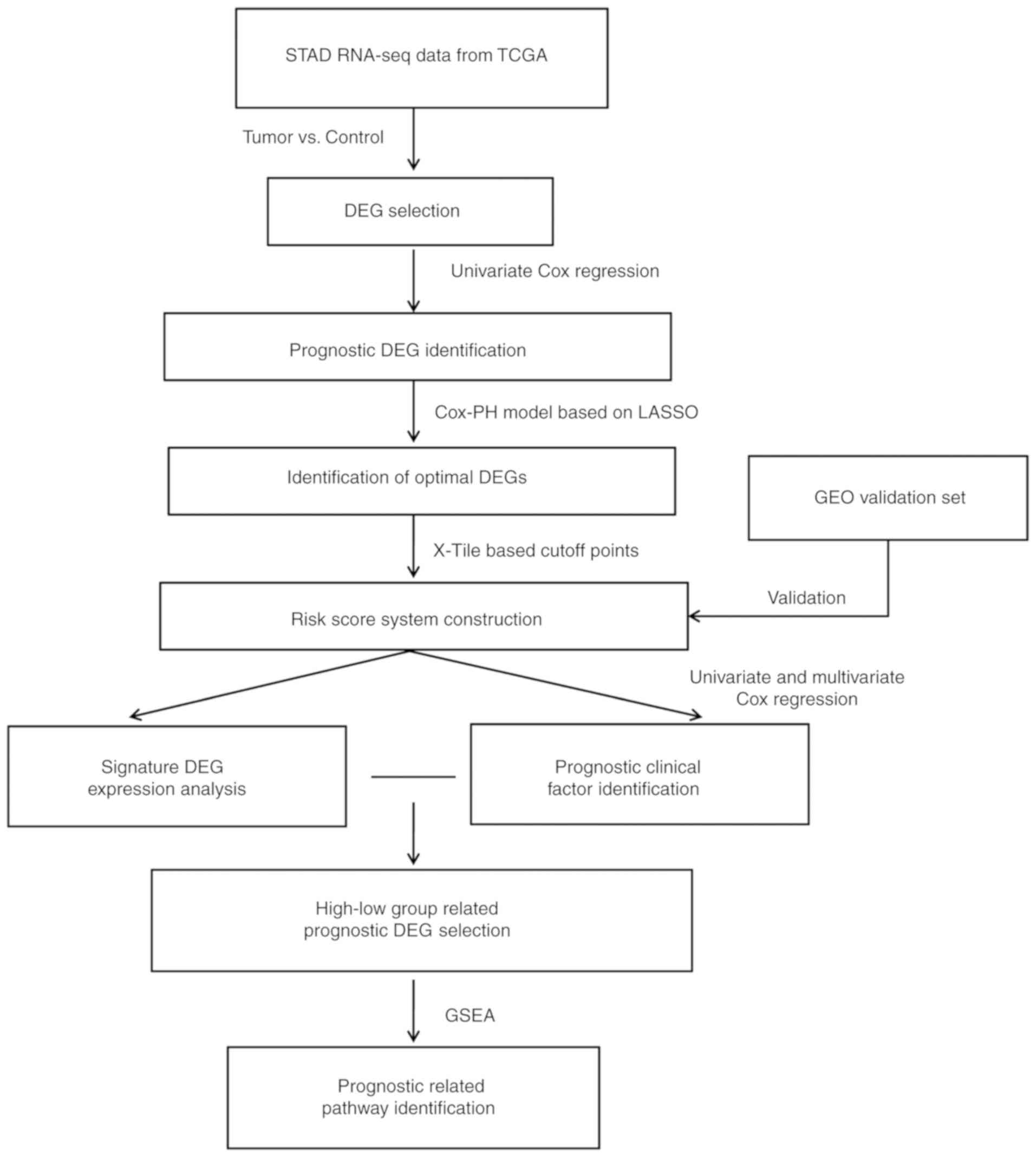
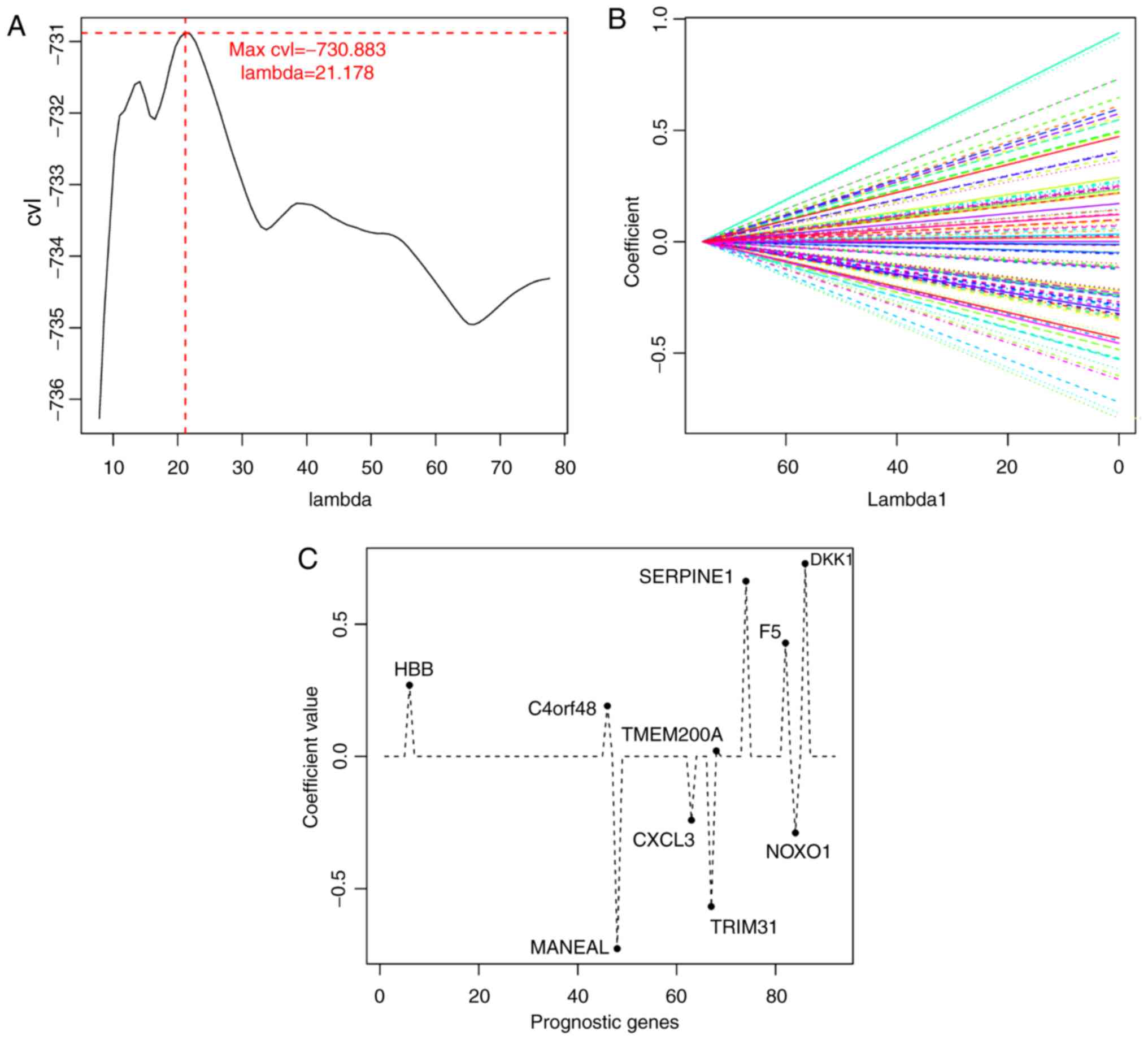
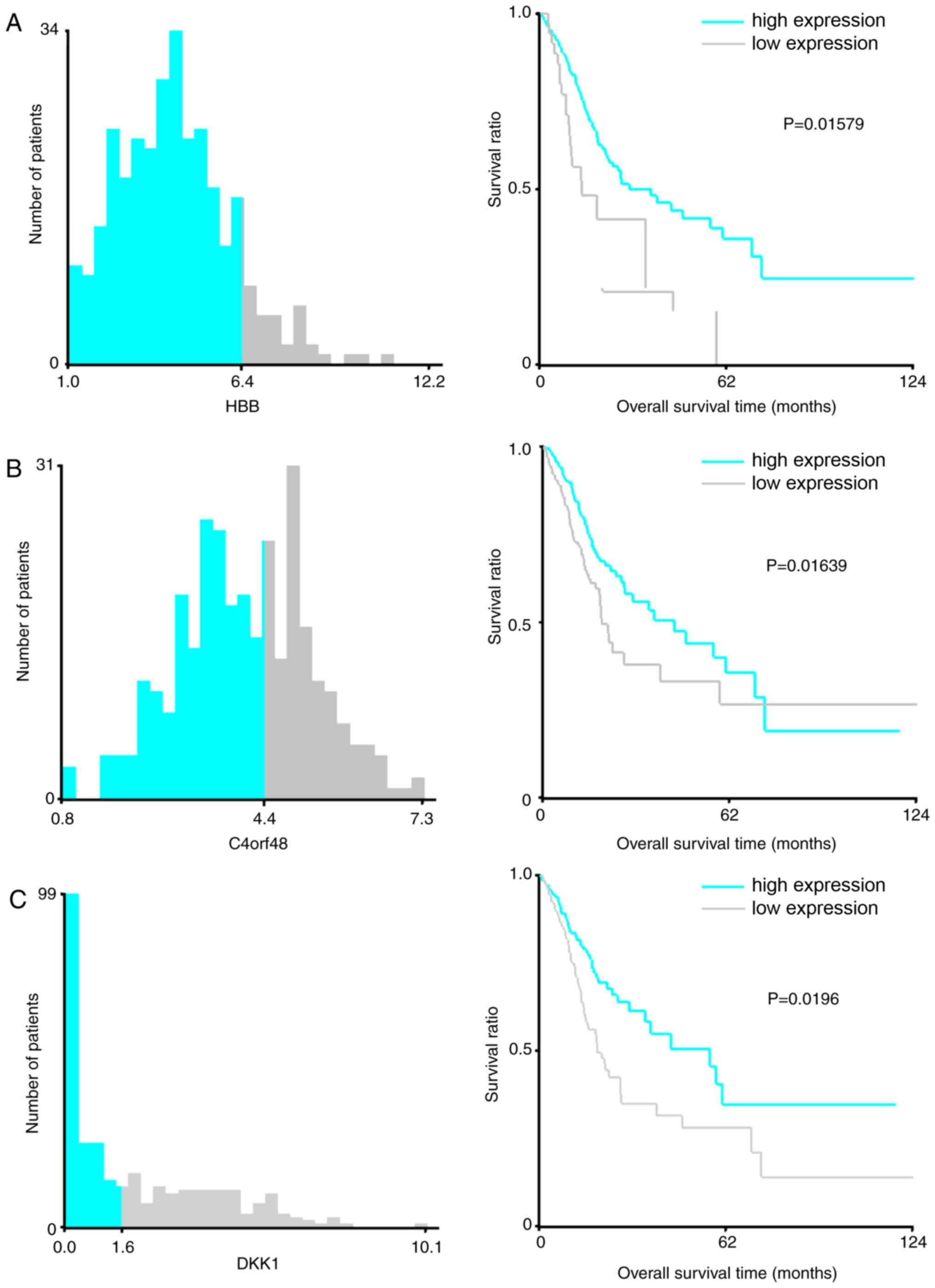
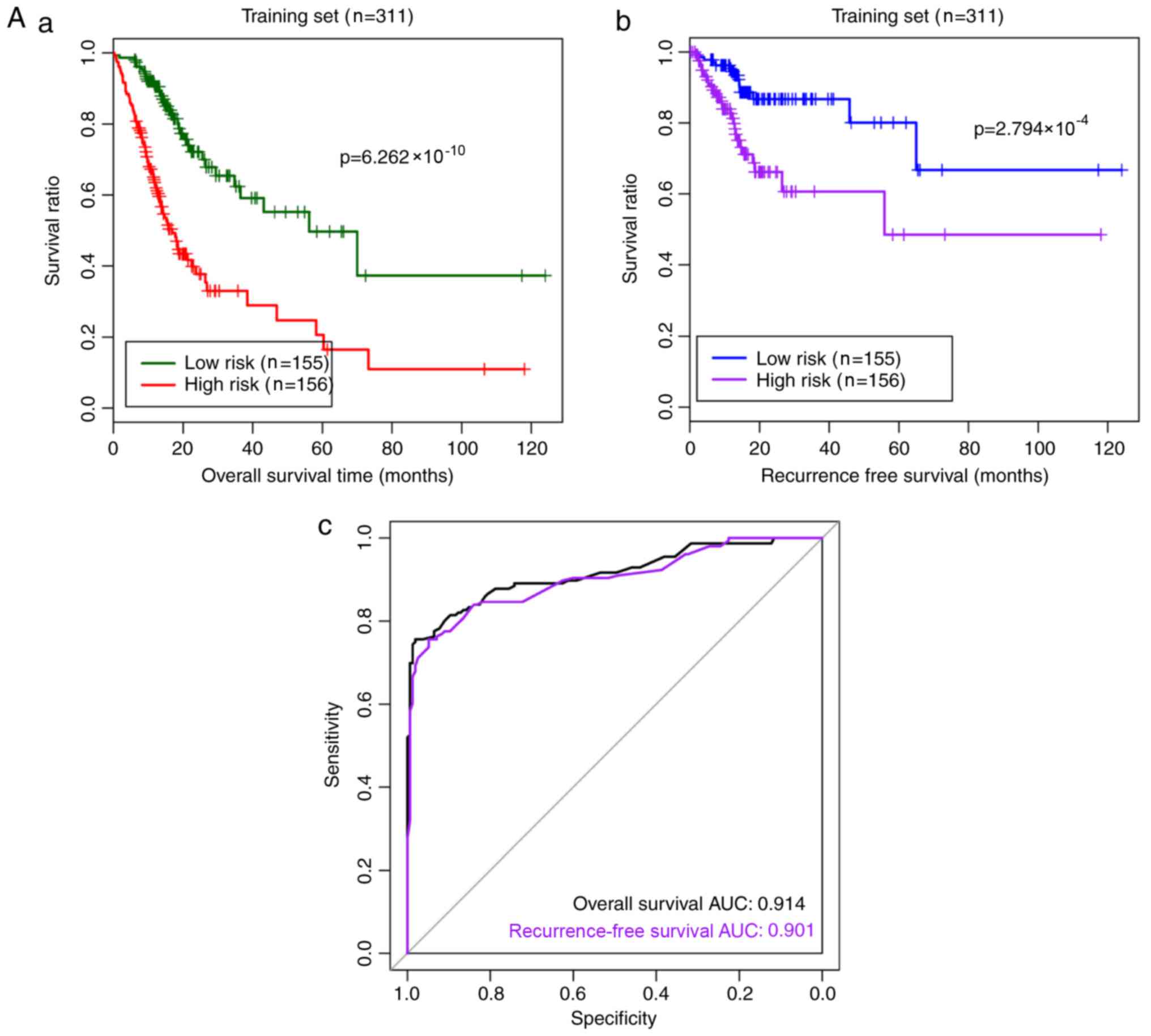
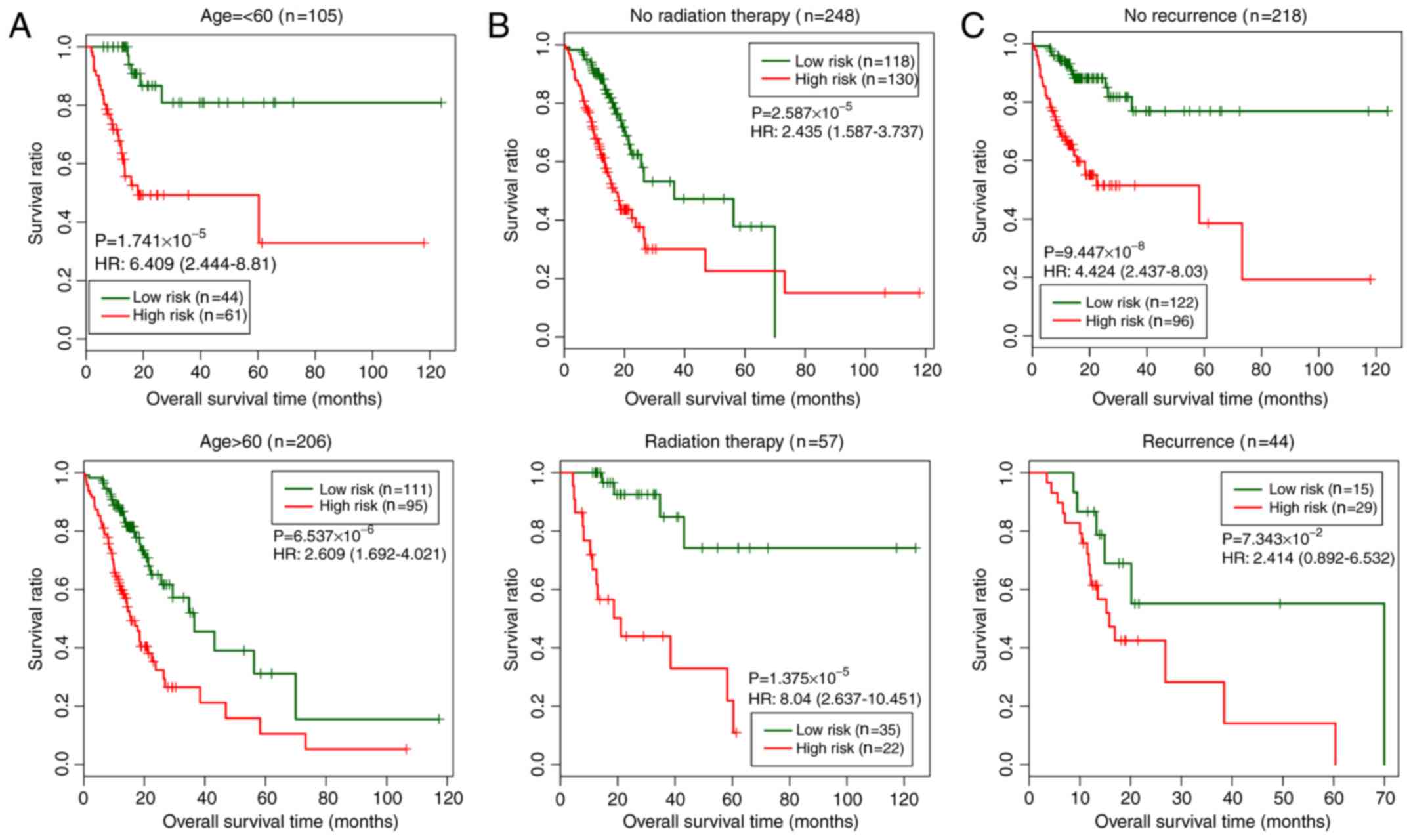
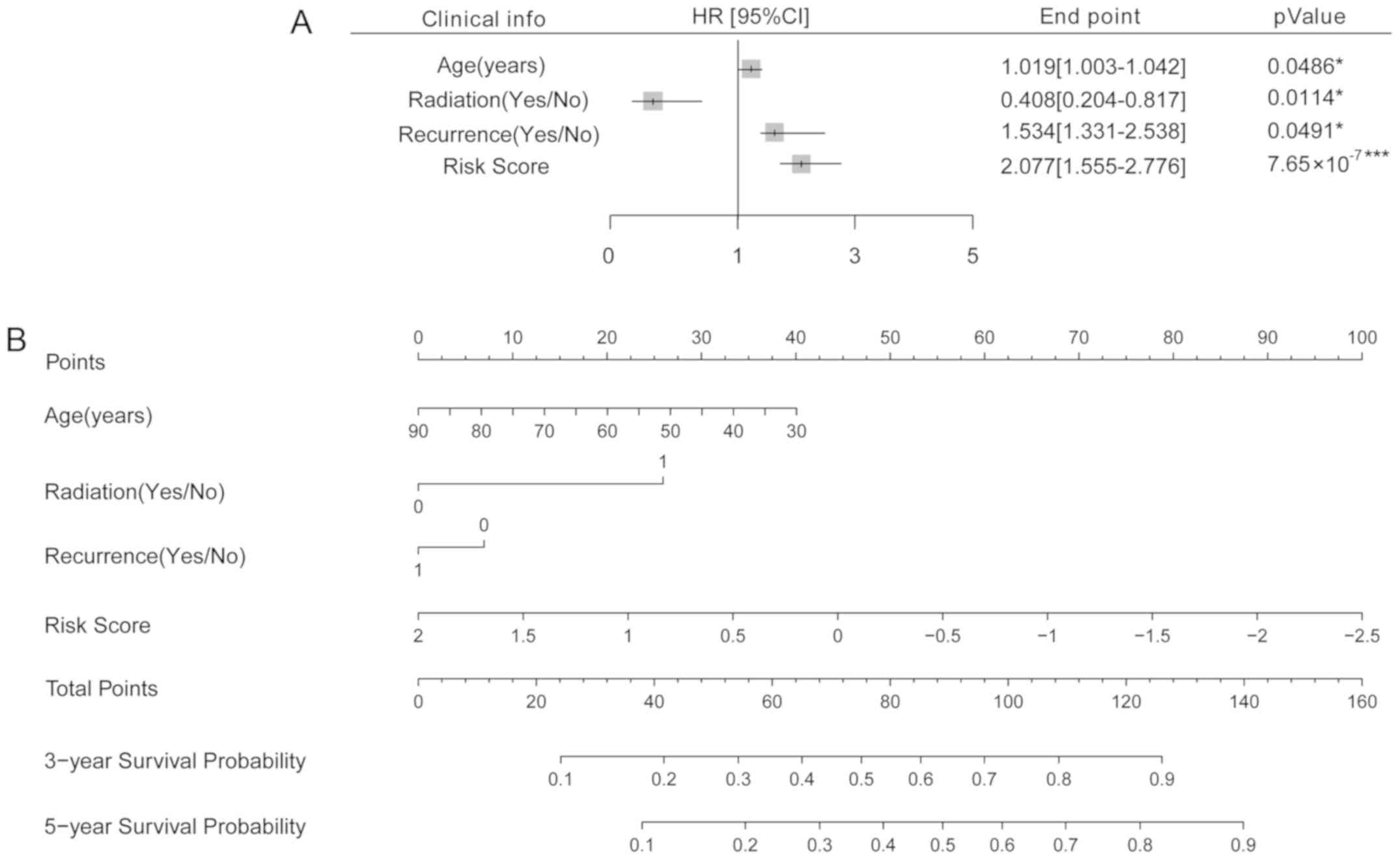
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rugge M, Fassan M and Graham DY:
Epidemiology of Gastric Cancer. Springer International Publishing;
New York, NY: pp. 23–34. 2015
|
|
3
|
Sanei MH, Mirmosayyeb O, Chehrei A, Ansari
