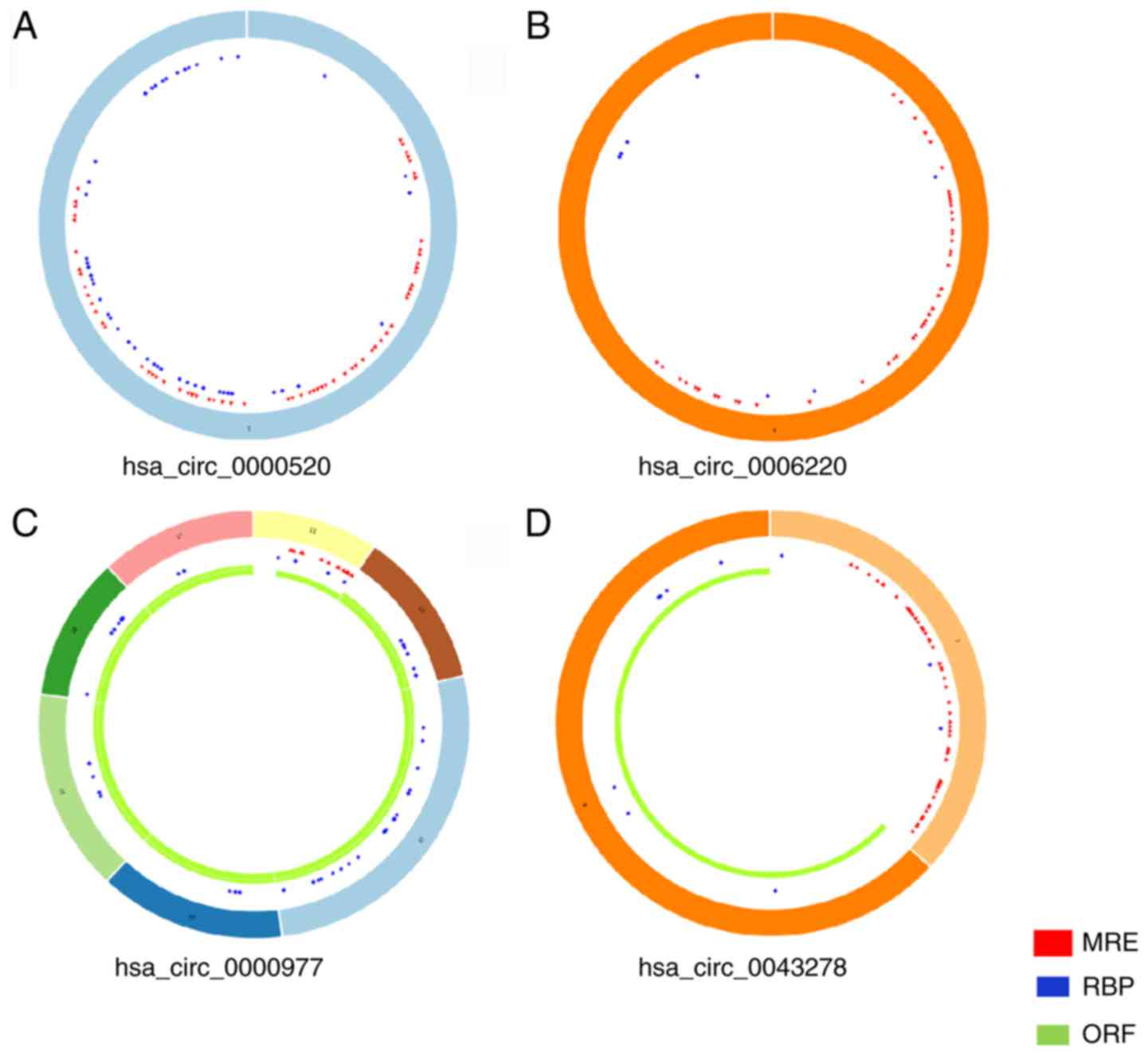
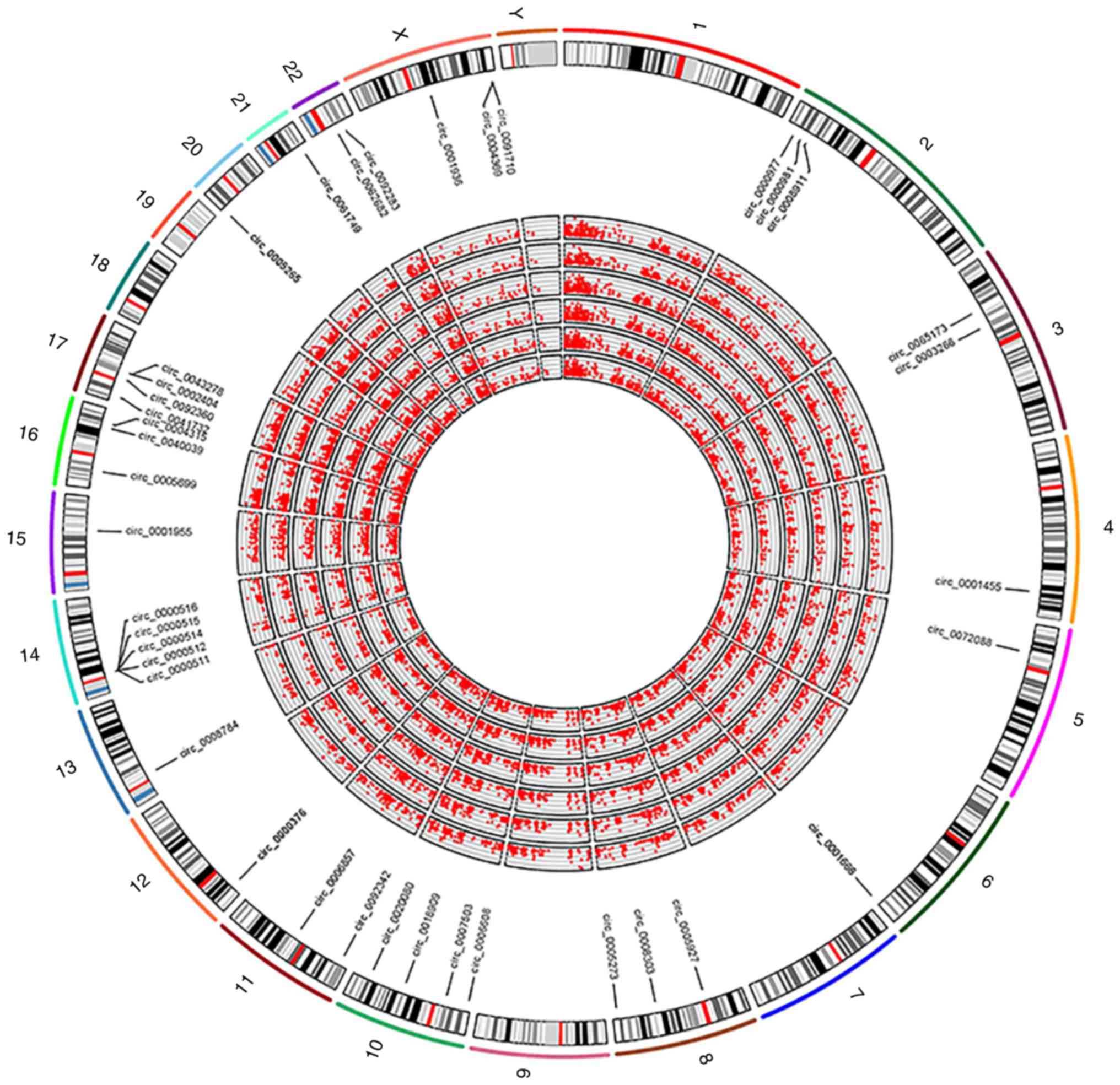
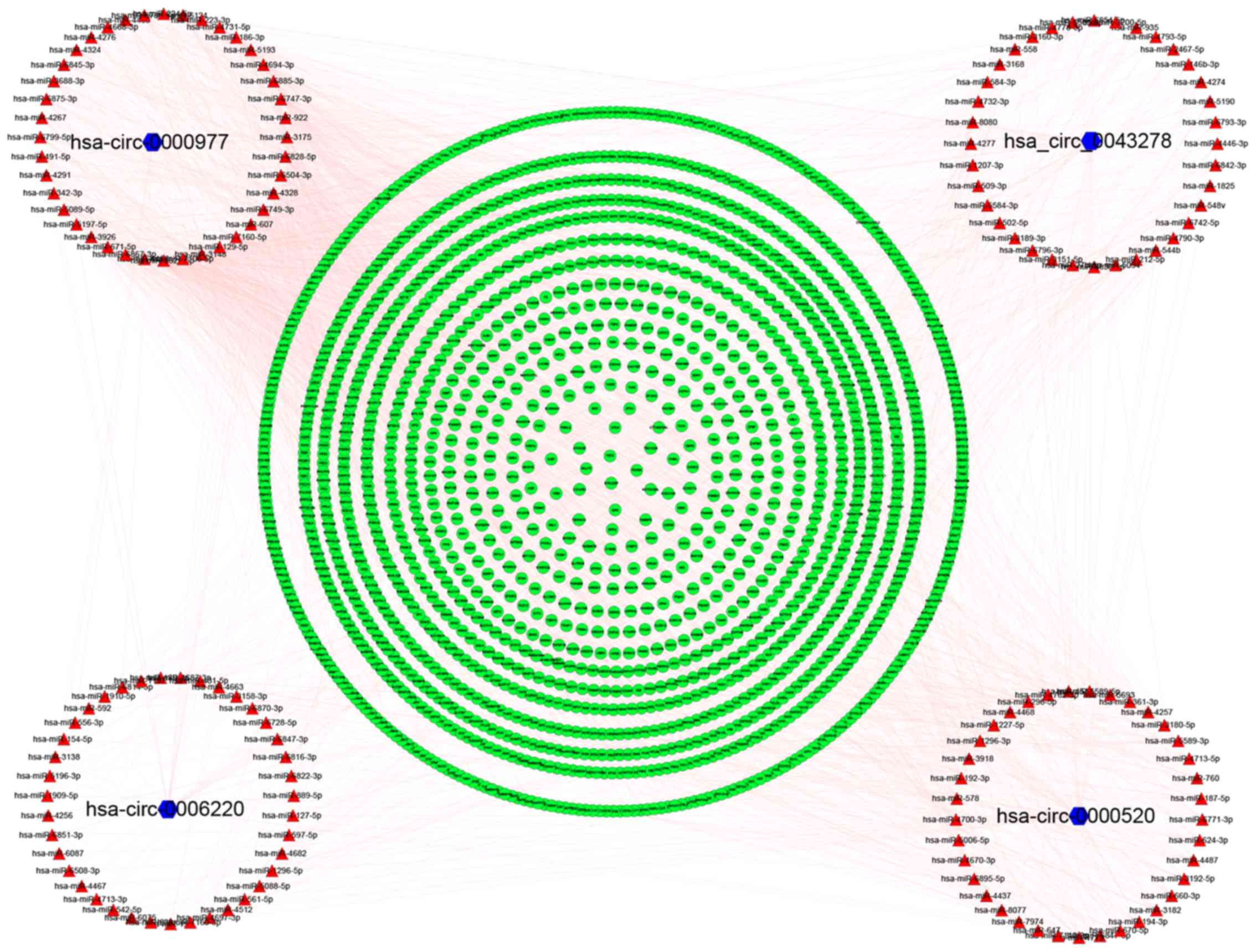
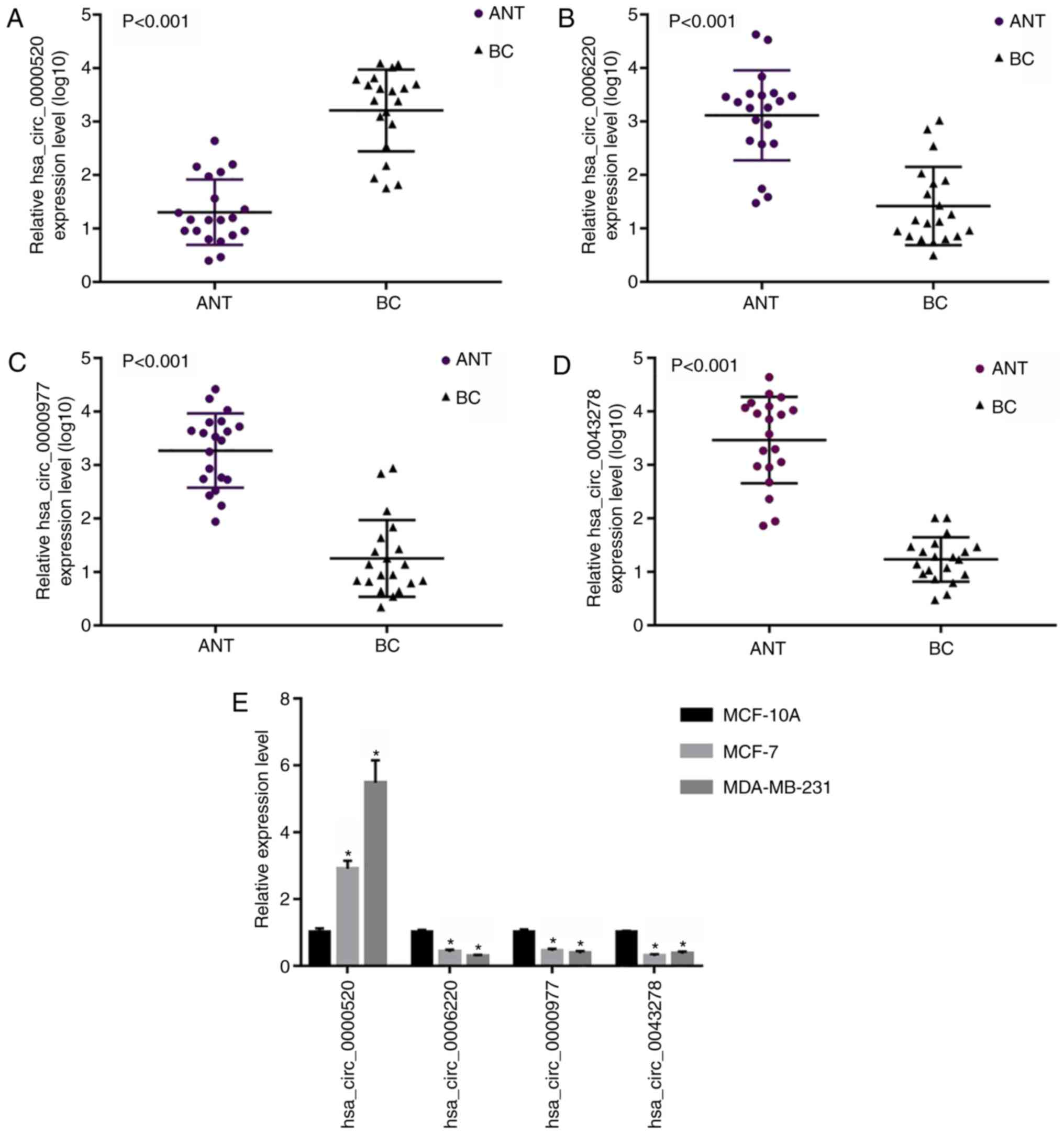
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo Y, Warren Andersen S, Shu XO,
Michailidou K, Bolla MK, Wang Q, Garcia-Closas M, Milne RL, Schmidt
MK, Chang-Claude J, et al: Genetically predicted body mass index
and breast cancer risk: Mendelian randomization analyses of data
from 145,000 women of european descent. PLoS Med. 13:e10021052016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chlebowski RT, Manson JE, Anderson GL,
Cauley JA, Aragaki AK, Stefanick ML, Lane DS, Johnson KC,
Wactawski-Wende J, Chen C, et al: Estrogen plus progestin and
