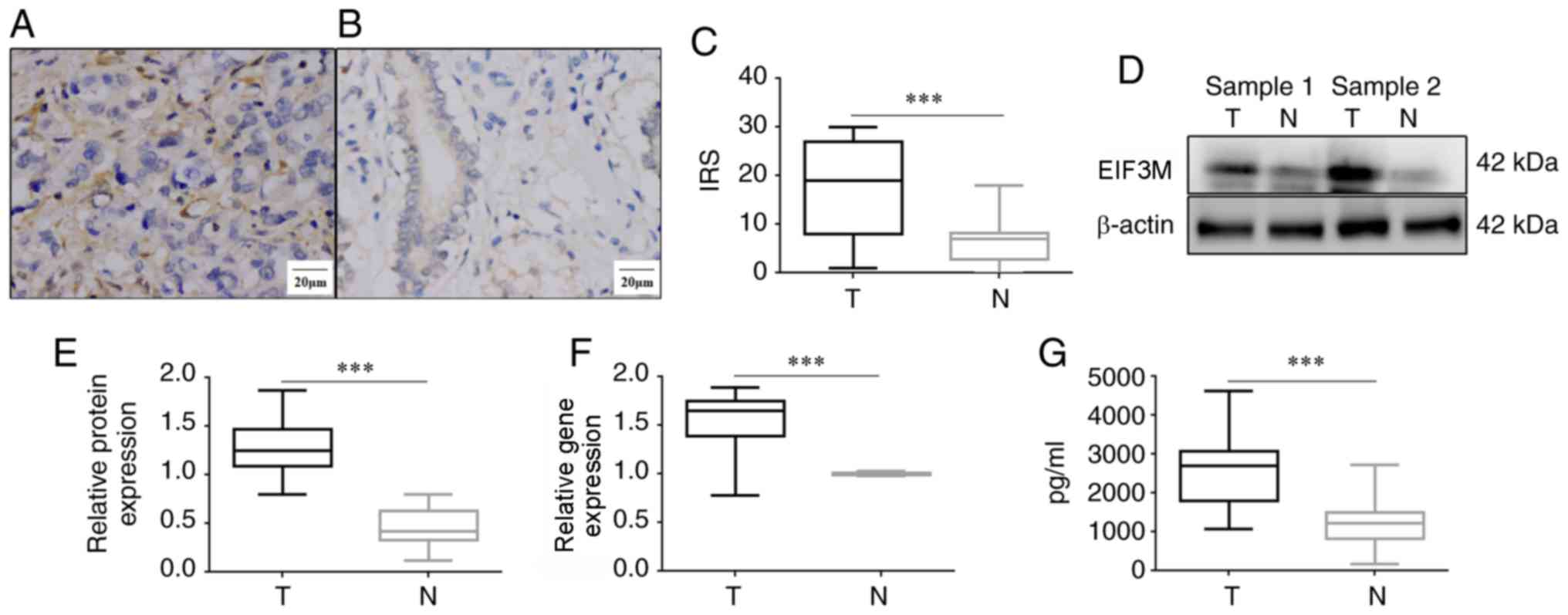
EIF3M was expressed in the cytoplasm in both colon
adenocarcinoma and normal colon tissues (Fig. 1A and B). The positive rate of EIF3M
in colon adenocarcinoma was higher than that in normal colon
tissues (62.20 vs. 29.27%; P<0.001). The mean IRS of EIF3M in
colon adenocarcinoma was significantly higher than that in normal
colon tissues (17.28±10.05 vs. 6.53±4.87; P<0.001; Fig. 1C). The levels of EIF3M mRNA and
protein in freeze-thawed tumors were higher than those in
corresponding normal tissues (P<0.001; Fig. 1D-F). In addition, the average level
of EIF3M in the serum supernatants of 20 patients with colon
adenocarcinoma was significantly higher compared with that in 80
healthy controls (2625.3±986.4 vs. 1203.3±493.5 pg/ml; t=9.17;
P<0.001).
Association between EIF3M and
clinicopathological parameters in colon adenocarcinoma
According to the IRS values, 51 patients were
classed into the ‘positive group’ (IRS >10) and used for
subsequent analysis. As presented in Table I, positive expression of EIF3M was
associated with tumor size (P=0.002) and Dukes' stage (P<0.001),
whilst there was no association found between EIF3M expression and
other parameters, including age, sex, location, differentiation,
CEA, CA19-9 and CA12-5.
 | Table I.Association of EIF3M expression in
patients with colon adenocarcinoma with clinicopathological
variables. |
Table I.