J and Saberi E: 5-year survival in gastric adenocarcinoma with
epithelial and stromal versican expression. Iran J Pathol.
14:26–32. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He XK and Sun LM: The increasing trend in
the incidence of gastric cancer in the young population, not only
in young Hispanic men. Gastric Cancer. 20:10102017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Merchant SJ, Kim J, Choi AH, Sun V, Chao J
and Nelson R: A rising trend in the incidence of advanced gastric
cancer in young Hispanic men. Gastric Cancer. 20:226–234. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Z, Lei H, Luo M, Wang Y, Dong L, Ma Y,
Liu C, Song W, Wang F, Zhang J, et al: DNA methylation
downregulated mir-10b acts as a tumor suppressor in gastric cancer.
Gastric Cancer. 18:43–54. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ng EK, Chong WW, Jin H, Lam EK, Shin VY,
Yu J, Poon TC, Ng SS and Sung JJ: Differential expression of
microRNAs in plasma of patients with colorectal cancer: A potential
marker for colorectal cancer screening. Gut. 58:1375–1381. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shikata K, Doi Y, Yonemoto K, Arima H,
Ninomiya T, Kubo M, Tanizaki Y, Matsumoto T, Iida M and Kiyohara Y:
Population-based prospective study of the combined influence of
cigarette smoking and Helicobacter pylori infection on gastric
cancer incidence: The Hisayama Study. Am J Epidemiol.
168:1409–1415. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamagata H, Kiyohara Y, Aoyagi K, Kato I,
Iwamoto H, Nakayama K, Shimizu H, Tanizaki Y, Arima H, Shinohara N,
et al: Impact of Helicobacter pylori infection on gastric cancer
incidence in a general Japanese population: The Hisayama study.
Arch Intern Med. 160:1962–1968. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shikata K, Kiyohara Y, Kubo M, Yonemoto K,
Ninomiya T, Shirota T, Tanizaki Y, Doi Y, Tanaka K, Oishi Y, et al:
A prospective study of dietary salt intake and gastric cancer
incidence in a defined Japanese population: The Hisayama study. Int
J Cancer. 119:196–201. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iida M, Ikeda F, Hata J, Hirakawa Y, Ohara
T, Mukai N, Yoshida D, Yonemoto K, Esaki M, Kitazono T, et al:
Development and validation of a risk assessment tool for gastric
cancer in a general Japanese population. Gastric Cancer.
