Introduction
Hepatocellular carcinoma (HCC) is the fourth most
common type of malignancy and has the third highest rate of
cancer-associated mortality of digestive system tumors worldwide,
according to statistics from 2019 (1). The incidence of HCC is high in East
Asia/Southeast Asia and Africa (2).
Currently, the most common treatment for HCC is surgical resection,
microwave ablation, transarterial chemoembolization, percutaneous
radiofrequency ablation and liver transplantation (3). Sorafenib, the chemotherapy drug for
HCC, is highly toxic (3,4). The majority of patients already have
advanced tumors when diagnosed (5).
Tumor phenotype, genetic heterogeneity, multifocal occurrence due
to intrahepatic metastasis and high recurrence and metastasis rates
adversely affect the treatment and prognosis of patients with HCC
(6,7). Tumor biomarkers are being discovered at
an increasing rate and may reveal disease mechanisms, aid in the
diagnosis of cancer type and stage, facilitate monitoring of tumor
progression, provide potential targets for novel therapies
(8) and identify candidates for
various treatments (9). Therefore,
identification of novel biomarkers is essential for effective
therapy in patients with liver cancer.
The Cancer Genome Atlas (TCGA) database has been
used to identify novel protein molecules differentially expressed
between cancerous and non-cancerous tissues (10). Fibrinogen C domain-containing 1
(FIBCD1) is a protein belonging to the fibrinogen-related domain
(FReD) superfamily. To date, 541 FReDs have been found in mammals,
21 of which have also been identified in humans (11,12). The
FIBCD1 protein is made up of 461 amino acids (13) and is an acetyl receptor that combines
with the acetyl sites of chitin at the FReD (14). The FReD superfamily consists mainly
of soluble proteins and is widely distributed (15). FIBCD1 was initially found to be
mainly present in epithelial cells in the intestine and salivary
gland ducts (16).
FIBCD1 is highly-expressed in certain digestive
system tumors, but its expression pattern in HCC has not been
investigated yet. The role of FIBCD1 expression in the prognosis of
HCC can be explored in bioinformatics analysis databases, such as
TCGA or Oncomine. However, for clinical work in a hospital, fresh
tissues are less common than pathological paraffin tissues, which
are easier to store and manipulate. Thus, immunohistochemistry
(IHC) for protein level detection is conducive to clinical research
and further application (17). In
the present study, bioinformatics tools and tissue microarray
(TMA)-IHC were used to analyze FIBCD1 expression in HCC and normal
samples. Associations between FIBCD1 and the clinicopathological
and prognostic aspects of HCC were also examined.
Materials and methods
Bioinformatics analysis using the
Oncomine database
The Oncomine database (http://www.oncomine.org) is currently the world's
largest oncogene chip database and integrated data mining platform.
The database has a high number of gene expression datasets and
sample data from a large number of cancer tissues and normal
tissues (18,19). In the present study, the Oncomine
database was used to evaluate FIBCD1 mRNA expression in HCC samples
at a threshold of P<0.05. ‘FIBCD1,’ ‘mRNA,’ ‘HCC’ and ‘cancer
vs. normal analysis’ were the search queries used to obtain data.
Two analyses from Wurmbach et al (20) were obtained, including ‘Liver Cancer
Precursor’ and ‘Liver Cell Dysplasia vs. Normal’ (cirrhotic
tissues, n=13; dysplastic nodules, n=17; HCCs, n=35; normal
tissues, n=10).
Tumor specimens and
clinicopathological information
The present study was approved by the Human Research
Ethics Committee at the Affiliated Hospital of Nantong University
(Nantong, China). All experimental methods and related protocols
were performed in accordance with the regulations of the Affiliated
Hospital of Nantong University. All participating patients provided
written informed consent for use of their tissues and for the
publication of the present study. Formalin-fixed, paraffin-embedded
tumor samples from 563 patients (range, 23–79 years), including 495
samples from patients with primary HCC, 32 chronic hepatitis
tissues, 14 hepatic cavernous hemangioma tissues and 22 liver
cirrhosis samples were collected. The tumor samples were matched
with 495 peritumoral tissues (adjacent normal tissues; >2 cm
from the tumors' edges). These 563 patients underwent surgery at
the Affiliated Hospital of Nantong University between January 2005
and December 2007. Clinical information, including sex, age, tumor
diameter, α-fetoprotein (AFP), tumor number, tumor node metastasis
(TNM) stage, degree of differentiation, hepatitis B virus
infection, vascular invasion and liver cirrhosis, was recorded. AFP
is mainly synthesized in the liver of rodents and human embryos
(21) and is the most specific
marker of primary liver cancer (22). Disease stage was determined according
to the 8th edition of the TNM Classification of Malignant Tumors
guidelines (23). The period from
diagnosis until death (from HCC only) was defined as overall
survival (OS). The longest follow-up period was 99 months and 343
patients died during the study. None of these patients underwent
any preoperative radiotherapy, chemotherapy or other special
treatment for cancer.