Association of EIF3M expression in
patients with colon adenocarcinoma with clinicopathological
variables.
|
|
| EIF3M
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Variables | n | Positive, n
(%) | Negative, n
(%) | χ2 | P-value |
|---|
| Total | 82 | 51 (62.20) | 31 (37.80) |
|
|
| Age, years |
|
|
| 0.049 | 0.825 |
|
≤55 | 49 | 30 (58.82) | 19 (61.29) |
|
|
|
>55 | 33 | 21 (41.18) | 12 (38.71) |
|
|
| Sex |
|
|
| 0.032 | 0.858 |
|
Male | 36 | 22 (43.14) | 14 (45.16) |
|
|
|
Female | 46 | 29 (56.86) | 17 (54.84) |
|
|
| Tumor size, cm |
|
|
| 10.038 | 0.002 |
| ≤5 | 48 | 23 (45.10) | 25 (80.65) |
|
|
|
>5 | 34 | 28 (54.90) | 6 (19.35) |
|
|
| Location |
|
|
| 0.848 | 0.357 |
| Right
colon | 37 | 21 (41.18) | 16 (51.61) |
|
|
| Left
colon | 45 | 30 (58.82) | 15 (48.39) |
|
|
|
Differentiation |
|
|
| 5.597 | 0.061 |
| I | 13 | 7 (13.73) | 6 (19.35) |
|
|
| II | 40 | 21 (41.18) | 19 (61.30) |
|
|
|
III | 29 | 23 (45.09) | 6 (19.35) |
|
|
| Dukes' stage |
|
|
| 14.366 | <0.001 |
| B | 47 | 21 (41.18) | 26 (83.87) |
|
|
| C | 35 | 30 (58.82) | 5 (16.13) |
|
|
| Serum CEA,
ng/ml |
|
|
| 3.319 | 0.068 |
|
<5 | 32 | 16 (31.37) | 16 (51.61) |
|
|
| ≥5 | 50 | 35 (68.63) | 15 (48.39) |
|
|
| Serum CA19-9,
U/ml |
|
|
| 3.701 | 0.054 |
|
<37 | 69 | 46 (90.20) | 23 (74.19) |
|
|
|
≥37 | 13 | 5 (9.80) | 8 (25.81) |
|
|
| Serum CA12-5,
U/ml |
|
|
| 2.416 | 0.120 |
|
<35 | 55 | 31 (60.78) | 24 (77.42) |
|
|
|
≥35 | 27 | 20 (39.22) | 7 (22.58) |
|
|
Association between EIF3M expression
and OS
In the Kaplan-Meier analysis, the OS rate of the
‘positive group’ was significantly lower than the ‘negative group’
(P=0.006; Fig. 2A). Based on
univariate Cox regression analysis, EIF3M, tumor size,
differentiation, Dukes' stage, serum CEA and serum CA12-5 were
associated with OS (P<0.01; Table
II). Based on multivariate analysis, EIF3M expression
(P=0.003), differentiation (P<0.001), Dukes stage (P<0.001)
and serum CEA (P<0.001) were independent prognostic factors for
OS rate for patients with colon adenocarcinoma (Table III).
 | Table II.Prognostic role of EIF3M expression
and clinicopathological factors for OS and DFS of patients with
colon cancer by univariate Cox regression analysis. |
Table II.
Prognostic role of EIF3M expression
and clinicopathological factors for OS and DFS of patients with
colon cancer by univariate Cox regression analysis.
|
| OS | DFS |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| EIF3M expression,
high vs. low | 2.21 | 1.22–4.02 | 0.009 | 2.17 | 1.27–3.70 | 0.004 |
| Age, >55 vs. ≤55
years | 1.57 | 0.89–2.76 | 0.119 | 1.43 | 0.79–2.56 | 0.238 |
| Sex, male vs.
female | 1.26 | 0.84–1.90 | 0.264 | 1.37 | 0.93–2.02 | 0.111 |
| Tumor size, >5
vs. <5 cm | 1.36 | 1.10–1.69 | 0.004 | 1.24 | 0.82–1.88 | 0.308 |
| Location, left vs.
right | 1.63 | 0.73–3.63 | 0.233 | 1.37 | 0.74–2.53 | 0.316 |
| Differentiation,
III vs. I and II | 1.80 | 1.48–2.19 | <0.001 | 1.52 | 1.27–1.80 | <0.001 |
| Dukes' stage, C vs.
