Introduction
Lung cancer remains one of the most serious global
health burdens and represents a dominant cause of cancer-associated
mortality (1). Among all cases of
lung cancer, non-small cell lung cancer (NSCLC) is the most
frequent type, accounting for ~80% of all cases (2). NSCLC consists of squamous cell
carcinoma, adenocarcinoma, adenosquamous cell carcinoma and large
cell carcinoma (3). The majority of
patients with NSCLC are diagnosed with advanced tumors, and the
5-year survival rates in NSCLC range from 73% in stage IA disease
to 13% in stage IV disease, worldwide (4). Despite great progress in therapeutic
approaches for cancer treatment, such as surgery, chemotherapy,
radiotherapy and targeted therapy, the prognosis and outcomes
remain poor (5). To improve the
management of NSCLC, efforts should be made in the development of
novel and efficient strategies for diagnosis, prognosis and
treatment.
Several studies have demonstrated that tumor
initiation and development involve changes in the expression levels
of numerous key molecules (6–8).
MicroRNAs (miRNAs/miRs) are a group of these key molecules that
have been investigated in various types of cancer (9,10). These
small RNAs have no capacity of protein coding, but exert regulatory
roles in gene expression at the post-transcriptional level
(11). In addition, miRNAs are
involved in the regulation of various cell processes, such as cell
proliferation, migration, invasion, differentiation, cell cycle and
apoptosis (12,13). In cancer research, a large number of
miRNAs have been reported to be involved in tumor progression,
which means that these functional miRNAs may be considered as
potential therapeutic targets for cancer treatment (14,15).
Furthermore, the clinical significance of miRNAs has attracted
increasing attention in the diagnosis and prognosis of human
malignancies, including NSCLC (16).
miR-665 has been reported to be closely associated with pivotal
signaling pathways in lung cancer pathogenesis (17), and miR-665 expression in
extracellular vesicles is elevated in NSCLC (18). However, the clinical significance and
biological function of miR-665 in NSCLC remain unclear.
To improve the prognosis and treatment of NSCLC, the
present study aimed to investigate the expression profile and
prognostic value of miR-665 in patients with NSCLC. In addition,
cell experiments were carried out to uncover the potential
functional role of miR-665 in NSCLC tumor progression. The results
of the present study may provide a novel biomarker and a
therapeutic target for the treatment of this malignancy.
Materials and methods
Patients and tissue collection
The present study was approved by the Ethics
Committee of Shouguang People's Hospital (Shouguang, China) and
written informed consent was obtained from all patients prior to
the study start. A total of 128 patients diagnosed with NSCLC were
recruited between January 2010 and December 2013 at Shouguang
People's Hospital. The inclusion criteria were as follows: i)
Patients underwent surgical resection and were pathologically
diagnosed with NSCLC at the Shouguang People's Hospital, ii)
patients did not receive any preoperative antitumor therapy and
iii) patients had complete clinicopathological data and follow-up
information. Tumor tissues and adjacent normal tissues were
collected from patients following surgical excision, snap frozen in
liquid nitrogen and stored at −80°C. The demographic and
clinicopathological patient characteristics are listed in Table I. A 5-year follow-up survey was
performed, and the survival status of patients was recorded.
 | Table I.Association of miR-665 with
clinicopathological features of patients with non-small cell lung
cancer. |
Table I.
Association of miR-665 with
clinicopathological features of patients with non-small cell lung
cancer.
|
|
| miR-665
expression |
|
|---|
|
|
|
|
|
|---|
| Features | Total no.