RT-qPCR
Total RNA was extracted from 35 pairs of
fresh-frozen tissues (tumor and adjacent normal tissues) collected
from 35 patients (25 males and 10 females; range, 42–71 years) who
provided written informed consent for use of their tissues with
HCC, from July to December in 2017, at the Affiliated Hospital of
Nantong University. An RNeasy Mini kit and QiaShredders (Qiagen,
Ltd.) were applied to isolate and purify total RNA from the
tissues. In accordance with the manufacturer's protocol, cDNA was
generated from total RNA using a reverse transcription kit
(RevertAid Reverse Transcriptase RT kit; cat. no. K1691; Fermentas;
Thermo Fisher Scientific, Inc.). qPCR was performed using the
QuantiTect SYBR-Green PCR mixture on a Bio-Rad iCycler (Bio-Rad
Laboratories, Inc.). The primer sequences for FIBCD1 were as
follows: Forward, 5′-GTGTGGGGTTCCGTTCTC-3′ and reverse,
5′-CCAGTGGTGCCAAGTCAA-3′. 18S rRNA (Thermo Fisher Scientific, Inc.)
was used as the endogenous control and the primer sequences are as
follows: Forward, 5′-TGCAGCGCACCGATGG-3′ and reverse,
5′-GAGGTTGGTGAGGGAGATCG-3′. The thermocycling conditions were as
follows: Initial denaturation at 95°C for 6 min, followed by 35
cycles of 30 sec at 95°C and 1 min at 60°C. The levels of FIBCD1
mRNA were analyzed using the 2−ΔΔCq method (24). All experiments were repeated 3
times.
TMAs and IHC
Core tissue biopsies (0.2 cm in diameter) obtained
from paraffin-embedded blocks were arranged in new paraffin blocks
using a Tissue Microarray system (cat. no. UT06; Quick-Ray; UNITMA,
Co., Ltd.). The samples were then sliced into 4-µm wide sections
for IHC analysis. The sections were stained with polyclonal rabbit
anti-FIBCD1 antibody (1:100 dilution; Atlas antibodies AB; cat. no.
HPA053898) overnight at 4°C, and then incubated with biotinylated
anti-rabbit secondary antibody (1:2,000 dilution; cat. no.
ZDR-5306; OriGene Technologies, Inc.) for 2 h at room
temperature.
The intensity and percentages of FIBCD1 staining on
each chip were scored by 2 trained observers. The intensity scores
were defined as: 0, negative; 1, weakly positive; 2, moderately
positive; and 3, strongly positive. Percentage scores of FIBCD1
positive staining were defined as 0–100%. The final score was
calculated as percentage score × intensity score. X-tile software
v3.6.1 (25) was used to determine
the cut-off point for FIBCD1 expression data. The point was
identified based on the maximum χ2 value, which was
estimated by log-rank χ2 statistics according to OS. A
cut-off value score of 110 was used to define the expression level;
111–300 was regarded as high and 0–110 was low or none.
Expression of FIBCD1 in various types
of cancer in bioinformatics databases
Gene Expression Profiling Interactive Analysis
(GEPIA) (http://gepia.cancer-pku.cn/index.html), a novel
interactive web server, can be used to analyze RNA sequencing
expression data from 9,736 tumors and 8,587 normal samples from
TCGA database and the Genotype Tissue Expression project with a
standard processing pipeline (26).
GEPIA offers customizable features such as tumor and normal
differential expression models. Datasets containing samples of
liver HCC (LIHC; tumor, n=369 and normal, n=50), lung
adenocarcinoma (LUAD; tumor, n=482 and normal, n=59), mesothelioma
(MESO; tumor, n=87) and uveal melanoma (UVM; tumor, n=79) were
investigated. In addition, the correlations of FIBCD1 expression
and the expression of human hepatocyte growth factor (HGF) and
recombinant heat shock 70 kDa protein 4 (HSPA4) in liver tissues
were further analyzed using Pearson's correlation coefficient test
in the GEPIA database.
Kaplan-Meier plotter analysis
Kaplan-Meier plotter is an online survival analysis
database (http://kmplot.com/analysis/) that
includes transcriptomic data of 364 patients with liver cancer.
This tool was used to analyze the prognostic significance of FIBCD1
using 4 as the cut-off value obtained from the database by
selecting the ‘auto select best cut-off’ option for the
dichotomization of FIBCD1 expression level.
Statistical analysis
All data were analyzed using SPSS software version
21.0 (IBM Corp.). The χ2 test was used to investigate
the association between FIBCD1 expression and clinicopathological
features. Wilcoxon signed-rank non-parametric test was used to
analyze the difference between the paired HCC tissues and adjacent
normal tissues. Kaplan-Meier curves and the log-rank test were used
to assess the survival of patients with HCC. Univariate analysis
and multivariate Cox regression was used to evaluate factors
associated with prognosis. P<0.05 was considered to indicate a
statistically significant difference.