B | 2.23 | 1.24–4.00 | 0.007 | 2.05 | 1.11–3.76 | 0.022 |
| Serum CEA, ≥5 vs.
<5 ng/ml | 1.94 | 1.59–2.35 | <0.001 | 1.83 | 1.44–2.33 | <0.001 |
| Serum CA19-9, ≥37
vs. <37 U/ml | 1.49 | 0.83–2.69 | 0.182 | 2.24 | 0.66–7.57 | 0.194 |
| Serum CA12-5, ≥35
vs. <35 U/ml | 2.36 | 1.60–3.49 | <0.001 | 1.64 | 1.29–2.08 | <0.001 |
 | Table III.Prognostic role of EIF3M expression
and clinicopathological factors for OS and DFS of patients with
colon cancer by multivariate Cox regression analysis. |
Table III.
Prognostic role of EIF3M expression
and clinicopathological factors for OS and DFS of patients with
colon cancer by multivariate Cox regression analysis.
|
| OS | DFS |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| EIF3M expression,
high vs. low | 2.46 | 1.37–4.43 |
0.003 | 2.02 | 1.36–2.99 | 0.001 |
| Differentiation,
III vs. I and II | 2.72 | 1.52–4.89 | <0.001 | 1.98 | 1.18–3.30 | 0.009 |
| Dukes' stage, C vs.
B | 3.07 | 2.08–4.54 | <0.001 | 2.42 | 1.45–4.03 | 0.001 |
| Serum CEA, ≥5 vs.
<5 ng/ml | 3.19 | 1.78–5.73 | <0.001 | 1.67 | 1.13–2.47 | 0.011 |
Association between EIF3M expression
and DFS
The DFS rate of patients with positive expression of
EIF3M was significantly lower than that of patients with negative
expression (P=0.002; Fig. 2B). Based
on univariate Cox regression analysis, EIF3M, differentiation,
Dukes' stage, serum CEA and serum CA12-5 were all significantly
associated with DFS (P<0.05; Table
II). In multivariate Cox regression analysis, EIF3M expression
(P=0.001), differentiation (P=0.009), Dukes' stage (P=0.001) and
serum CEA (P=0.011) were independent prognostic factors for DFS
(Table III).
EIF3M expression in colon cancer using
Oncomine
All 6 datasets from the Oncomine database showed a
higher expression of EIF3M in colon/colorectal carcinoma tissues
compared with normal tissues (P<0.001; Fig. 3A). The Ki Colon dataset also
highlighted the top 15 genes related to EIF3M by co-expression
analysis (Fig. 3B). These genes were
all expressed to significantly higher levels in colon
adenocarcinoma compared with normal tissues, and exhibited a strong
co-expression correlation (correlation index, 0.675–0.876).
Information and the functions of these 15 co-expression genes are
listed in Table IV. In addition,
EIF3M expression was higher in colon adenocarcinoma compared with
that in normal colon tissues, colon squamous cell carcinomas or
gastrointestinal stromal tumors (Fig.
3B).
 | Table IV.Information and functions of the top
15 genes associated with EIF3M expression using the Ki Colon
dataset. |
Table IV.
Information and functions of the top
15 genes associated with EIF3M expression using the Ki Colon
dataset.