(n=128) | Low (n=60) | High (n=68) | P-value |
|---|
| Age, years |
|
|
| 0.383 |
|
≤60 | 42 | 22 | 20 |
|
|
>60 | 86 | 38 | 48 |
|
| Sex |
|
|
| 0.469 |
|
Female | 47 | 24 | 23 |
|
|
Male | 81 | 36 | 45 |
|
| Smoking |
|
|
| 0.872 |
| No | 46 | 22 | 24 |
|
|
Yes | 82 | 38 | 44 |
|
| Tumor size
(cm) |
|
|
| 0.773 |
| ≤3 | 58 | 28 | 30 |
|
|
>3 | 70 | 32 | 38 |
|
|
Differentiation |
|
|
| 0.855 |
|
Well/moderate | 80 | 37 | 43 |
|
|
Poor | 48 | 23 | 25 |
|
| Lymph node
metastasis |
|
|
| 0.035a |
| No | 62 | 35 | 27 |
|
|
Yes | 66 | 25 | 41 |
|
| TNM stage |
|
|
| 0.009a |
|
I–II | 61 | 36 | 25 |
|
|
III–IV | 67 | 24 | 43 |
|
Cell culture and transfection
The NSCLC A549, H1299 and H522 cell lines, and the
human bronchial epithelial 16HBE cell line were obtained from The
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences. All cells were cultured in RPMI 1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.), and 100 IU/ml penicillin and 100
µg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at
37°C in a humidified atmosphere containing 5% CO2.
To regulate miR-665 expression in vitro, cell
transfection was performed using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The synthesized miR-665 mimic and
inhibitor (50 nM) were used to upregulate and downregulate the
expression levels of miR-665 in tumor cells. In addition, a
negative control sequence (miR-NC, 50 nM) transfected in the tumor
cells was used as a control, and cells transfected with the
transfection reagent only were set as the mock group. All vectors
were synthesized by Shanghai GenePharma Co., Ltd. with the
following sequences: miR-665 mimic, 5′-ACCAGGAGGCUGAGGCCCCU-3′;
miR-665 inhibitor, 5′-AGGGGCCUCAGCCUCCUGGU-3′; and miR-NC,
5′-UUCUCCGAACGUGUCACGU-3′. Subsequent experiments were performed 48
h post-transfection.
RNA extraction and reverse
transcription (RT)
Total RNA from tissues and cells was extracted using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The purity and
concentration of the RNA were measured using a NanoDrop 2000
(Thermo Fisher Scientific, Inc.). Total RNA was reversed
transcribed into cDNA using the PrimeScript RT reagent kit (Takara
Bio, Inc.), with the following reaction conditions: 42°C for 30 min
and 85°C for 5 sec.
RT-quantitative (q)PCR
The expression levels of miR-665 were measured using
RT-qPCR, which was carried out using a SYBR Green I Master Mix kit
(Invitrogen; Thermo Fisher Scientific, Inc.) and the 7300 Real-Time
PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The following primer sequences were used for qPCR: miR-665 forward,
5′-GCCGAGACCAGGAGGCTGA-3′ and reverse, 5′-CTCAACTGGTGTCGTGGA-3′;
and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. The following thermocycling conditions
were used for qPCR: Initial denaturation at 95°C for 10 min; 40
cycles of 95°C for 30 sec, 60°C for 15 sec and 72°C for 15 sec; and
a final extension at 72°C for 10 min. Relative miR-665 expression
was calculated using the 2−ΔΔCq method (19) and normalized to U6.
Cell proliferation assay
To explore the effect of miR-665 on tumor cell
proliferation, the MTT assay was performed to estimate NSCLC cell
proliferation after 48 h of transfection. Cells at a density of
4×105 cells/well were seeded into 48-well plates and
cultured at 37°C for 72 h. The MTT solution (5 mg/ml) was added
into the wells at 0, 24, 48 and 72 h, followed by 4 h of
incubation. Following MTT incubation, the purple formazan crystals
were dissolved in DMSO for 1 h and cell proliferation was
subsequently analyzed at a wavelength of 570 nm, using a microplate
reader (Thermo Fisher Scientific, Inc.).