Results
FIBCD1 mRNA and protein expression in
HCC
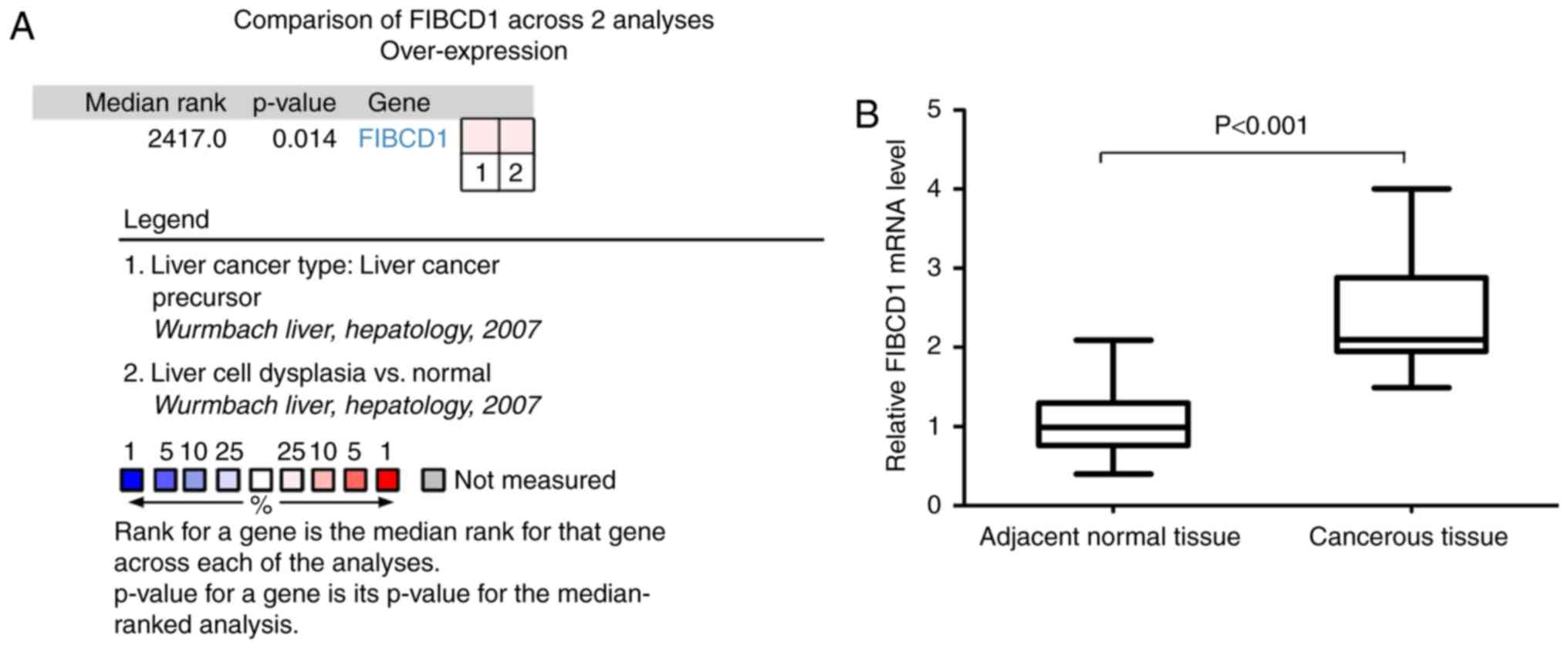
The Oncomine database was utilized to assess FIBCD1
mRNA data. The mRNA levels of FIBCD1 were significantly increased
in HCC in the 2 datasets (Fig. 1A).
RT-qPCR revealed that the mean ± SEM FIBCD1 mRNA expression in
cancerous tissues and adjacent normal tissues was 2.37±0.10 and
1.08±0.07, respectively. The level of FIBCD1 mRNA expression in
cancerous tissues was higher than that in adjacent normal tissues
(P<0.001; Fig. 1B).
The TMA-IHC results showed that FIBCD1 is primarily
localized to the nucleus of hepatocytes (Fig. 2). However, 3 chronic hepatitis
tissues and 2 cirrhotic tissues showed positive FIBCD1 staining
localized in the cytoplasm and cell membrane. However, this
staining did not reach statistical significance, so only the
tissues that were stained positively in the nucleus are discussed.
Furthermore, FIBCD1 was expressed in 244/495 (49.3%) HCC tissues
compared with 40/495 (8.0%) adjacent normal, 2/32 (6.3%) chronic
hepatitis, 1/14 (7.1%) hepatic cavernous hemangioma and 3/22
(13.6%) liver cirrhosis tissues (Fig.
2; Table I). High FIBCD1 protein
expression was most frequent in HCC tissues (χ2, 224.27;
P<0.001).
 | Table I.Fibrinogen C domain-containing 1
expression in liver tissues from benign and malignant diseases. |
Table I.
Fibrinogen C domain-containing 1
expression in liver tissues from benign and malignant diseases.
|
Characteristics | n | Low or none
expression, n (%) | High expression, n
(%) |
|---|
| Chronic
hepatitis | 32 | 30 (93.8) | 2 (6.3) |
| Hepatic cavernous
hemangioma | 14 | 13 (92.9) | 1 (7.1) |
| Liver
cirrhosis | 22 | 19 (86.4) | 3 (13.6) |
| Hepatocellular
carcionoma | 495 | 251 (50.7) | 244 (49.3) |
| Adjacent
normal | 495 | 455 (92.0) | 40 (8.0) |
FIBCD1 protein expression and clinical
characteristics of patients with HCC
FIBCD1 protein expression level was associated with
tumor diameter (P=0.002), tumor number (P=0.001), TNM stage
(P<0.001), primary tumor (T; P<0.001), lymph node metastases
(N; P=0.002), distant metastases (M; P=0.023), differentiation
degree (P=0.003), vascular invasion (P<0.001) and liver
cirrhosis (P=0.011), but not with sex, age, AFP and hepatitis B
virus infection (P>0.05; Table
II).
 | Table II.Association of FIBCD1 expression and
clinicopathological characteristics of patients with hepatocellular
carcinoma. |
Table II.