breast cancer incidence and mortality in the women's health
initiative observational study. J Natl Cancer Inst. 105:526–535.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lambertini M, Santoro L, Del Mastro L,
Nguyen B, Livraghi L, Ugolini D, Peccatori FA and Azim HA Jr:
Reproductive behaviors and risk of developing breast cancer
according to tumor subtype: A systematic review and meta-analysis
of epidemiological studies. Cancer Treat Rev. 49:65–76. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rice MS, Eliassen AH, Hankinson SE, Lenart
EB, Willett WC and Tamimi RM: Breast cancer research in the Nurses'
health studies: Exposures across the life course. Am J Public
Health. 106:1592–1598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu L, Shen Y, Peng X, Zhang S, Wang M, Xu
G, Zheng X, Wang J and Lu C: Aberrant promoter methylation of
cancer-related genes in human breast cancer. Oncol Lett.
12:5145–5155. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang SM, Dowhan DH and Muscat G:
Epigenetic Arginine Methylation in Breast Cancer: Emerging
therapeutic strategies. J Mol Endocrinol. 62:R223–R237. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Salzman J: Circular RNA expression: Its
potential regulation and function. Trends Genet. 32:309–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu H, Guo S, Li W and Yu P: The circular
RNA Cdr1as, via miR-7 and its targets, regulates insulin
transcription and secretion in islet cells. Sci Rep. 5:124532015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei
P, Liu H, Xu J, Xiao F, Zhou H, et al: A peptide encoded by
circular form of LINC-PINT suppresses oncogenic transcriptional
elongation in glioblastoma. Nat Commun. 9:44752018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B
and Guo J: Global circular RNA expression profile of human gastric
cancer and its clinical significance. Cancer Med. 6:1173–1180.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thomas M, Lieberman J and Lal A:
Desperately seeking microRNA targets. Nat Struct Mol Biol.
