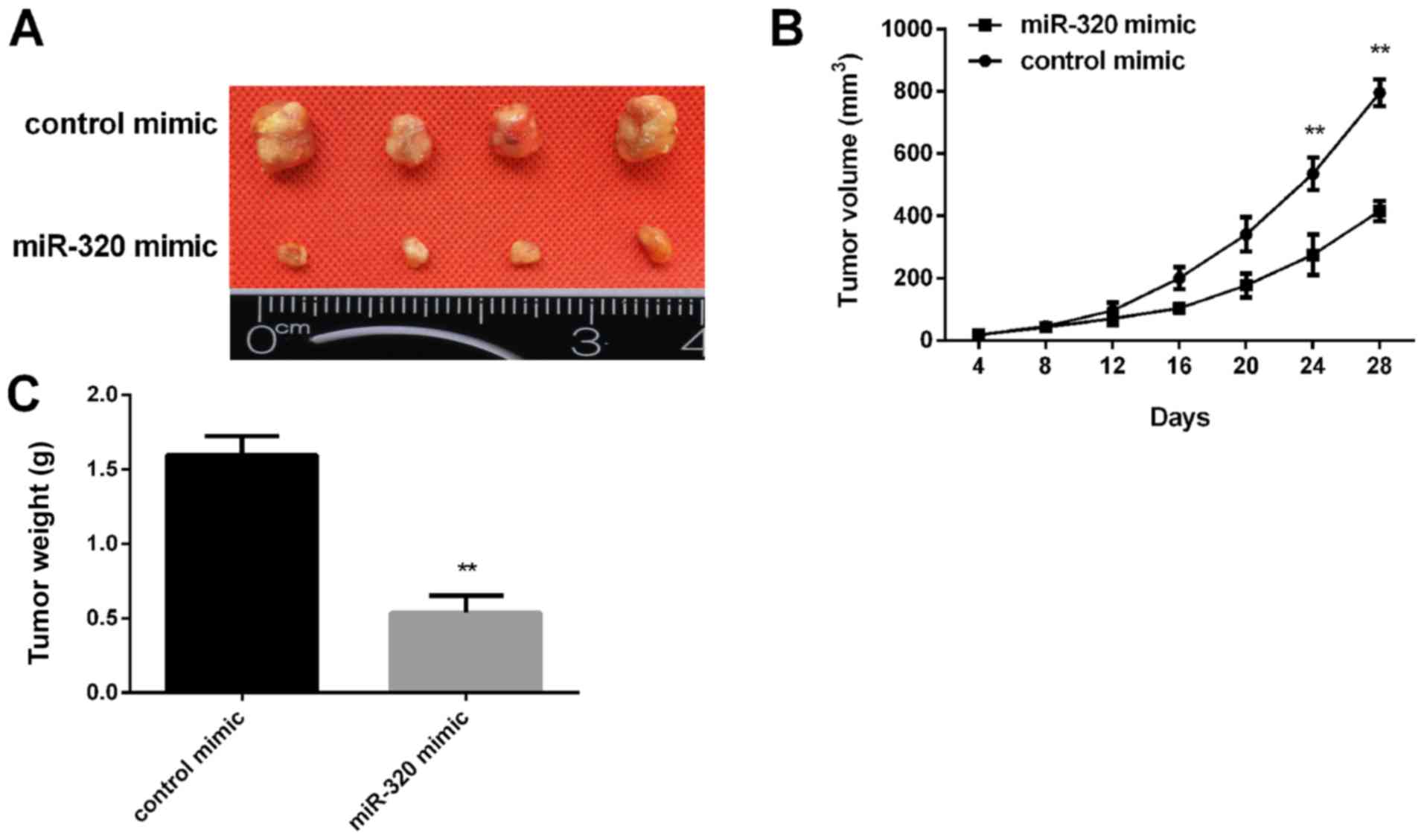
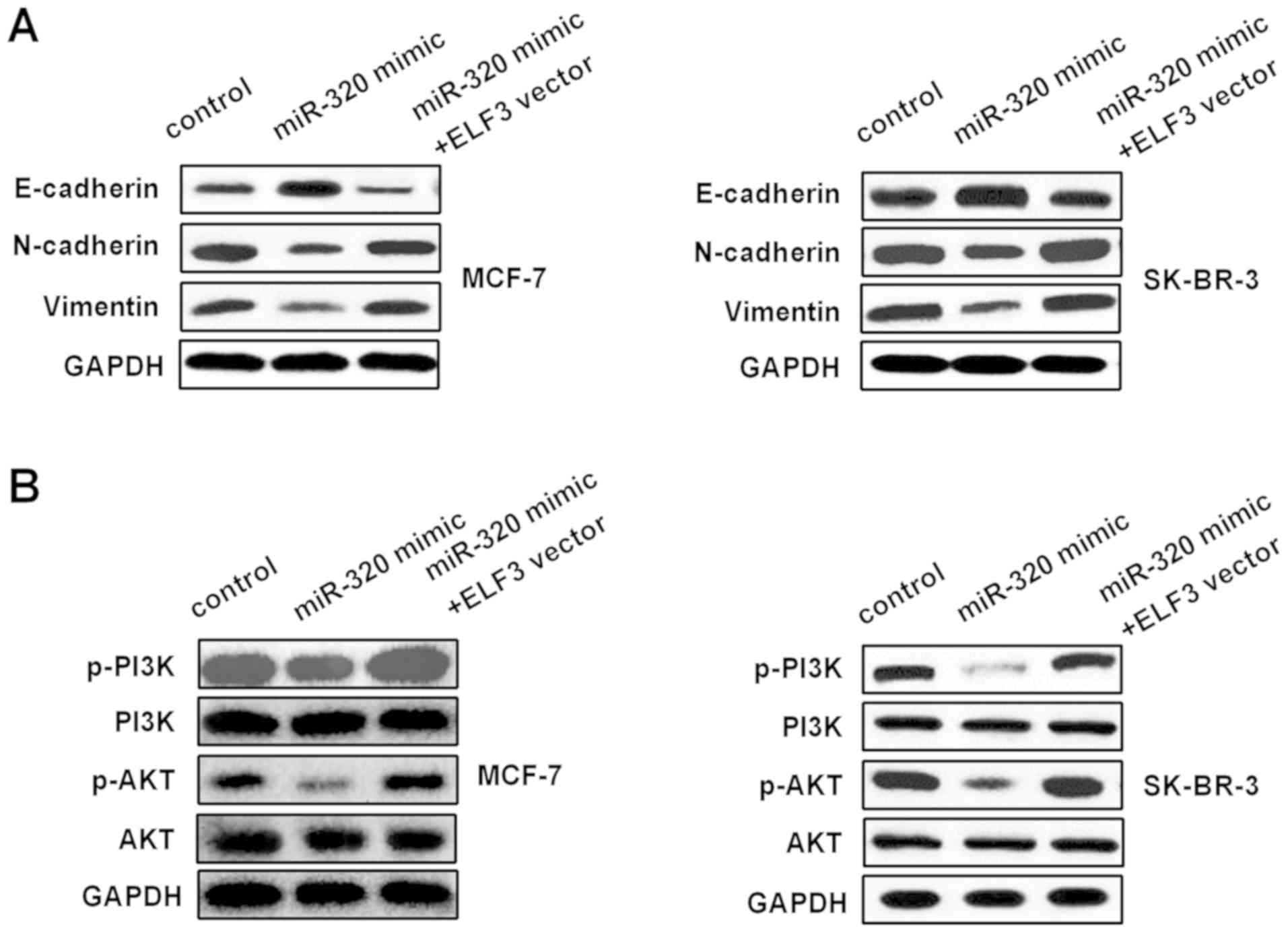
|
1
|
Hong W and Dong E: The past, present and
future of breast cancer research in China. Cancer Lett. 351:1–5.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ren L, Li Y, Zhao Q, Fan L, Tan B, Zang A
and Yang H: miR-519 regulates the proliferation of breast cancer
cells via targeting human antigen R. Oncol Lett. 19:1567–1576.