Association of FIBCD1 expression and
clinicopathological characteristics of patients with hepatocellular
carcinoma.
|
|
| FIBCD1 expression,
n (%) |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Cases, n | Low or none | High | Pearson
χ2 | P-value |
|---|
| Total patients | 495 | 251 | 244 |
|
|
| Sex |
|
|
| 0.092 | 0.761 |
|
Male | 139 | 72 (51.8) | 67 (48.2) |
|
|
|
Female | 356 | 179 (50.3) | 177 (49.7) |
|
|
| Age, years |
|
|
| 0.030 | 0.862 |
|
≤60 | 379 | 193 (50.9) | 186 (49.1) |
|
|
|
>60 | 116 | 58 (50.0) | 58 (50.0) |
|
|
| Tumor diameter,
cm |
|
|
| 9.564 | 0.002a |
| ≤3 | 256 | 147 (57.4) | 109 (42.6) |
|
|
|
>3 | 239 | 104 (43.5) | 135 (56.5) |
|
|
| α-fetoprotein,
µg/l |
|
|
| 0.668 | 0.414 |
|
≤400 | 388 | 193 (49.7) | 195 (50.3) |
|
|
|
>400 | 107 | 58 (54.2) | 49 (45.8) |
|
|
| Tumor number |
|
|
| 10.222 |
0.001a |
|
Solitary | 449 | 238 (53.0) | 211 (47.0) |
|
|
|
Multiple | 46 | 13 (28.3) | 33 (71.7) |
|
|
| TNM stage |
|
|
| 61.562 |
<0.001a |
| I | 375 | 227 (60.5) | 148 (39.5) |
|
|
| II | 74 | 18 (24.3) | 56 (75.7) |
|
|
|
III | 31 | 5 (16.1) | 26 (83.9) |
|
|
| IV | 15 | 1 (6.7) | 14 (93.3) |
|
|
| T |
|
|
| 61.203 |
<0.001a |
| 1 | 375 | 227 (60.5) | 148 (39.5) |
|
|
| 2 | 74 | 18 (24.3) | 56 (75.7) |
|
|
| 3 | 30 | 4 (13.3) | 26 (86.7) |
|
|
| 4 | 16 | 2 (12.5) | 14 (87.5) |
|
|
| N |
|
|
| 9.883 | 0.002a |
| 0 | 482 | 250 (51.9) | 232 (48.1) |
|
|
| 1 | 13 | 1 (7.7) | 12 (92.3) |
|
|
| M |
|
|
| 5.196 | 0.023a |
| 0 | 490 | 251 (51.2) | 239 (48.8) |
|
|
| 1 | 5 | 0 (0.0) | 5 (100.0) |
|
|
|
Differentiation |
|
|
|
|
|
|
Well | 124 | 70 (56.5) | 54 (43.5) | 13.754 | 0.003a |
|
Moderate | 287 | 152 (53.0) | 135 (47.0) |
|
|
|
Poor | 74 | 28 (37.8) | 46 (62.2) |
|
|
|
Othersb | 10 | 1 (10.0) | 9 (90.0) |
|
|
| Hepatitis B virus
infection |
|
|
|
|
|
| No | 268 | 126 (47.0) | 142 (53.0) | 3.187 | 0.074 |
|
Yes | 227 | 125 (55.1) | 102 (44.9) |
|
|
| Vascular
invasion |
|
|
|
|
|
| No | 451 | 241 (53.4) | 210 (46.6) | 15.126 |
<0.001a |
|
Yes | 44 | 10 (22.7) | 34 (77.3) |
|
|
| Liver
cirrhosis |
|
|
|
|
|
| No | 264 | 148 (56.1) | 116 (43.9) | 6.487 | 0.011a |
|
Yes | 231 | 102 (44.6) | 128 (55.4) |
|
|
High FIBCD1 protein expression is
associated with poor prognosis in patients with HCC
In univariate analysis, poor OS time was associated
with high FIBCD1 expression (HR, 2.025; P<0.001), TNM stage (HR,
2.136; P<0.001), T (HR, 2.310; P<0.001), N (HR, 8.159;
P<0.001), M (HR, 5.111; P<0.001), vascular invasion (HR,
5.669; P<0.001) and liver cirrhosis (HR, 1.290; P=0.020)
(Table III). In multivariate
analysis, high FIBCD1 expression (HR, 1.625; P<0.001), TNM stage
(HR, 0.316; P=0.003), T (HR, 4.822; P<0.001), N (HR, 3.296;
P=0.014) and vascular invasion (HR, 2.343; P<0.001) were
independent prognostic factors (Table
III).
 | Table III.Univariate and multivariable analysis
of survival factors in patients with hepatocellular carcinoma. |
Table III.
Univariate and multivariable analysis
of survival factors in patients with hepatocellular carcinoma.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | P-value | 95% CI | HR | P-value | 95% CI |
|---|
| FIBCD1 expression,
high vs. low or none | 2.025 |
<0.001a | 1.633 | 2.512 | 1.625 |
<0.001a | 1.280 | 2.062 |
| Age, ≤60 vs. >60
years | 0.929 | 0.570 | 0.720 | 1.198 |
|
|
|
|
| Sex, male vs.