21:383–390. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pavlou M, Ambler G, Seaman SR, Guttmann O,
Elliott P, King M and Omar RZ: How to develop a more accurate risk
prediction model when there are few events. BMJ. 351:h38682015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hussein AA, Ghani KR, Peabody J, Sarle R,
Abaza R, Eun D, Hu J, Fumo M, Lane B, Montgomery JS, et al:
Development and validation of an objective scoring tool for
robot-assisted radical prostatectomy: Prostatectomy assessment and
competency evaluation. J Urol. 197:1237–1244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nimeri AA, Bautista J, Ibrahim M, Philip
R, Al Shaban T, Maasher A and Altinoz A: Mandatory risk assessment
reduces venous thromboembolism in bariatric surgery patients. Obes
Surg. 28:541–547. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Aminian A, Andalib A, Khorgami Z, Cetin D,
Burguera B, Bartholomew J, Brethauer SA and Schauer PR: Who should
get extended thromboprophylaxis after bariatric surgery?: A risk
assessment tool to guide indications for post-discharge
pharmacoprophylaxis. Ann Surg. 265:143–150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kastrinos F, Uno H, Ukaegbu C, Alvero C,
McFarland A, Yurgelun MB, Kulke MH, Schrag D, Meyerhardt JA, Fuchs
CS, et al: Development and validation of the PREMM5
model for comprehensive risk assessment of lynch syndrome. J Clin
Oncol. 35:2165–2172. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Charvat H, Sasazuki S, Inoue M, Iwasaki M,
Sawada N, Shimazu T, Yamaji T and Tsugane S; JPHC Study Group, :
Prediction of the 10-year probability of gastric cancer occurrence
in the Japanese population: The JPHC study cohort II. Int J Cancer.
138:320–331. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yuan SQ, Wu WJ, Qiu MZ, Wang ZX, Yang LP,
Jin Y, Yun JP, Gao YH, Li YH, Zhou ZW, et al: Development and
validation of a nomogram to predict the benefit of adjuvant
radiotherapy for patients with resected gastric cancer. J Cancer.
8:3498–3505. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang P, Wang Y, Hang B, Zou X and Mao JH:
A novel gene expression-based prognostic scoring system to predict
survival in gastric cancer. Oncotarget. 7:55343–55351.
2016.PubMed/NCBI
|
|
21
|
Tibshirani R: The lasso method for
variable selection in the Cox model. Stat Med. 16:385–395. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cristescu R, Lee J, Nebozhyn M, Kim KM,
Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al: Molecular
analysis of gastric cancer identifies subtypes associated with
distinct clinical outcomes. Nat Med. 21:449–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng
Z, Zhu G, Qi J, Ma H, Nian H and Wang Y: RNA-seq analyses of
multiple meristems of soybean: Novel and alternative transcripts,
evolutionary and functional implications. BMC Plant Biol.
14:1692014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Therneau T: A Package for Survival
Analysis in S. version 2.38. https://CRAN.R-project.org/package=survival2015
|
|
27
|
Thernea u, Terry M, Grambsc h and Patricia
M: Modeling Survival Data: Extending the Cox Model. Springer; New
York, NY: 2000, View Article : Google Scholar
|
|
28
|
Goeman JJ: L1 penalized estimation in the
Cox proportional hazards model. Biom J. 52:70–84. 2010.PubMed/NCBI
|
|
29
|
Bachoc F: Cross validation and maximum
likelihood estimations of hyper-parameters of gaussian processes
with model misspecification. Comput Stat Data Anal. 66:55–69. 2013.
View Article : Google Scholar
|
|
30
|
Camp RL, Dolled-Filhart M and Rimm DL:
X-tile: A new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Eng KH, Schiller E and Morrell K: On
representing the prognostic value of continuous gene expression
biomarkers with the restricted mean survival curve. Oncotarget.