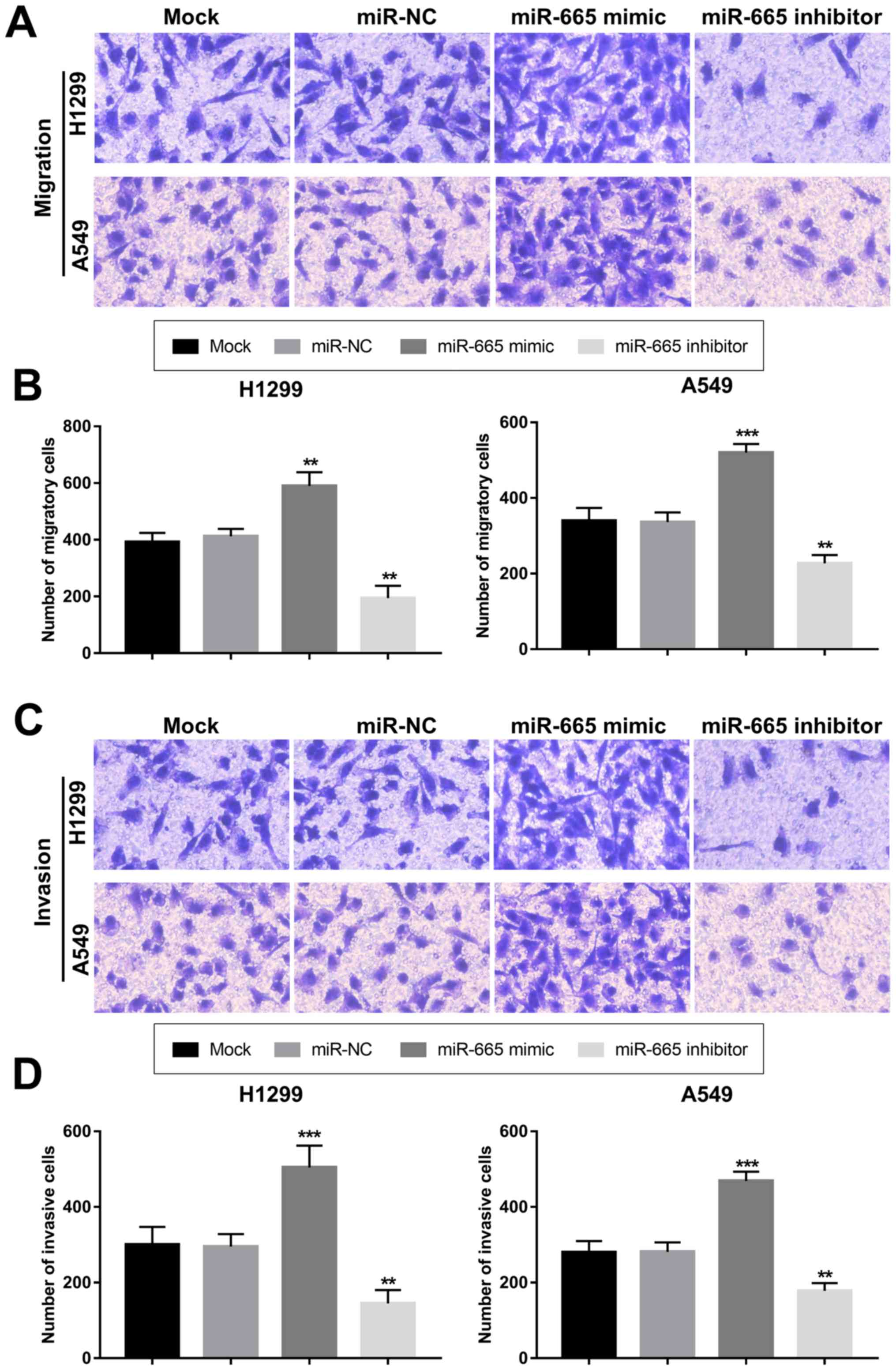
Cell migration and invasion
analysis
Transwell chambers (Corning Inc.) were used to
evaluate the abilities of cell migration and invasion. After 48 h
post-transfection, cells transfected with vectors or only
transfection reagent were plated in the upper chambers in
serum-free RPMI 1640 medium at a density of 5×104
cells/well, while the lower chambers were filled with RPMI 1640
medium supplemented with 10% FBS. The chambers were kept in a
humidified atmosphere containing 5% CO2 at 37°C for 48
h. For the invasion assay, the Transwell membranes were pre-coated
with Matrigel (Corning Inc.) at 37°C for 1 h. Following incubation,
cells in the upper chambers were removed using cotton swabs, while
migratory cells in the lower chambers were incubated with 0.1%
crystal violet (Sigma-Aldrich; Merck KGaA) for 10 min at room
temperature. Stained cells were counted in five randomly selected
fields using an inverted light microscope (magnification, ×200;
Olympus Corporation).