female | 0.921 | 0.484 | 0.730 | 1.161 |
|
|
|
|
| Tumor diameter, ≤3
vs. >3 cm | 0.925 | 0.472 | 0.748 | 1.144 |
|
|
|
|
| α-fetoprotein, ≤400
vs. >400 µg/l | 0.927 | 0.570 | 0.715 | 1.203 |
|
|
|
|
| Tumor number,
solitary vs. multiple | 1.116 | 0.540 | 0.786 | 1.583 |
|
|
|
|
| TNM stage, I vs. II
vs. III vs. IV | 2.136 |
<0.001a | 1.869 | 2.440 | 0.316 | 0.003a | 0.147 | 0.679 |
| T, 1 vs. 2 vs. 3
vs. 4 | 2.310 |
<0.001a | 2.010 | 2.656 | 4.822 |
<0.001a | 2.348 | 9.903 |
| N, 0 vs. 1 | 8.159 |
<0.001a | 4.561 | 14.595 | 3.296 | 0.014a | 1,276 | 8.511 |
| M, 0 vs. 1 | 5.111 |
<0.001a | 2.093 | 12.485 | 0.454 | 0.163 | 0.149 | 1.379 |
| Differentiation,
well vs. moderate vs. poor vs. clear cell type | 1.065 | 0.426 | 0.912 | 1.242 |
|
|
|
|
| Hepatitis B virus
infection, no vs. yes | 1.056 | 0.614 | 0.854 | 1.306 |
|
|
|
|
| Vascular invasion,
no vs. yes | 5.669 |
<0.001a | 4.052 | 7.930 | 2.343 |
<0.001a | 1.468 | 3.739 |
| Liver cirrhosis, no
vs. yes | 1.290 | 0.020a | 1.042 | 1.598 | 1.203 | 0.104 | 0.962 | 1.504 |
Kaplan-Meier survival curve, based on the cohort of
563 patients from our institution, demonstrated that high FIBCD1
expression (Fig. 3A), TNM stage
(Fig. 3B), T (Fig. 3C), N (Fig.
3D) and vascular invasion (Fig.
3E) were significantly associated with OS time. Hence, FIBCD1
expression may be a prognostic factor in HCC. The results from the
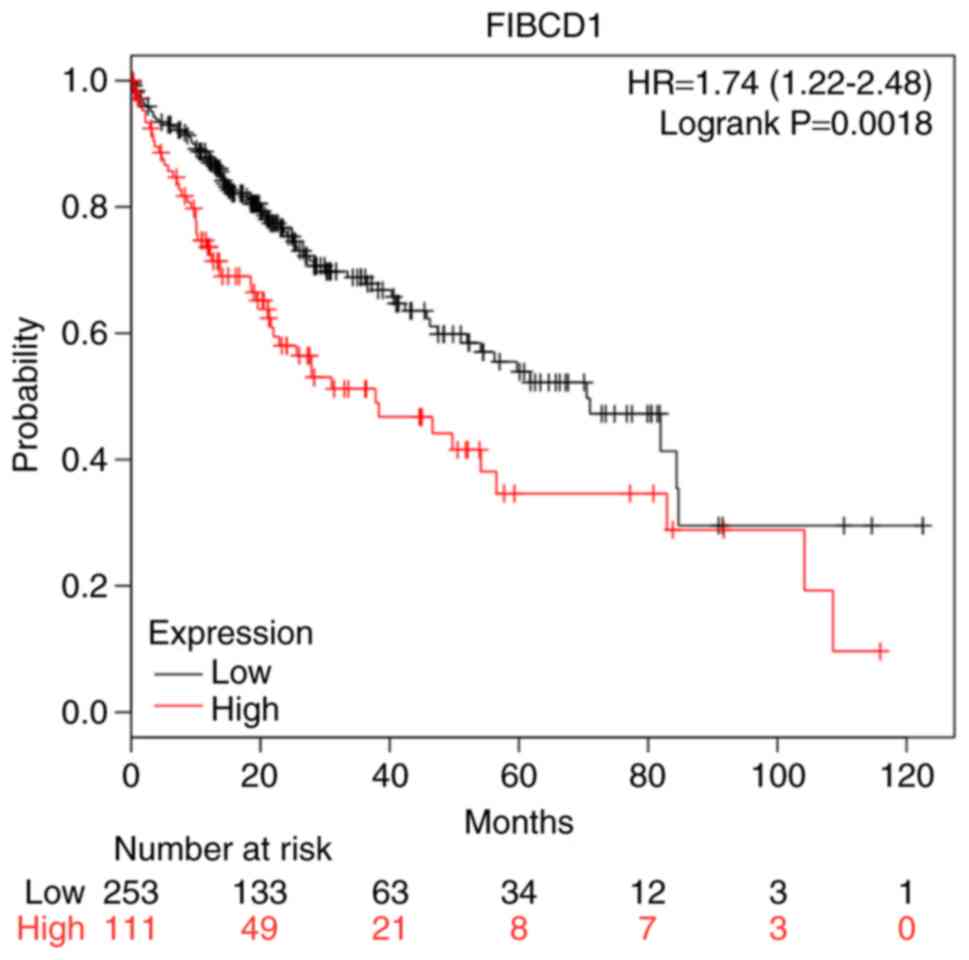
Kaplan-Meier plotter database further demonstrated that patients
with high FIBCD1 expression had a shorter OS time compared with
those with low or no FIBCD1 expression (P<0.001; Fig. 4), consistent with the aforementioned
results in Fig. 3.
Survival analysis of FIBCD1 using the
GEPIA database
The results obtained from the GEPIA database showed
that FIBCD1 was expressed in different types of cancers, such as
LIHC, LUAD, MESO and UVM. High FIBCD1 expression was found to
result in a reduced OS time for patients with LIHC (HR, 1.60;
P=0.016), LUAD (HR,1.40; P=0.024), MESO (HR, 2.10; P=0.0025) and
UVM (HR, 6.90; P=9.3×10−5) (Fig. 5).