17:1169–1174. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Swain AC and Mallick B: miRNA-mediated
‘tug-of-war’ model reveals ceRNA propensity of genes in cancers.
Mol Oncol. 12:855–868. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peng L, Yuan XQ and Li GC: The emerging
landscape of circular RNA ciRS-7 in cancer (Review). Oncol Rep.
33:2669–2674. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Presti M, Mazzon E, Basile MS, Petralia
MC, Bramanti A, Colletti G, Bramanti P, Nicoletti F and Fagone P:
Overexpression of macrophage migration inhibitory factor and
functionally-related genes, D-DT, CD74, CD44, CXCR2 and CXCR4, in
glioblastoma. Oncol Lett. 16:2881–2886. 2018.PubMed/NCBI
|
|
23
|
Fagone P, Mazzon E, Mammana S, Di Marco R,
Spinasanta F, Basile MS, Petralia MC, Bramanti P, Nicoletti F and
Mangano K: Identification of CD4+ T cell biomarkers for
predicting the response of patients with relapsing-remitting
multiple sclerosis to natalizumab treatment. Mol Med Rep.
20:678–684. 2019.PubMed/NCBI
|
|
24
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu JZ, Shao CC, Wang XJ, Zhao X, Chen JQ,
Ouyang YX, Feng J, Zhang F, Huang WH, Ying Q, et al: circTADA2As
suppress breast cancer progression and metastasis via targeting
miR-203a-3p/SOCS3 axis. Cell Death Dis. 10:1752019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xia S, Feng J, Chen K, Ma Y, Gong J, Cai
F, Jin Y, Gao Y, Xia L, Chang H, et al: CSCD: A database for
cancer-specific circular RNAs. Nucleic Acids Res. 46:D925–D929.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
30
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Su G, Morris JH, Demchak B and Bader GD:
Biological network exploration with Cytoscape 3. Curr Protoc
Bioinformatics. 47:8.13.1–24. 2014. View Article : Google Scholar
|
|
32
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kanehisa M, Goto S, Furumichi M, Tanabe M
and Hirakawa M: KEGG for representation and analysis of molecular
networks involving diseases and drugs. Nucleic Acids Res.
38:D355–D360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li M, Ding W, Tariq MA, Chang W, Zhang X,
Xu W, Hou L, Wang Y and Wang J: A circular transcript of ncx1 gene
mediates ischemic myocardial injury by targeting miR-133a-3p.
Theranostics. 8:5855–5869. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ludwig NF, Sperb-Ludwig F, Velho RV and
Schwartz IV: Next-generation sequencing corroborates a probable de
novo GNPTG variation previously detected by Sanger sequencing. Mol
Genet Metab Rep. 11:92–93. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong
F, Ren D, Ye X, Li C, Wang Y, et al: Circular RNAs function as
ceRNAs to regulate and control human cancer progression. Mol
Cancer. 17:792018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong
W, Li X, Li G, Zeng Z and Tang H: circGFRA1 and GFRA1 act as ceRNAs
in triple negative breast cancer by regulating miR-34a. J Exp Clin
Cancer Res. 36:1452017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen B, Wei W, Huang X and Xie X, Kong Y,
Dai D, Yang L, Wang J, Tang H and Xie X: circEPSTI1 as a prognostic
marker and mediator of triple-negative breast cancer progression.
Theranostics. 8:4003–4015. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang R, Xing L, Wang M, Chi H, Zhang L and
Chen J: Comprehensive analysis of differentially expressed profiles
of lncRNAs/mRNAs and miRNAs with associated ceRNA networks in
triple-negative breast cancer. Cell Physiol Biochem. 50:473–488.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu
X, Li Z, Wei J, Liu M and Li G: Circular RNA circ-DONSON
facilitates gastric cancer growth and invasion via NURF complex
dependent activation of transcription factor SOX4. Mol Cancer.