Luciferase reporter assay
According to the bioinformatics analysis using
TargetScan software (version 7.2; http://www.targetscan.org/vert_72), a potential target
gene was predicted for miR-665, known as protein tyrosine
phosphatase receptor type B (PTPRB). A luciferase reporter
assay was performed to confirm the interaction between miR-665 and
PTPRB in H1299 cells. The wild-type (WT) and mutant (MT)
3′-untranslated regions (UTRs) of PTPRB were synthesized and
separately cloned into the pmiR-GLO dual-luciferase vector
(Shanghai GenePharma Co., Ltd.). After 12 h of cell inoculation, at
a density of 5×104 cell/well, the combined vectors were
co-transfected into H1299 cells with either miR-665 mimic, miR-665
inhibitor or miR-NC using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.). Following incubation
at 37°C for 48 h, cells were collected, and firefly and
Renilla luciferase activities were detected using a
Dual-Luciferase Reporter assay system (Promega Corporation),
according to the manufacturer's protocol. Firefly luciferase
activity was normalized to Renilla luciferase activity.
Statistical analysis
Statistical analysis was performed using SPSS
software (version 21.0; IBM Corp.) and GraphPad Prism software
(version 5.0; GraphPad Software, Inc.) Data are presented as the
mean ± standard deviation and all experiments were performed in
triplicate. Differences between groups were analyzed using paired
Student's t-test or one-way ANOVA followed by Tukey's post-hoc
test. Associations between miR-665 and the clinical features of the
patients were assessed using a χ2 test. Survival
analysis was performed with the Kaplan-Meier method, and a Cox
regression analysis was conducted to confirm the prognostic value
of miR-665. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-665 expression in NSCLC
The present study investigated the expression
profile of miR-665 in both NSCLC tissue and cell samples. As shown
in Fig. 1A, miR-665 expression was
significantly upregulated in the NSCLC tissues compared with in
non-cancerous tissues. Furthermore, the expression of miR-665 in
patients with different lymph node metastasis (LM) status and TNM
stages was compared. The results shown in Fig. 1B indicated that patients with
positive LM had significantly higher miR-665 expression than those
with negative LM. Furthermore, significantly increased miR-665
expression was observed in patients with advanced TNM stage
compared with in patients with TNM stage I–II (Fig. 1C). In addition to the tissue samples,
a marked increase in the relative expression of miR-665 was also
found in the NSCLC cell lines compared with in the normal cell line
(Fig. 1D).
Association between miR-665 and the
clinicopathological characteristics of the patients
All demographic and clinical features are summarized
in Table I, including age, sex,
smoking history, tumor size, differentiation, LM and TNM stage. To
explore the association of miR-665 with the clinicopathological
data, the mean expression value of miR-665 (0.397) was used to
divide the patients into a low miR-665 expression group (n=60) and
a high miR-665 expression group (n=68). According to the
χ2 test, miR-665 expression was significantly associated
with LM and TNM stage. However, no other significant associations
between miR-665 expression and the remaining clinical features were
observed.
Aberrant expression of miR-665 is
independently associated with the overall survival of patients
The present study further evaluated the clinical
significance of deregulated miR-665 in the prognosis of NSCLC. The
survival curves in Fig. 2A show that
patients with low miR-665 expression exhibited improved overall
survival compared with those with high miR-665 expression. The
association of miR-665 with the overall survival in patients with
different TNM stages was further analyzed. This suggested that high
miR-665 expression was associated with a shorter survival time in
both TNM I–II stage groups (Fig. 2B)
and III–IV stage groups (Fig. 2C).
The aforementioned data indicated a potential association of
miR-665 with the overall survival of patients with NSCLC.
Furthermore, the results of the Cox regression analysis shown in
Table II revealed that miR-665
expression was independently associated with overall survival,
suggesting a prognostic value of miR-665 in patients with
NSCLC.
 | Table II.Cox regression analysis in patients
with non-small cell lung cancer. |
Table II.