Investigation of the correlation between the
expression of FIBCD1, HGF and HSPA4 showed that the level FIBCD1 in
liver tissues was positively correlated with HGF (r=0.20; P=0.041)
and HSPA4 (r=0.20; P=0.04) (Fig.
6).
Discussion
HCC is a common cancer in China; the incidence and
mortality rates have increased annually (27). Similar to other tumors, the
development of HCC is a multi-step process that is associated with
oncogene activation, tumor suppressor gene inactivation,
accumulation of mutations and epigenetic changes in regulatory
genes (28). HCC is associated with
many different gene mutations (29).
Early diagnosis of HCC is difficult and most patients have advanced
disease when they finally seek treatment (30). Targeted therapy is increasingly
favored for liver cancer (31), and
therapies that target epidermal growth factor receptor, vascular
endothelial growth factor receptor, fatty acid binding protein 4
and ERK1/2 are being developed or tested to treat HCC (32,33).
With the rapid development of molecular biology
techniques in recent years, research on tumor biomarkers of HCC has
made some progress, but the treatment and prognosis for patients
with HCC are not ideal. Thus, sensitive and effective HCC
biomarkers should be developed. FIBCD1 may be one of these putative
biomarkers.
The FIBCD1 gene, which is located on human
chromosome 9q34.1, is next to the homologues that encode M- and
L-ficolins (34,35). FIBCD1 can be oligomerized and
assembled into ~250 kDa tetramers with each chain containing a
cytoplasmic tail, a transmembrane helix and an extracellular
domain, consisting of a coiled region, a polycationic region and
the C-terminal FReD, which assembles proteins into tetramers linked
by disulfide bonds (13,14). The FIBCD1 tetramer assembles in a
calcium-dependent manner combined with acetylated compounds such as
sialic acid and chitin (36).
FIBCD1 is widely distributed in human tissues and
organs and may function as a pattern recognition receptor (PRR)
(35). FIBCD1 was first found to
bind to chitin, the second most abundant biopolymer in nature after
cellulose (37), to stimulate and
regulate the immune system in different ways, such as inducing
cytokine production, promoting leukocyte recruitment, and
activating macrophages (38). FIBCD1
was also the first membrane-binding FReD molecule found in
vertebrates, and it mediates the degradation of acetylated
components in vivo (13). In
dermal tissues, FIBCD1 acts as a PRR for dendritic cells,
macrophages, lymphocytes and other immune cells (39), and is also associated with
dermatophyte infection (40).
Furthermore, FIBCD1 plays a role against other bacterial
infections, including pneumonia and urinary tract infections
(35).
FIBCD1 is upregulated in the gastrointestinal tract
compared with that in other organs within the body, such as the
kidney, thymus, and the heart (35,41).
FIBCD1 upregulation predicts poor prognosis in patients with
gastric cancer (41). FIBCD1 has
high affinity for chitin fragments and can therefore control
intestinal exposure to chitin and significantly affect immune
responses to fungi and parasites (42). FIBCD1 is present in the apical
intestinal epithelium and may play a large role in the innate
immunity and homeostasis of the intestine (13).
FIBCD1 expression has recently been demonstrated in
cells derived from all 3 germ layers, including lymph tissues, the
thyroid gland and myocytes of the heart, and it is highly expressed
in the respiratory, gastrointestinal and urogenital tracts
(35). The expression pattern of
FIBCD1, together with its known binding characteristics, supports
its role in innate immunity (13).
The liver is not only the largest digestive and metabolic organ of
the body, but is also an important immune organ (43). HCC is the most common malignant tumor
of the liver (44).
In the present study, the expression of FIBCD1 in
HCC and the association with patient prognosis were investigated.
The RT-qPCR and TMA-IHC analyses confirmed that mRNA and protein
expression levels of FIBCD1 were increased in HCC compared with
normal tissues. High FIBCD1 expression was associated with certain
clinicopathological parameters, including large tumor diameter,
tumor number, advanced TNM stage, degree of differentiation,
vascular invasion and liver cirrhosis. High FIBCD1 expression,
along with vascular invasion and TNM stage, predicts poor prognosis
and increased mortality for patients with HCC. FIBCD1 mediates the
endocytosis of intracellular binding ligands that are released into
the surrounding environment after degradation; FIBCD1 is then
simultaneously recycled back to the plasma membrane (14). FIBCD1 has 2 potential phosphorylation
sites, including chondroitin and dermatan sulfate in its
cytoplasmic domain (14). Thus,
FIBCD1 may be a signaling protein, but its signal transduction
pathway remains unclear. In the present study, the results obtained
using the GEPIA database demonstrated that the correlations between
FIBCD1 expression and HGF, and FIBCD1 expression and HSPA4 were
positive. Activation of the HGF/c-Met axis facilitates the
proliferation and migration of HCC cells (45). HSPA4 was found to be upregulated in
HCC, and may be associated with the early recurrence and poor OS of
HCC (46,47). From the results of the current study,
it can be inferred that high FIBCD1 expression may promote the
occurrence and development of HCC, but understanding how it
interacts with HGF and HSPA4 requires further investigation.
The present study has certain limitations. As the
current study is retrospective, the sample size and quality of
specimens was limited. Additionally, the detection methods used to
determine FIBCD1 expression in HCC were limited as RT-qPCR and IHC
may not be accurate and comprehensive. RNAscope in situ
hybridization maybe more suitable for detecting FIBCD1 mRNA.
Furthermore, the biological mechanisms of FIBCD1 action have not
been studied in HCC. Future larger-scale studies with newer
techniques for investigating FIBCD1 expression are required.