| Genes | Chromosomal
location | Ensembl
version | Locus type | Functions |
|---|
| IPO7 | 11p15.4 |
ENSG00000205339 | Gene with protein
product | Ribosome biogenesis
related proteins; carcinogenesis (28) |
| SULT4A1 | 22q13.31 |
ENSG00000130540 | Gene with protein
product | Neuronal-associated
genes; carcinogenesis (29) |
| GNE | 9p13.3 |
ENSG00000159921 | Gene with protein
product | GNE myopathy
(30) |
| NOB1 | 16q22.1 |
ENSG00000141101 | Gene with protein
product | Carcinogenesis in
colorectal cancer (31) |
| EIF2S2 | 20q11.22 |
ENSG00000125977 | Gene with protein
product | Essential for cell
proliferation; regulators of oncogenesis (32) |
| UBA2 | 19q13.11 |
ENSG00000126261 | Gene with protein
product | Carcinogenesis
(33) |
| ATIC | 2q35 |
ENSG00000138363 | Gene with protein
product | Polymorphisms in
cancer (34) |
| CCT6B | 17q12 |
ENSG00000132141 | Gene with protein
product | Implicated in
cancer (35) |
| CCT6P1 | 7q11.21 |
ENSG00000228409 | Pseudogene | Associated with
sickle cell disease (36) |
| DARS | 2q21.3 |
ENSG00000115866 | Gene with protein
product | Reinitiating DNA
replication (37) |
| CCDC34 | 11p14.1 |
ENSG00000109881 | Gene with protein
product | Carcinogenesis in
colorectal cancer (38) |
| RNASEH2B | 13q14.3 |
ENSG00000136104 | Gene with protein
product | Its deletion
related to cancer occurrence (39) |
| SNRPE | 1q32.1 |
ENSG00000182004 | Gene with protein
product | Poor prognostic
indicator (40) |
| CDK7 | 5q13.2 |
ENSG00000134058 | Gene with protein
product | Transcriptional
cyclin-dependent kinase; carcinogenesis (41) |
| SNRPB2 | 20p12.1 |
ENSG00000125870 | Gene with protein
product | mRNA splicing;
carcinogenesis(42) |
Discussion
Colon cancer is the third most common malignancy and
has become a great public health concern (1). In order to explore novel and valuable
biomarkers for colon cancer, the present study investigated EIF3M
expression and evaluated its clinicopathological and prognostic
roles in patients with colon cancer. EIF3M is one of the most
pivotal subunits of the EIF3 complex and accelerates protein
synthesis and ribosomal recycling (6). A previous study revealed the elevated
expression of EIF3M and other core subunits is indispensable to
carcinogenesis. Goh et al (11) reported that EIF3M was upregulated in
colon cancer and colon cancer cell lines. After knocking down EIF3M
expression in the HCT-116 colon cancer cell line, proliferation was
reduced and the apoptosis rate was promoted due to a prolonging of
the sub-G0/G1 stage of the cell cycle (11). In concordance with previous studies,
the current study proved that the expression of EIF3M in colon
adenocarcinoma was significantly higher when compared with normal
tissues. In addition, EIF3M expression was associated with tumor
size and Dukes' stage. In Kaplan-Meier analysis and Cox regression
analysis, the role of EIF3M in negatively influencing prognosis of
patients with colon adenocarcinoma was confirmed. Therefore,
positive EIF3M expression may indicate that patients with colon
adenocarcinoma may be at a later Dukes' stage and have a worse
prognosis.
Dukes' stage was first established in 1935, and is a
traditional medical clinical classification, including stages A to
D (13). Patients at stage A weren't
treated with chemotherapy and patients at stage D had not received
a radical resection. To more accurately assess the role of EIF3M in
colon carcinoma, the current study only enrolled patients at stage
B or C. Tumors at stage B invaded the serosa without any lymph node
metastasis, whilst tumors at stage C invaded the serosa with lymph
node metastasis. The influence of EIF3M expression on lymph node
metastasis was not assessed separately. In this study, positive
EIF3M expression was higher in colon adenocarcinoma patients at
stage C than those at stage B. In addition, Dukes' stage was an
independent factor of OS and DFS, which suggested that colon
adenocarcinoma patients with positive EIF3M expression in tumors
had a worse prognosis. Hence, further research is required to
identify if EIF3M could be used as a therapeutic target for
anticancer drugs. As serum levels of EIF3M could be found in the
blood, further studies are needed to evaluate the role of EIF3M in
monitoring of colon cancer.
A study by Goh et al (11) has demonstrated the molecular
mechanism of EIF3M in colon cancer. MIF and MT2 expression was
found to be downregulated when EIF3M was knocked down in the
HCT-116 colon cancer cell line (11). MIF not only plays a critical role in
inflammation and immunity by deregulating the inhibitory effect of
glucocorticoids, but is also associated with tumorigenesis and
tumor growth (19,20). MT2 is a member of the metallothionein
family that performs a plethora of metal ion-related events in
stress responses, tumorigenesis, neurodegeneration and inflammation
(21). A previous study demonstrated
that MT2 is involved in cell migration, proliferation and
angiogenesis and could be a novel regulator of vascular endothelial
growth factor C expression (22). In
addition, EIF3M knockdown interfered with cell cycle regulation and
induced cell apoptosis by degradation of cell division cycle 25
homolog A (22).