Cox regression analysis in patients
with non-small cell lung cancer.
|
| Multivariate
analysis |
|---|
|
|
|
|---|
| Variables | HR | 95% CI | P-value |
|---|
| miR-665 | 2.053 | 1.062–3.969 | 0.032a |
| Age | 0.973 | 0.494–1.914 | 0.936 |
| Sex | 1.041 | 0.554–1.955 | 0.901 |
| Smoking | 0.975 | 0.521–1.825 | 0.938 |
| Tumor size | 1.409 | 0.735–2.701 | 0.302 |
|
Differentiation | 1.005 | 0.535–1.887 | 0.988 |
| Lymph node
metastasis | 0.822 | 0.454–1.488 | 0.517 |
| TNM stage | 1.816 | 0.932–3.540 | 0.080 |
Overexpression of miR-665 contributes
to proliferation, migration and invasion of NSCLC cells
The present study aimed to uncover the functional
role of miR-665 in NSCLC progression. By transfecting H1299 and
A549 cells with a miR-665 mimic or a miR-665 inhibitor, the
expression of miR-665 was upregulated or downregulated,
respectively (Fig. 3A and B). The
MTT cell proliferation assays revealed that the overexpression of
miR-665 in tumor cells led to increased cell proliferation, while
the knockdown of miR-665 resulted in suppressed cell proliferation
(Fig. 3C and D). In accordance with
the proliferation results, the tumor cell migration and invasion
were also enhanced by the upregulation of miR-665 and inhibited by
the downregulation of miR-665 (Fig.
4).
PTPRB is a direct target gene of
miR-665 in NSCLC cells
Using bioinformatics prediction, a complementary
sequence of miR-665 was identified in the 3′-UTR of PTPRB
(Fig. 5A). The subsequent luciferase
reporter assay results shown in Fig.
5B indicated that the luciferase activity was markedly
decreased by miR-665 overexpression, but was promoted by the
knockdown of miR-665 in cells with WT 3′-UTR PTPRB. However,
no significant changes in the luciferase activity were observed in
cells transfected with the MT 3′-UTR PTPRB.
Discussion
Numerous studies have highlighted the critical roles
of the aberrant expression of miRNAs in various diseases,
especially in human malignancies (20–22).
These functional miRNAs are involved in the occurrence and
development of tumors, and a number of them have been identified as
efficient therapeutic targets for targeted cancer therapy (23). For example, miR-381 has been
identified as a tumor suppressor in the progression of pancreatic
cancer, demonstrated by the inhibited tumor cell proliferation,
migration and invasion, and enhanced cell apoptosis following
miR-381 overexpression (15).
Increased expression of miR-1307-3p detected in the hepatocellular
carcinoma tissues has been demonstrated to enhance tumor growth and
metastasis by regulating disabled homolog 2-interacting protein
(24). Additionally, deregulated
miR-501 has been demonstrated to be a potential therapeutic target
in gastric cancer, and exosomal transfer of miR-501 could regulate
the development of this malignancy (25). In addition to the functional roles of
miRNAs, their clinical significance has also received increasing
attention for the diagnosis and prognosis of various types of
cancer, including NSCLC (26). For
instance, Sun et al (27)
showed that the serum expression levels of miR-30a-5p are
downregulated in patients with colorectal carcinoma, and that it
could represent a candidate diagnostic and prognostic biomarker.
Decreased miR-424-5p expression in hepatocellular carcinoma tissues
is independently associated with poor overall survival of the
patients, highlighting its value in cancer prognosis (28). In NSCLC, some aberrant miRNAs with
dramatic clinical significance have also been identified, such as
miR-25 (16) and miR-411 (29).
In the current study, the expression levels of
miR-665 were significantly upregulated in the NSCLC tissues and
cell lines compared with in the corresponding normal controls, and
this increased expression was closely associated with the LM and
TNM stage of the patients. Thus, it was hypothesized that miR-665
may serve as an onco-miR and may be involved in the development of
NSCLC. Given the dysregulation of miR-665, a survival analysis was
performed to evaluate the prognostic significance of miR-665.
Patients with high miR-665 levels had shorter survival times
compared with those with low miR-665 expression, suggesting that
high miR-665 expression was associated with poor overall survival.