2020.PubMed/NCBI
|
|
3
|
DeSantis CE, Ma J, Goding Sauer A, Newman
LA and Jemal A: Breast cancer statistics, 2017, racial disparity in
mortality by state. CA Cancer J Clin. 67:439–448. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zuo TT, Zheng RS, Zeng HM, Zhang SW and
Chen WQ: Female breast cancer incidence and mortality in China,
2013. Thorac Cancer. 8:214–218. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dinami R, Buemi V, Sestito R, Zappone A,
Ciani Y, Mano M, Petti E, Sacconi A, Blandino G, Giacca M, et al:
Epigenetic silencing of miR-296 and miR-512 ensures hTERT dependent
apoptosis protection and telomere maintenance in basal-type breast
cancer cells. Oncotarget. 8:95674–95691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao J and Jiang GQ: miR-4282 inhibits
proliferation, invasion and metastasis of human breast cancer by
targeting Myc. Eur Rev Med Pharmacol Sci. 22:8763–8771.
2018.PubMed/NCBI
|
|
7
|
Lieb V, Weigelt K, Scheinost L, Fischer K,
Greither T, Marcou M, Theil G, Klocker H, Holzhausen HJ, Lai X, et
al: Serum levels of miR-320 family members are associated with
clinical parameters and diagnosis in prostate cancer patients.
Oncotarget. 9:10402–10416. 2017.PubMed/NCBI
|
|
8
|
Zhang H and Lu W: LncRNA SNHG12 regulates
gastric cancer progression by acting as a molecular sponge of
miR-320. Mol Med Rep. 17:2743–2749. 2018.PubMed/NCBI
|
|
9
|
Bai JW, Wang X, Zhang YF, Yao GD and Liu
H: MicroRNA-320 inhibits cell proliferation and invasion in breast
cancer cells by targeting SOX4. Oncol Lett. 14:7145–7152.
2017.PubMed/NCBI
|
|
10
|
Oettgen P, Alani RM, Barcinski MA, Brown
L, Akbarali Y, Boltax J, Kunsch C, Munger K and Libermann TA:
Isolation and characterization of a novel epithelium-specific
transcription factor, ESE-1, a member of the ets family. Mol Cell
Biol. 17:4419–4433. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Neve RM, Parmar H, Amend C, Chen C,
Rizzino A and Benz CC: Identification of an epithelial-specific
enhancer regulating ESX expression. Gene. 367:118–125. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang JL, Chen ZF, Chen HM, Wang MY, Kong
X, Wang YC, Sun TT, Hong J, Zou W, Xu J and Fang JY: Elf3 drives
β-catenin transactivation and associates with poor prognosis in
colorectal cancer. Cell Death Dis. 5:e12632014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Longoni N, Sarti M, Albino D, Civenni G,
Malek A, Ortelli E, Pinton S, Mello-Grand M, Ostano P, D'Ambrosio
G, et al: ETS transcription factor ESE1/ELF3 orchestrates a
positive feedback loop that constitutively activates NF-κB and
drives prostate cancer progression. Cancer Res. 73:4533–4547. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, Yu Z, Huo S, Chen Z, Ou Z, Mai J,
Ding S and Zhang J: Overexpression of ELF3 facilitates cell growth
and metastasis through PI3K/Akt and ERK signaling pathways in
non-small cell lung cancer. Int J Biochem Cell Biol. 94:98–106.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iwai S, Amekawa S, Yomogida K, Sumi T,
Nakazawa M, Yura Y, Nishimune Y and Nozaki M: ESE-1 inhibits the
invasion of oral squamous cell carcinoma in conjunction with MMP-9
suppression. Oral Dis. 14:144–149. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yeung TL, Leung CS, Wong KK,
Gutierrez-Hartmann A, Kwong J, Gershenson DM and Mok SC: ELF3 is a
negative regulator of epithelial-mesenchymal transition in ovarian
cancer cells. Oncotarget. 8:16951–16963. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ke K and Lou T: MicroRNA-10a suppresses
breast cancer progression via PI3K/Akt/mTOR pathway. Oncol Lett.