It has been shown that other EIF3 subunits are also
associated with colon cancer. EIF3A is overexpressed in colon
tumors, and elevated EIF3A expression inhibits Caco-2 cell
differentiation (23). EIF3B
knockdown inhibits the proliferation and increases the apoptotic
rate of SW1116 colon cancer cells (24). EIF3C gene knockdown suppresses the
proliferation of RKO colon cancer cells; the cell cycle is arrested
at G0/G1 stage and apoptosis is also induced (25). Knockdown of EIF3D inhibits
proliferation of HCT116 colon cancer cells via modulating AMPKα,
glycogen synthase kinase 3β and JNK (8). Knockdown of EIF3E reduces the
proliferation and clonality of HCT116 cells and promotes cell
apoptosis (26). In addition, high
EIF3E expression might predict poor prognosis in colon cancer
(26). Qi et al (27) demonstrated that EIF3I was a
proto-oncogene in colon carcinoma, and worked by activating the
β-catenin signaling pathway and upregulating cyclooxygenase 2. This
body of research shows that translation initiation factors
including EIF3A, EIF3B, EIF3C, EIF3D, EIF3E, EIF3I and EIF3M
promote the formation of colon neoplasms. The present study
demonstrated that EIF3M could be an indicator of poor prognosis in
patients with colon adenocarcinoma. The prognostic role of all the
subunits of the EIF3 complex should be investigated in the future.
In conclusion, elevated EIF3M expression in patients with colon
adenocarcinoma was associated with larger tumor size, Dukes' stage
C, and worse OS and DFS rates. Therefore, upregulated EIF3M is a
putative candidate biomarker for poor prognosis in colon
adenocarcinoma patients.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Kunshan
Science and Technology Program of Social Development (grant no.
KS1659) and the Binhai Science and Technology Program (grant no.
BHKY201801).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QHW, MZ and FL conceived and designed the study.
QHW, MZ, MHZ and XY performed the experiments and wrote the
manuscript. QHW, XJG and FC analyzed the data. All authors read and
approved the manuscript and agreed to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
The present study was approved by the Kunshan First
People's Hospital Ethics Committee (Kunshan, China) and written
informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EIF3
|
eukaryotic initiation factor 3
|
|
IRS
|
immunoreactivity score
|
|
OS
|
overall survival
|
|
DFS
|
disease free survival
|
|
AMPKα
|
AMP-activated protein kinase α
|
|
MIF
|
migration inhibitory factor
|
|
MT2
|
metallothionein 2
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Papagiorgis PC, Zizi AE, Tseleni S,
Oikonomakis IN and Nikiteas NI: The pattern of epidermal growth
factor receptor variation with disease progression and
aggressiveness in colorectal cancer depends on tumor location.
Oncol Lett. 3:1129–1135. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bae JM, Kim JH, Cho NY, Kim TY and Kang
GH: Prognostic implication of the CpG island methylator phenotype
in colorectal cancers depends on tumour location. Br J Cancer.
109:1004–1012. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cate JH: Human eIF3: From ‘blobology’ to
biological insight. Philos Trans R Soc Lond B Biol Sci. 372(pii):
201601762017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee AS, Kranzusch PJ, Doudna JA and Cate
JH: eIF3d is an mRNA cap-binding protein that is required for
specialized translation initiation. Nature. 536:96–99. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gomes-Duarte A, Lacerda R, Menezes J and
Romão L: eIF3: A factor for human health and disease. RNA Biol.