18:452019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nair AA, Niu N, Tang X, Thompson KJ, Wang
L, Kocher JP, Subramanian S and Kalari KR: Circular RNAs and their
associations with breast cancer subtypes. Oncotarget.
7:80967–80979. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Coscujuela Tarrero L, Ferrero G, Miano V,
De Intinis C, Ricci L, Arigoni M, Riccardo F, Annaratone L,
Castellano I, Calogero RA, et al: Luminal breast cancer-specific
circular RNAs uncovered by a novel tool for data analysis.
Oncotarget. 9:14580–14596. 2018.PubMed/NCBI
|
|
46
|
Zhang HD, Jiang LH, Hou JC, Zhou SY, Zhong
SL, Zhu LP, Wang DD, Yang SJ, He YJ, Mao CF, et al: Circular RNA
hsa_circ_0072995 promotes breast cancer cell migration and invasion
through sponge for miR-30c-2-3p. Epigenomics. 10:1229–1242. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412.
2017.PubMed/NCBI
|
|
48
|
Wu Z, Huang W, Wang X, Wang T, Chen Y,
Chen B, Liu R, Bai P and Xing J: Circular RNA CEP128 acts as a
sponge of miR-145-5p in promoting the bladder cancer progression
via regulating SOX11. Mol Med. 24:402018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wei H, Pan L, Tao D and Li R: Circular RNA
circZFR contributes to papillary thyroid cancer cell proliferation
and invasion by sponging miR-1261 and facilitating C8orf4
expression. Biochem Biophys Res Commun. 503:56–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang L, Tong X, Zhou Z, Wang S, Lei Z,
Zhang T, Liu Z, Zeng Y, Li C, Zhao J, et al: Circular RNA
hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced
epithelial-mesenchymal transition and metastasis by controlling
TIF1γ in non-small cell lung cancer. Mol Cancer. 17:1402018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R
and Li H: The emerging functions and roles of circular RNAs in
cancer. Cancer Lett. 414:301–309. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li S, Teng S, Xu J, Su G, Zhang Y, Zhao J,
Zhang S, Wang H, Qin W, Lu ZJ, et al: Microarray is an efficient
tool for circRNA profiling. Brief Bioinform. 20:1420–1433. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lan X, Xu J, Chen C, Zheng C, Wang J, Cao
J, Zhu X and Ge M: the landscape of circular RNA expression
profiles in papillary thyroid carcinoma based on RNA sequencing.
Cell Physiol Biochem. 47:1122–1132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Petralia MC, Mazzon E, Fagone P, Russo A,
Longo A, Avitabile T, Nicoletti F, Reibaldi M and Basile MS:
Characterization of the pathophysiological role of CD47 in uveal
melanoma. Molecules. 24(pii): E24502019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Basile MS, Mazzon E, Russo A, Mammana S,
Longo A, Bonfiglio V, Fallico M, Caltabiano R, Fagone P, Nicoletti
F, et al: Differential modulation and prognostic values of
immune-escape genes in uveal melanoma. PLoS One. 14:e02102762019.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Fagone P, Caltabiano R, Russo A, Lupo G,
Anfuso CD, Basile MS, Longo A, Nicoletti F, De Pasquale R, Libra M
and Reibaldi M: Identification of novel chemotherapeutic strategies
for metastatic uveal melanoma. Sci Rep. 7:445642017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Fagone P, Mangano K, Mammana S, Pesce A,
Pesce A, Caltabiano R, Giorlandino A, Portale TR, Cavalli E,
Lombardo GA, et al: Identification of novel targets for the
diagnosis and treatment of liver fibrosis. Int J Mol Med.
36:747–752. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Fagone P, Mazzon E, Cavalli E, Bramanti A,
Petralia MC, Mangano K, Al-Abed Y, Bramati P and Nicoletti F:
Contribution of the macrophage migration inhibitory factor
superfamily of cytokines in the pathogenesis of preclinical and
human multiple sclerosis: In silico and in vivo evidences. J
Neuroimmunol. 322:46–56. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fagone P, Nunnari G, Lazzara F, Longo A,
Cambria D, Distefano G, Palumbo M, Nicoletti F, Malaguarnera L and
Di Rosa M: Induction of OAS gene family in HIV monocyte infected
patients with high and low viral load. Antiviral Res. 131:66–73.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Basile MS, Fagone P, Mangano K, Mammana S,
Magro G, Salvatorelli L, Li Destri G, La Greca G, Nicoletti F,
Puleo S and Pesce A: KCNMA1 expression is downregulated in
colorectal cancer via epigenetic mechanisms. Cancers (Basel).