In summary, FIBCD1 expression is increased in HCC
tissues. High FIBCD1 expression is associated with poor prognosis
in patients with HCC. Hence, FIBCD1 has value as a prognostic
predictor and a potential target for HCC therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Construction
of Clinical Medical Center for tumor biological samples in Nantong
(grant no. HS2016004).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YW and MS performed data analyses and wrote the
manuscript. JL, YL, CJ, HZ and WW contributed significantly to data
analyses and manuscript revision. YW conceived and designed the
study. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Human Research
Ethics Committee at the Affiliated Hospital of Nantong University
(Nantong, China). All experimental methods and related protocols
were performed according to the regulations of the Affiliated
Hospital of Nantong University. Signed informed consent was
obtained from the patients or their guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu KY, Wang LT, Hsu SH and Wang SN:
Homeobox genes and hepatocellular carcinoma. Cancers (Basel).
11(pii): E6212019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xue F, Liu Y, Chu H, Wen Y, Yan L, Tang Q,
Xiao E, Zhang D and Zhang H: eIF5A2 is an alternative pathway for
cell proliferation in cetuximab-treated epithelial hepatocellular
carcinoma. Am J Transl Res. 8:4670–4681. 2016.PubMed/NCBI
|
|
4
|
Xue F, Liu Y, Zhang H, Wen Y, Yan L, Tang
Q, Xiao E and Zhang D: Let-7a enhances the sensitivity of
hepatocellular carcinoma cells to cetuximab by regulating STAT3
expression. Onco Targets Ther. 9:7253–7261. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Banini BA and Sanyal AJ: The use of cell
free DNA in the diagnosis of HCC. Hepatoma Res. 5(pii):
342019.PubMed/NCBI
|
|
6
|
Amicone L and Marchetti A:
Microenvironment and tumor cells: Two targets for new molecular
therapies of hepatocellular carcinoma. Transl Gastroenterol
Hepatol. 3:242018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Waller LP, Deshpande V and Pyrsopoulos N:
Hepatocellular carcinoma: A comprehensive review. World J Hepatol.
7:2648–2663. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mordente A, Meucci E, Martorana GE and
Silvestrini A: Cancer biomarkers discovery and validation: State of
the art, problems and future perspectives. Adv Exp Med Biol.
867:9–26. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cetin B, Gumusay O, Cengiz M and Ozet A:
Advances of molecular targeted therapy in gastric cancer. J
Gastrointest Cancer. 47:125–134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deng M, Brägelmann J, Schultze JL and
Perner S: Web-TCGA: An online platform for integrated analysis of
molecular cancer data sets. BMC Bioinformatics. 17:722016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zuliani-Alvarez L and Midwood KS:
Fibrinogen-related proteins in tissue repair: How a unique domain
with a common structure controls diverse aspects of wound healing.
Adv Wound Care (New Rochelle). 4:273–285. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thomsen T, Schlosser A, Holmskov U and
Sorensen GL: Ficolins and FIBCD1: Soluble and membrane bound
pattern recognition molecules with acetyl group selectivity. Mol
Immunol. 48:369–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schlosser A, Thomsen T, Moeller JB,
Nielsen O, Tornoe I, Mollenhauer J, Moestrup SK and Holmskov U:
Characterization of FIBCD1 as an acetyl group-binding receptor that
binds chitin. J Immunol. 183:3800–3809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shrive AK, Moeller JB, Burns I, Paterson
JM, Shaw AJ, Schlosser A, Sorensen GL, Greenhough TJ and Holmskov
U: Crystal structure of the tetrameric fibrinogen-like recognition
domain of fibrinogen C domain containing 1 (FIBCD1) protein. J Biol
Chem. 289:2880–2887. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Doolittle RF, McNamara K and Lin K:
Correlating structure and function during the evolution of
fibrinogen-related domains. Protein Sci. 21:1808–1823. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rossi V, Bally I, Thielens NM, Esser AF
and Arlaud GJ: Baculovirus-mediated expression of truncated modular
fragments from the catalytic region of human complement serine
protease C1s. Evidence for the involvement of both complement
control protein modules in the recognition of the C4 protein
substrate. J Biol Chem. 273:1232–1239. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peschl P, Ramberger M, Höftberger R,
Jöhrer K, Baumann M, Rostásy K and Reindl M: Methodological
challenges in protein microarray and immunohistochemistry for the
discovery of novel autoantibodies in paediatric acute disseminated
encephalomyelitis. Int J Mol Sci. 18(pii): E6792017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao T, Pan W, Sun X and Shen H: Increased
expression of TET3 predicts unfavorable prognosis in patients with
ovarian cancer-a bioinformatics integrative analysis. J Ovarian
Res. 12:1012019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wurmbach E, Chen YB, Khitrov G, Zhang W,
Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et
al: Genome-wide molecular profiles of HCV-induced dysplasia and
hepatocellular carcinoma. Hepatology. 45:938–947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Terentiev AA and Moldogazieva NT:
Alpha-fetoprotein: A renaissance. Tumour Biol. 34:2075–2091. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sauzay C, Petit A, Bourgeois AM, Barbare
JC, Chauffert B, Galmiche A and Houessinon A: Alpha-foetoprotein
(AFP): A multi-purpose marker in hepatocellular carcinoma. Clin
Chim Acta. 463:39–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Abdel-Rahman O: Assessment of the
discriminating value of the 8th AJCC stage grouping for
hepatocellular carcinoma. HPB (Oxford). 20:41–48. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Camp RL, Dolled-Filhart M and Rimm DL:
X-tile: A new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Parikh ND, Fu S, Rao H, Yang M, Li Y,
Powell C, Wu E, Lin A, Xing B, Wei L and Lok ASF: Risk assessment
of hepatocellular carcinoma in patients with hepatitis C in China
and the USA. Dig Dis Sci. 62:3243–3253. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanda M, Sugimoto H and Kodera Y: Genetic
and epigenetic aspects of initiation and progression of
hepatocellular carcinoma. World J Gastroenterol. 21:10584–10597.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu J and Gao DZ: Distinction immune genes
of hepatitis-induced heptatocellular carcinoma. Bioinformatics.