15:26–34. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Haybaeck J, O'Connor T, Spilka R, Spizzo
G, Ensinger Ch, Mikuz G, Brunhuber T, Vogetseder A, Theurl I,
Salvenmoser W, et al: Overexpression of p150, a part of the large
subunit of the eukaryotic translation initiation factor 3, in colon
cancer. Anticancer Res. 30:1047–1055. 2010.PubMed/NCBI
|
|
8
|
Yu X, Zheng B and Chai R:
Lentivirus-mediated knockdown of eukaryotic translation initiation
factor 3 subunit D inhibits proliferation of HCT116 colon cancer
cells. Biosci Rep. 34:e001612014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zeng L, Wan Y, Li D, Wu J, Shao M, Chen J,
Hui L, Ji H and Zhu X: The m subunit of murine translation
initiation factor EIF3Maintains the integrity of the eIF3 complex
and is required for embryonic development, homeostasis, and organ
size control. J Biol Chem. 288:30087–30093. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Valášek LS, Zeman J, Wagner S, Beznosková
P, Pavlíková Z, Mohammad MP, Hronová V, Herrmannová A, Hashem Y and
Gunišová S: Embraced by eIF3: Structural and functional insights
into the roles of eIF3 across the translation cycle. Nucleic Acids
Res. 45:10948–10968. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goh SH, Hong SH, Hong SH, Lee BC, Ju MH,
Jeong JS, Cho YR, Kim IH and Lee YS: eIF3m expression influences
the regulation of tumorigenesis-related genes in human colon
cancer. Oncogene. 30:398–409. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brill E, Gobble R, Angeles C,
Lagos-Quintana M, Crago A, Laxa B, Decarolis P, Zhang L, Antonescu
C, Socci ND, et al: ZIC1 overexpression is oncogenic in
liposarcoma. Cancer Res. 70:6891–6901. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jorissen RN, Gibbs P, Christie M, Prakash
S, Lipton L, Desai J, Kerr D, Aaltonen LA, Arango D, Kruhøffer M,
et al: Metastasis-associated gene expression changes predict poor
outcomes in patients with dukes stage B and C colorectal cancer.
Clin Cancer Res. 15:7642–7651. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Skrzypczak M, Goryca K, Rubel T, Paziewska
A, Mikula M, Jarosz D, Pachlewski J, Oledzki J and Ostrowski J:
Modeling oncogenic signaling in colon tumors by multidirectional
analyses of microarray data directed for maximization of analytical
reliability. PLoS One. 5(pii): e130912010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Graudens E, Boulanger V, Mollard C,
Mariage-Samson R, Barlet X, Grémy G, Couillault C, Lajémi M,
Piatier-Tonneau D, Zaborski P, et al: Deciphering cellular states
of innate tumor drug responses. Genome Biol. 7:R192006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ki DH, Jeung HC, Park CH, Kang SH, Lee GY,
Lee WS, Kim NK, Chung HC and Rha SY: Whole genome analysis for
liver metastasis gene signatures in colorectal cancer. Int J
Cancer. 121:2005–2012. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hong Y, Downey T, Eu KW, Koh PK and Cheah
PY: A ‘metastasis-prone’ signature for early-stage mismatch-repair
proficient sporadic colorectal cancer patients and its implications
for possible therapeutics. Clin Exp Metastasis. 27:83–90. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Calandra T and Bucala R: Macrophage
migration inhibitory factor (MIF): A glucocorticoid
counter-regulator within the immune system. Crit Rev Immunol.
37:359–370. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bach JP, Deuster O, Balzer-Geldsetzer M,
Meyer B, Dodel R and Bacher M: The role of macrophage inhibitory
factor in tumorigenesis and central nervous system tumors. Cancer.
115:2031–2040. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Atrian S and Capdevila M:
Metallothionein-protein interactions. Biomol Concepts. 4:143–160.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schuermann A, Helker CS and Herzog W:
Metallothionein 2 regulates endothelial cell migration through
transcriptional regulation of vegfc expression. Angiogenesis.
18:463–475. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu Z, Dong Z, Yang Z, Chen Q, Pan Y, Yang
Y, Cui P, Zhang X and Zhang JT: Role of eIF3a (eIF3 p170) in
intestinal cell differentiation and its association with early
development. Differentiation. 75:652–661. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Z, Chen J, Sun J, Cui Z and Wu H: RNA
interference-mediated silencing of eukaryotic translation
initiation factor 3, subunit B (EIF3B) gene expression inhibits
proliferation of colon cancer cells. World J Surg Oncol.