14:5994–6000. 2017.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ke SB, Qiu H, Chen JM, Shi W and Chen YS:
MicroRNA-202-5p functions as a tumor suppressor in colorectal
carcinoma by directly targeting SMARCC1. Gene. 676:329–335. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Quan H, Li B and Yang J: MicroRNA-504
functions as a tumor suppressor in hepatocellular carcinoma through
inhibiting Frizzled-7-mediated-Wnt/β-catenin signaling. Biomed
Pharmacother. 107:754–762. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duan S, Dong X, Hai J, Jiang J, Wang W,
Yang J, Zhang W and Chen C: MicroRNA-135a-3p is downregulated and
serves as a tumour suppressor in ovarian cancer by targeting CCR2.
Biomed Pharmacother. 107:712–720. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rohini M, Gokulnath M, Miranda PJ and
Selvamurugan N: miR-590-3p inhibits proliferation and promotes
apoptosis by targeting activating transcription factor 3 in human
breast cancer cells. Biochimie. 154:10–18. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan L, Yu MC, Gao GL, Liang HW, Zhou XY,
Zhu ZT, Zhang CY, Wang YB and Chen X: miR-125a-5p functions as a
tumour suppressor in breast cancer by downregulating BAP1. J Cell
Biochem. 119:8773–8783. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li C, Zhang J, Ma Z, Zhang F and Yu W:
miR-19b serves as a prognostic biomarker of breast cancer and
promotes tumor progression through PI3K/AKT signaling pathway. Onco
Targets Ther. 11:4087–4095. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Taylor MA, Sossey-Alaoui K, Thompson CL,
Danielpour D and Schiemann WP: TGF-β upregulates miR-181a