6:36308–36318. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Iasonos A, Schrag D, Raj GV and Panageas
KS: How to build and interpret a nomogram for cancer prognosis. J
Clin Oncol. 26:1364–1370. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Piel FB, Howes RE, Patil AP, Nyangiri OA,
Gething PW, Bhatt S, Williams TN, Weatherall DJ and Hay SI: The
distribution of haemoglobin C and its prevalence in newborns in
Africa. Sci Rep. 3:16712013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Thom CS, Dickson CF, Gell DA and Weiss MJ:
Hemoglobin variants: Biochemical properties and clinical
correlates. Cold Spring Harb Perspect Med. 3:a0118582013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Endele S, Nelkenbrecher C, Bördlein A,
Schlickum S and Winterpacht A: C4ORF48, a gene from the
Wolf-Hirschhorn syndrome critical region, encodes a putative
neuropeptide and is expressed during neocortex and cerebellar
development. Neurogenetics. 12:155–163. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Deeb A: Diabetes mellitus secondary to
acute pancreatitis in a child with Wolf-Hirschhorn syndrome. Case
Rep Endocrinol. 2017:38924672017.PubMed/NCBI
|
|
39
|
Grotewold L and Rüther U: The Wnt
antagonist Dickkopf-1 is regulated by Bmp signaling and c-Jun and
modulates programmed cell death. EMBO J. 21:966–975. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tian EM, Zhan F, Walker R, Rasmussen E, Ma
Y, Barlogie B and Shaughnessy JD Jr: The role of the Wnt-signaling
antagonist DKK1 in the development of osteolytic lesions in
multiple myeloma. N Engl J Med. 349:2483–2494. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Providence KM, White LA, Tang J, Gonclaves
J, Staiano-Coico L and Higgins PJ: Epithelial monolayer wounding
stimulates binding of USF-1 to an E-box motif in the plasminogen
activator inhibitor type 1 gene. J Cell Sci. 115:3767–3777. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Turchi L, Chassot AA, Rezzonico R, Yeow K,
Loubat A, Ferrua B, Lenegrate G, Ortonne JP and Ponzio G: Dynamic
characterization of the molecular events during in vitro epidermal
wound healing. J Investig Dermatol. 119:56–63. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
French D, Hamilton LH, Mattano LA Jr,
Sather HN, Devidas M, Nachman JB and Relling MV; Children's
Oncology Group, : A PAI-1 (SERPINE1) polymorphism predicts
osteonecrosis in children with acute lymphoblastic leukemia: A
report from the Children's Oncology Group. Blood. 111:4496–4499.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Providence KM, Higgins SP, Mullen A,
Battista A, Samarakoon R, Higgins CE, Wilkins-Port CE and Higgins
PJ: SERPINE1 (PAI-1) is deposited into keratinocyte migration
‘trails’ and required for optimal monolayer wound repair. Arch
Dermatol Res. 300:303–310. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Herebian D, Alhaddad B, Seibt A,
Schwarzmayr T, Danhauser K, Klee D, Harmsen S, Meitinger T, Strom
TM, Schulz A, et al: Coexisting variants in OSTM1 and MANEAL cause
a complex neurodegenerative disorder with NBIA-like brain
abnormalities. Eur J Hum Genet. 25:1092–1095. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Moll F, Walter M, Rezende F, Helfinger V,
Vasconez E, De Oliveira T, Greten FR, Olesch C, Weigert A, Radeke
HH and Schröder K: NoxO1 controls proliferation of colon epithelial
cells. Front Immunol. 9:9732018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Oshima H, Ishikawa T, Yoshida GJ, Naoi K,
Maeda Y, Naka K, Ju X, Yamada Y, Minamoto T, Mukaida N, et al:
TNF-α/TNFR1 signaling promotes gastric tumorigenesis through
induction of Noxo1 and Gna14 in tumor cells. Oncogene.
33:3820–3829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gui SL, Teng LC, Wang SQ, Liu S, Lin YL,
Zhao XL, Liu L, Sui HY, Yang Y, Liang LC, et al: Overexpression of
CXCL3 can enhance the oncogenic potential of prostate cancer. Int
Urol Nephrol. 48:701–709. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sugiura T and Miyamoto K: Characterization
of TRIM31, upregulated in gastric adenocarcinoma, as a novel RBCC
protein. J Cell Biochem. 105:1081–1091. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
See AL, Chong PK, Lu SY and Lim YP: CXCL3
is a potential target for breast cancer metastasis. Curr Cancer
Drug Targets. 14:294–309. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sugiura T: The cellular level of TRIM31,
an RBCC protein overexpressed in gastric cancer, is regulated by
multiple mechanisms including the ubiquitin-proteasome system. Cell
Biol Int. 35:657–661. 2013. View Article : Google Scholar
|
|
52
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Neuzillet C, Tijeras-Raballand A, Cohen R,
Cros J, Faivre S, Raymond E and de Gramont A: Targeting the TGFβ
pathway for cancer therapy. Pharmacol Ther. 147:22–31. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zou X and Blank M: Targeting p38 MAP
kinase signaling in cancer through post-translational
modifications. Cancer Lett. 384:19–26. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Waddell T, Verheij M, Allum W, Cunningham
D, Cervantes A and Arnold D; European Society for Medical Oncology
(ESMO); European Society of Surgical Oncology (ESSO); European
Society of Radiotherapy Oncology (ESTRO), : Gastric cancer:
ESMO-ESSO-ESTRO clinical practice guidelines for diagnosis,
treatment and follow-up. Ann Oncol. 24 (Suppl 6):vi57–vi63. 2013.
View Article : Google Scholar : PubMed/NCBI
|