28:3191–3194. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Finn RS: Advanced HCC: Emerging molecular
therapies. Minerva Gastroenterol Dietol. 58:25–34. 2012.PubMed/NCBI
|
|
31
|
Marquardt JU, Galle PR and Teufel A:
Molecular diagnosis and therapy of hepatocellular carcinoma (HCC):
An emerging field for advanced technologies. J Hepatol. 56:267–275.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang K, Fan Y, Chen G, Wang Z, Kong D and
Zhang P: MEK-ERK inhibition potentiates WAY-600-induced anti-cancer
efficiency in preclinical hepatocellular carcinoma (HCC) models.
Biochem Biophys Res Commun. 474:330–337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhong CQ, Zhang XP, Ma N, Zhang EB, Li JJ,
Jiang YB, Gao YZ, Yuan YM, Lan SQ, Xie D and Cheng SQ: FABP4
suppresses proliferation and invasion of hepatocellular carcinoma
cells and predicts a poor prognosis for hepatocellular carcinoma.
Cancer Med. 7:2629–2640. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Thomsen T, Moeller JB, Schlosser A,
Sorensen GL, Moestrup SK, Palaniyar N, Wallis R, Mollenhauer J and
Holmskov U: The recognition unit of FIBCD1 organizes into a
noncovalently linked tetrameric structure and uses a hydrophobic
funnel (S1) for acetyl group recognition. J Biol Chem.
285:1229–1238. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
von Huth S, Moeller JB, Schlosser A,
Marcussen N, Nielsen O, Nielsen V, Sorensen GL and Holmskov U:
Immunohistochemical localization of fibrinogen C domain containing
1 on epithelial and mucosal surfaces in human tissues. J Histochem
Cytochem. 66:85–97. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bueter CL, Specht CA and Levitz SM: Innate
sensing of chitin and chitosan. PLoS Pathog. 9:e10030802013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kurita K: Chitin and chitosan: Functional
biopolymers from marine crustaceans. Mar Biotechnol (NY).
8:203–226. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Elieh Ali Komi D, Sharma L and Dela Cruz
CS: Chitin and its effects on inflammatory and immune responses.
Clin Rev Allergy Immunol. 54:213–223. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tong PL, Roediger B, Kolesnikoff N, Biro
M, Tay SS, Jain R, Shaw LE, Grimbaldeston MA and Weninger W: The
skin immune atlas: three-dimensional analysis of cutaneous
leukocyte subsets by multiphoton microscopy. J Invest Dermatol.
135:84–93. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Abdel-Rahman SM: Genetic predictors of
susceptibility to dermatophytoses. Mycopathologia. 182:67–76. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jiang C, Zhu J, Zhou P, Zhu H, Wang W, Jin
Q and Li P: Overexpression of FIBCD1 is predictive of poor
prognosis in gastric cancer. Am J Clin Pathol. 149:474–483. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Garlatti V, Belloy N, Martin L, Lacroix M,
Matsushita M, Endo Y, Fujita T, Fontecilla-Camps JC, Arlaud GJ,
Thielens NM and Gaboriaud C: Structural insights into the innate
immune recognition specificities of L- and H-ficolins. EMBO J.
26:623–633. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao
R, Modak M, Carotta S, Haslinger C, Kind D, et al: Landscape and
dynamics of single immune cells in hepatocellular carcinoma. Cell.
179:829–845 e20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang Y, Liu X, Liu G, Wang X, Hu R and
Liang X: PIG11 over-expression predicts good prognosis and induces
HepG2 cell apoptosis via reactive oxygen species-dependent
mitochondrial pathway. Biomed Pharmacother. 108:435–442. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu Y, Tan J, Ou S, Chen J and Chen L:
MicroRNA-101-3p suppresses proliferation and migration in
hepatocellular carcinoma by targeting the HGF/c-Met pathway. Invest
New Drugs. Mar 30–2019.(Epub ahead of print). View Article : Google Scholar
|
|
46
|
Ma C, Xu T, Sun X, Zhang S, Liu S, Fan S,
Lei C, Tang F, Zhai C, Li C, et al: Network pharmacology and
bioinformatics approach reveals the therapeutic mechanism of action
of baicalein in hepatocellular carcinoma. Evid Based Complement
Alternat Med. 2019:75183742019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang Z, Zhuang L, Szatmary P, Wen L, Sun
H, Lu Y, Xu Q and Chen X: Upregulation of heat shock proteins
(HSPA12A, HSP90B1, HSPA4, HSPA5 and HSPA6) in tumour tissues is
associated with poor outcomes from HBV-related early-stage
hepatocellular carcinoma. Int J Med Sci. 12:256–263. 2015.
View Article : Google Scholar : PubMed/NCBI
|