expression to promote breast cancer metastasis. J Clin Invest.
123:150–163. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luo L, Yang R, Zhao S, Chen Y, Hong S,
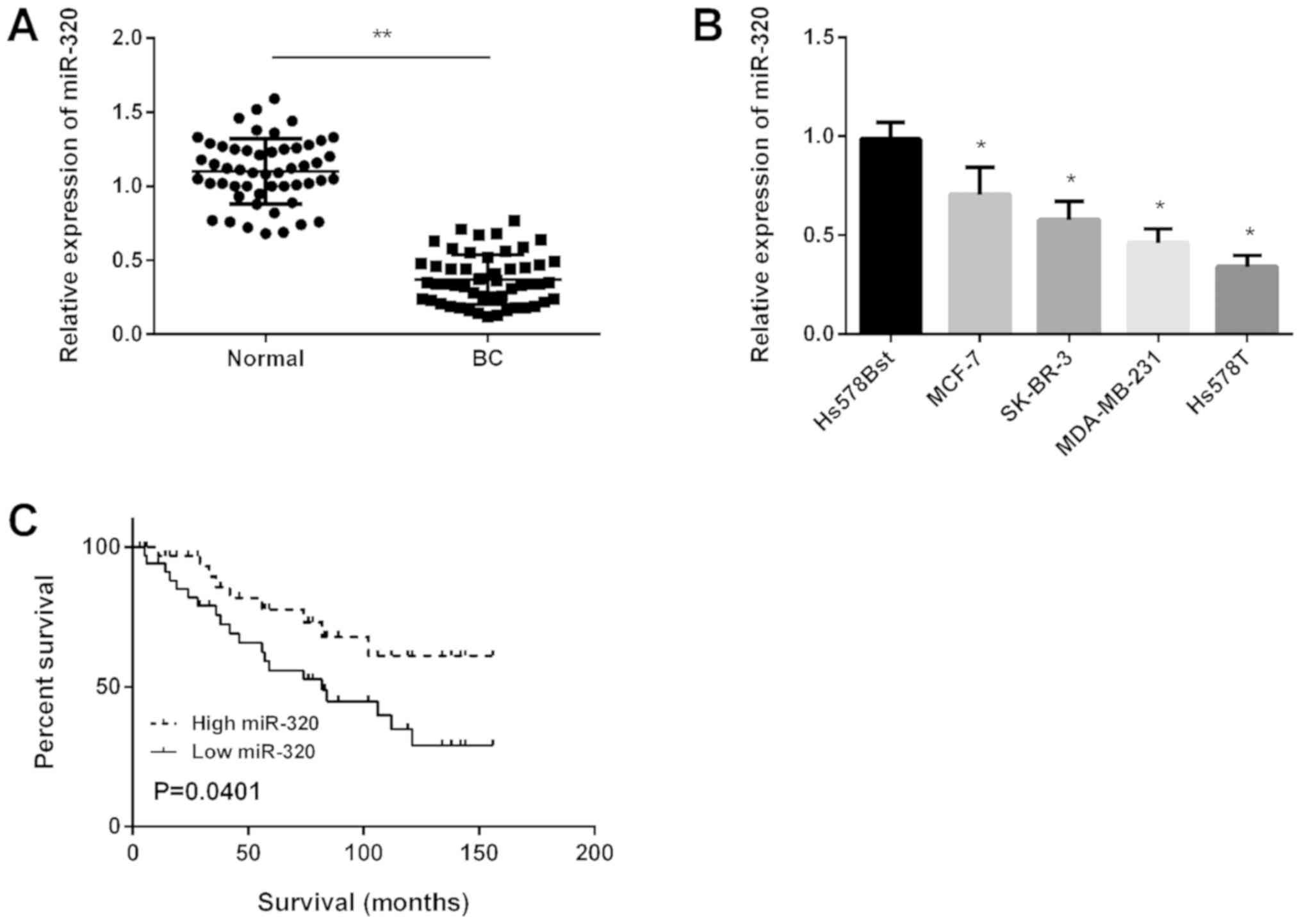
Wang K, Wang T, Cheng J, Zhang T and Chen D: Decreased miR-320
expression is associated with breast cancer progression, cell
migration, and invasiveness via targeting Aquaporin 1. Acta Biochim
Biophys Sin (Shanghai). 50:473–480. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Palumbo T, Faucz FR, Azevedo M, Xekouki P,
Iliopoulos D and Stratakis CA: Functional screen analysis reveals
miR-26b and miR-128 as central regulators of pituitary
somatomammotrophic tumor growth through activation of the PTEN-AKT
pathway. Oncogene. 32:1651–1659. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li T, Ma J, Han X, Jia Y, Yuan H, Shui S