11(pii): E2452019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yuan C, Zhou L, Zhang L, Yin K, Peng J,
Sha R, Zhang S, Xu Y, Sheng X, Wang Y, et al: Identification and
integrated analysis of key differentially expressed circular RNAs
in ER-positive subtype breast cancer. Epigenomics. 11:297–321.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Baer M, Nilsen TW, Costigan C and Altman
S: Structure and transcription of a human gene for H1 RNA, the RNA
component of human RNase P. Nucleic Acids Res. 18:97–103. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Barlev NA, Emelyanov AV, Castagnino P,
Zegerman P, Bannister AJ, Sepulveda MA, Robert F, Tora L,
Kouzarides T, Birshtein BK and Berger SL: A novel human Ada2
homologue functions with Gcn5 or Brg1 to coactivate transcription.
Mol Cell Biol. 23:6944–6957. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wu Y, Xie Z, Chen J, Chen J, Ni W, Ma Y,
Huang K, Wang G, Wang J, Ma J, et al: Circular RNA circTADA2A
promotes osteosarcoma progression and metastasis by sponging
miR-203a-3p and regulating CREB3 expression. Mol Cancer. 18:732019.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bammert L, Jonas S, Ungricht R and Kutay
U: Human AATF/Che-1 forms a nucleolar protein complex with NGDN and
NOL10 required for 40S ribosomal subunit synthesis. Nucleic Acids
Res. 44:9803–9820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhao F, Han Y, Liu Z, Zhao Z, Li Z and Jia
K: circFADS2 regulates lung cancer cells proliferation and invasion
via acting as a sponge of miR-498. Biosci Rep. 38(pii):
BSR201805702018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
He JH, Li YG, Han ZP, Zhou JB, Chen WM, Lv
YB, He ML, Zuo JD and Zheng L: The CircRNA-ACAP2/Hsa-miR-21-
5p/Tiam1 regulatory feedback circuit affects the proliferation,
migration, and invasion of colon cancer SW480 cells. Cell Physiol
Biochem. 49:1539–1550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Xue J, Liu Y, Luo F, Lu X, Xu H, Liu X, Lu
L, Yang Q, Chen C, Fan W and Liu Q: Circ100284, via miR-217
regulation of EZH2, is involved in the arsenite-accelerated cell
cycle of human keratinocytes in carcinogenesis. Biochim Biophys
Acta Mol Basis Dis. 1863:753–763. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Huang S, Li X, Zheng H, Si X, Li B, Wei G,
Li C, Chen Y, Chen Y, Liao W, et al: Loss of
super-enhancer-regulated CircRNA Nfix induces cardiac regeneration
after myocardial infarction in adult mice. Circulation.
139:2857–2876. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhao J, Zou H, Han C, Ma J, Zhao J and
Tang J: Circlular RNA BARD1 (Hsa_circ_0001098) overexpression in
breast cancer cells with TCDD treatment could promote cell
apoptosis via miR-3942/BARD1 axis. Cell Cycle. 17:2731–2744. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
74
|
Mehrian-Shai R, Chen CD, Shi T, Horvath S,
Nelson SF, Reichardt JK and Sawyers CL: Insulin growth
factor-binding protein 2 is a candidate biomarker for PTEN status
and PI3K/Akt pathway activation in glioblastoma and prostate
cancer. Proc Natl Acad Sci USA. 104:5563–5568. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhen N, Gu S, Ma J, Zhu J, Yin M, Xu M,
Wang J, Huang N, Cui Z, Bian Z, et al: CircHMGCS1 promotes
hepatoblastoma cell proliferation by regulating the IGF signaling
pathway and glutaminolysis. Theranostics. 9:900–919. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Xu L, Feng X, Hao X, Wang P, Zhang Y,
Zheng X, Li L, Ren S, Zhang M and Xu M: CircSETD3
(Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting
hepatocellular carcinoma growth. J Exp Clin Cancer Res. 38:982019.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Nanta R, Shrivastava A, Sharma J, Shankar
S and Srivastava RK: Inhibition of sonic hedgehog and PI3K/Akt/mTOR
pathways cooperate in suppressing survival, self-renewal and
tumorigenic potential of glioblastoma-initiating cells. Mol Cell
Biochem. 454:11–23. 2019. View Article : Google Scholar : PubMed/NCBI
|