10:1192012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song N, Wang Y, Gu XD, Chen ZY and Shi LB:
Effect of siRNA-mediated knockdown of eIF3c gene on survival of
colon cancer cells. J Zhejiang Univ Sci B. 14:451–459. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z, Lin S, Jiang T, Wang J, Lu H, Tang
H, Teng M and Fan J: Overexpression of eIF3e is correlated with
colon tumor development and poor prognosis. Int J Clin Exp Pathol.
7:6462–6474. 2014.PubMed/NCBI
|
|
27
|
Qi J, Dong Z, Liu J and Zhang JT: EIF3i
promotes colon oncogenesis by regulating COX-2 protein synthesis
and β-catenin activation. Oncogene. 33:4156–4163. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Monteleone F, Taverna S, Alessandro R and
Fontana S: SWATH-MS based quantitative proteomics analysis reveals
that curcumin alters the metabolic enzyme profile of CML cells by
affecting the activity of miR-22/IPO7/HIF-1α axis. J Exp Clin
Cancer Res. 37:1702018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun Z, Li H, Shu XH, Shi H, Chen XY, Kong
QY, Wu ML and Liu J: Distinct sulfonation activities in
resveratrol-sensitive and resveratrol-insensitive human
glioblastoma cells. FEBS J. 279:2381–2392. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu Y, Yuan L, Guo Y, Lu A, Zheng W, Xu H,
Yang Y, Hu P, Gu S, Wang B and Deng H: Identification of a GNE
homozygous mutation in a Han-Chinese family with GNE myopathy. J
Cell Mol Med. 22:5533–5538. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He XW, Feng T, Yin QL, Jian YW and Liu T:
NOB1 is essential for the survival of RKO colorectal cancer cells.
World J Gastroenterol. 21:868–877. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gatza ML, Silva GO, Parker JS, Fan C and
Perou CM: An integrated genomics approach identifies drivers of
proliferation in luminal-subtype human breast cancer. Nat Genet.
46:1051–1059. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang B, Fan X, Zhang D, Liu H and Fan C:
Identifying UBA2 as a proliferation and cell cycle regulator in
lung cancer A549 cells. J Cell Biochem. 120:12752–12761. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Choi R, Sohn I, Kim MJ, Woo HI, Lee JW, Ma
Y, Yi ES, Koo HH and Lee SY: Pathway genes and metabolites in
thiopurine therapy in Korean children with acute lymphoblastic
leukaemia. Br J Clin Pharmacol. 85:1585–1597. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Love C, Sun Z, Jima D, Li G, Zhang J,
Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, et al: The
genetic landscape of mutations in Burkitt lymphoma. Nat Genet.
44:1321–1325. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu FF, Tu TT, Zhang HF, Hu F, Huang L,
Deng LF, Guo M, Wei Q and Li K: Coexpression network analysis of
platelet genes in sickle cell disease. Platelets. 30:1022–1029.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Katayama T, Kasho K and Kawakami H: The
DnaA cycle in Escherichia coli: Activation, function and
inactivation of the initiator protein. Front Microbiol. 8:24962017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Geng W, Liang W, Fan Y, Ye Z and Zhang L:
Overexpression of CCDC34 in colorectal cancer and its involvement
in tumor growth, apoptosis and invasion. Mol Med Rep. 17:465–473.
2018.PubMed/NCBI
|
|
39
|
Zimmermann M, Murina O, Reijns MAM,
Agathanggelou A, Challis R, Tarnauskaitė Ž, Muir M, Fluteau A,
Aregger M, McEwan A, et al: CRISPR screens identify genomic
ribonucleotides as a source of PARP-trapping lesions. Nature.
559:285–289. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tapak L, Saidijam M, Sadeghifar M,
Poorolajal J and Mahjub H: Competing risks data analysis with
high-dimensional covariates: An application in bladder cancer.
Genomics Proteomics Bioinformatics. 13:169–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Y, Zhang T, Kwiatkowski N, Abraham
BJ, Lee TI, Xie S, Yuzugullu H, Von T, Li H, Lin Z, et al:
CDK7-dependent transcriptional addiction in triple-negative breast
cancer. Cell. 163:174–186. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rozanov DV, Savinov AY, Williams R, Liu K,
Golubkov VS, Krajewski S and Strongin AY: Molecular signature of
MT1-MMP: Transactivation of the downstream universal gene network
in cancer. Cancer Res. 68:4086–896. 2008. View Article : Google Scholar : PubMed/NCBI
|