and Guo D: MicroRNA-320 enhances radiosensitivity of glioma through
down-regulation of sirtuin type 1 by directly targeting forkhead
box protein M1. Transl Oncol. 11:205–212. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao W, Sun Q, Yu Z, Mao S, Jin Y, Li J,
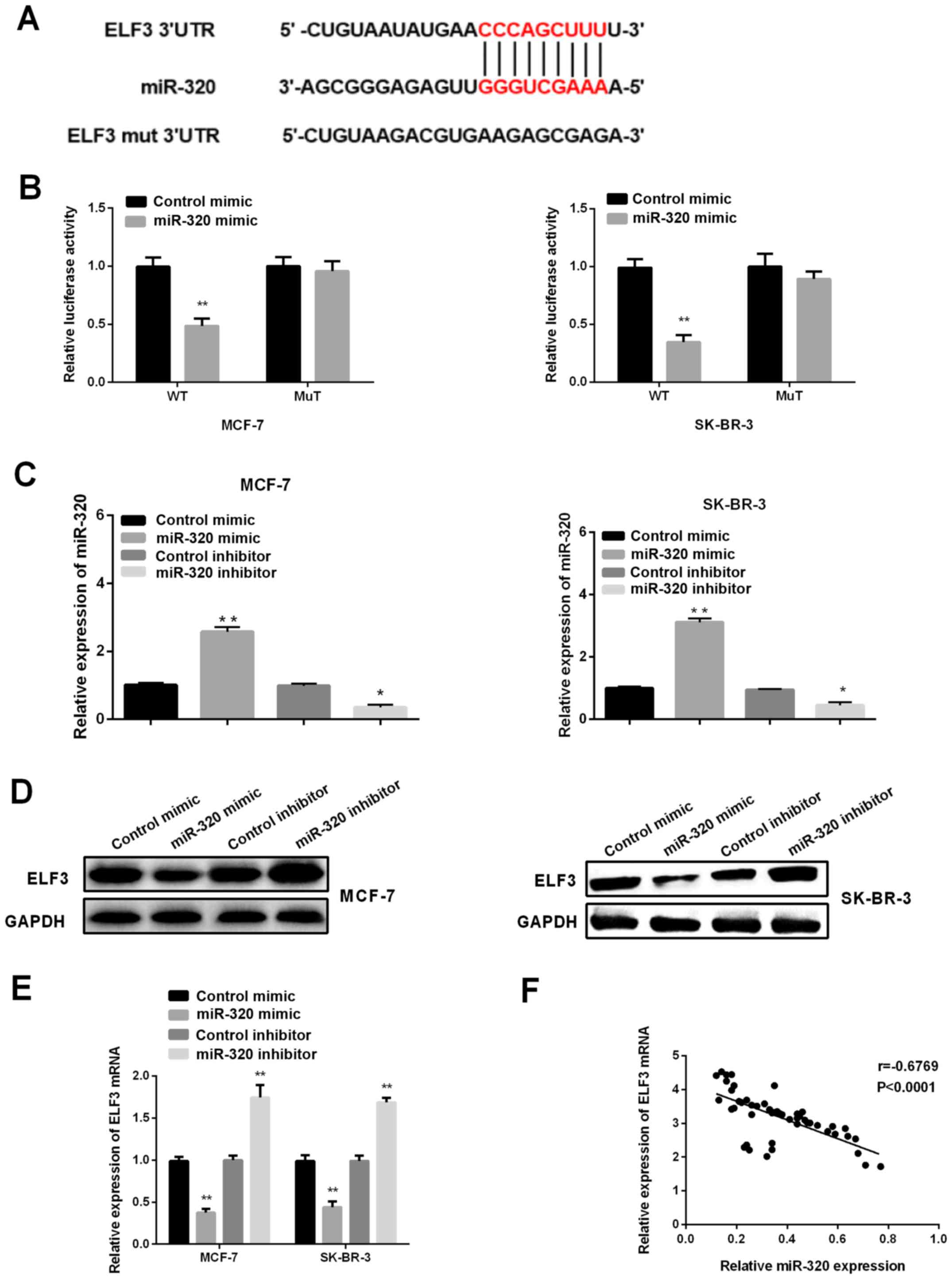
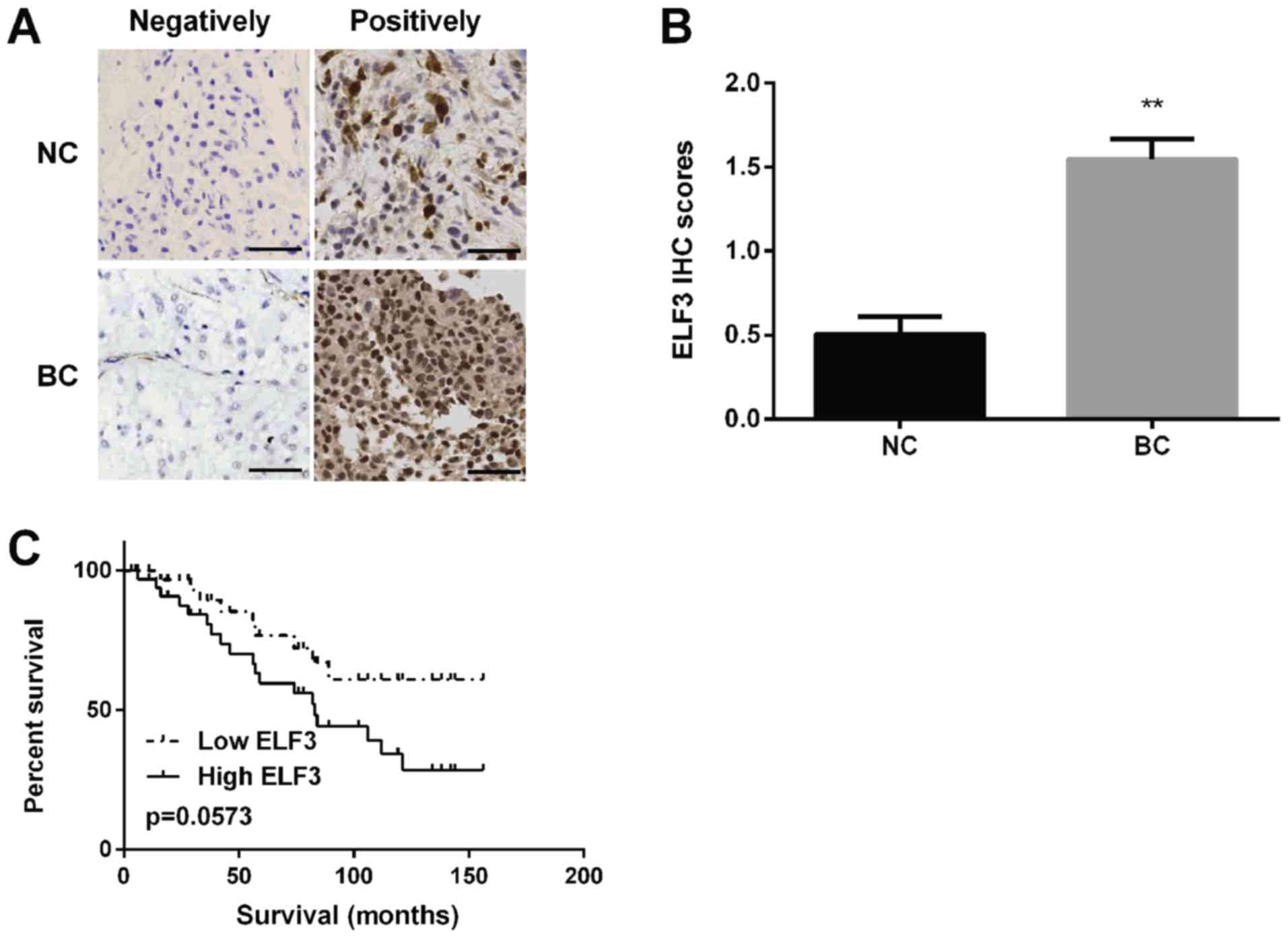
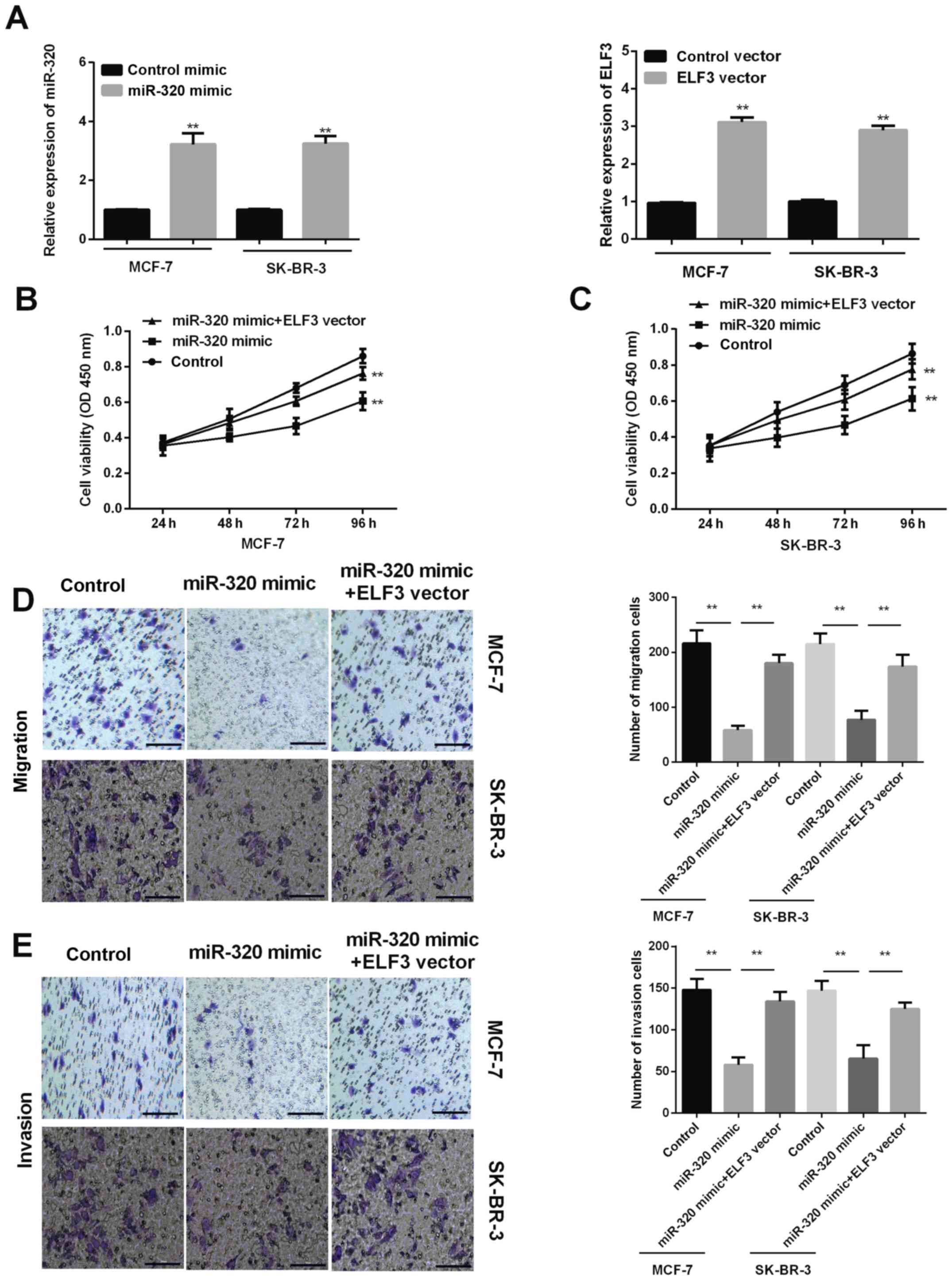
Jiang Z, Zhang Y, Chen M, Chen P, et al: miR-320a-3p/ELF3 axis
regulates cell metastasis and invasion in non-small cell lung
cancer via PI3K/Akt pathway. Gene. 670:31–37. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng L, Xu M, Xu J, Wu K, Fang Q, Liang
Y, Zhou S, Cen D, Ji L, Han W and Cai X: ELF3 promotes
epithelial-mesenchymal transition by protecting ZEB1 from
miR-141-3p-mediated silencing in hepatocellular carcinoma. Cell
Death Dis. 9:3872018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fang J, Ding M, Yang L, Liu LZ and Jiang
BH: PI3K/PTEN/AKT signaling regulates prostate tumor angiogenesis.
Cell Signal. 19:2487–2497. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dong Y, Liang G, Yuan B, Yang C, Gao R and
Zhou X: MALAT1 promotes the proliferation and metastasis of
osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol.
36:1477–1486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qiu N, He YF, Zhang SM, Zhan YT, Han GD,
Jiang M, He WX, Zhou J, Liang HL, Ao X, et al: Cullin7 enhances
resistance to trastuzumab therapy in Her2 positive breast cancer
via degrading IRS-1 and downregulating IGFBP-3 to activate the
PI3K/AKT pathway. Cancer Lett. 464:25–36. 2019. View Article : Google Scholar : PubMed/NCBI
|