In addition, by dividing the patients into groups based on TNM
stages, it was revealed that high miR-665 expression predicted poor
overall survival of patients with both early and advanced TNM
stages. Furthermore, the Cox regression analysis demonstrated that
miR-665 was an independent prognostic indicator in patients with
NSCLC.
Numerous studies have indicated that miRNAs are
involved in the pathogenesis of human cancer by regulating tumor
cell processes, such as proliferation, migration and invasion
(30–32). Therefore, the present study carried
out further cellular experiments to uncover the functional role of
miR-665 in NSCLC progression. When regulating miR-665 expression
in vitro, the overexpression of miR-665 in NSCLC cells
resulted in enhanced cell proliferation, migration and invasion,
while the knockdown of miR-665 led to the opposite results.
Overall, miR-665 may serve as an enhancer of NSCLC tumorigenesis.
Although a promoting effect of miR-665 on tumor cell proliferation
was observed, an association between miR-665 expression and tumor
size was not detected. This might be due to the small sample size
that limited the accuracy of the clinical data. Thus, further
investigations with a larger study cohort are necessary. The
regulatory effects of miR-665 on tumor cell processes have been
previously investigated in other malignancies. In hepatocellular
carcinoma cells, miR-665 can promote cell migration, invasion and
proliferation by targeting PTPRB (33). In breast cancer, tumor cell migration
and invasion are enhanced by the overexpression of miR-665 by
promoting the epithelial-mesenchymal transition process (34). By contrast, miR-665 has been
demonstrated to suppress cell proliferation and migration in
ovarian cancer cells (35). This
controversy on the role of miR-665 may be due to the different
cancer types. Although the cell experimental data indicated an
oncogenic role of miR-665 in NSCLC progression, further studies are
required to confirm the biological function of miR-665 by
investigating its effects on NSCLC cell cycle distribution and
apoptosis, as well as its functional role in tumorigenesis in
vivo.
Although the present study provided novel insights
on the clinical significance and functional role of miR-665 in
NSCLC, the molecular mechanisms underlying the role of miR-665
remain unclear and warrant further investigations. A study by Hu
et al (33) demonstrated that
miR-665 exerts promoting effects on hepatocellular carcinoma cell
proliferation, migration and invasion by directly targeting PTPRB
and through the Hippo signaling pathway. Interestingly, PTPRB has
been found to suppress NSCLC cell proliferation and invasion by
inhibiting the phosphorylation of the proto-oncogene
tyrosine-protein kinase Src (36).
In the present study, the binding site of miR-665 was predicted in
the 3′-UTR of PTPRB, confirming that PTPRB was a
direct target gene of miR-665 in NSCLC. Thus, the miR-665/PTPRB
axis may be involved in the regulation of NSCLC progression through
the Src and/or Hippo signaling pathways. However, the regulatory
effects of miR-665 on PTPRB and the aforementioned signaling
pathways in NSCLC were not examined, which represents one of the
limitations of the present study and warrants further confirmation
to better understand these molecular mechanisms. In addition, other
limitations included a relatively small sample size and the lack of
cell cycle analysis results in tumor cells with deregulated miR-665
expression. Further studies are required to confirm the role of
miR-665 in NSCLC by using a larger sample size and analyzing its
regulatory effect on tumor cell biological function.
In conclusion, the results of the present study
revealed that upregulated miR-665 expression was associated with
the LM and TNM stage of patients with NSCLC, and could be used as a
potential prognostic biomarker. NSCLC cell proliferation, migration
and invasion were promoted by the overexpression of miR-665,
indicating that methods to decrease miR-665 expression could be
used as novel therapeutic strategies for NSCLC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JX, DL and QX designed the present study, drafted
and revised the initial manuscript for important intellectual
content, and acquired and analyzed the data. XZ, WX, ZQ and GL
performed the experiments and analyzed the data.
Ethics approval and consent to
participate
The Ethics Committee of Shouguang People's Hospital
approved the present study. Written informed consent was obtained
from all patients prior to enrollment in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Siegel RL and Jemal A: Lung
cancer statistics. Adv Exp Med Biol. 893:1–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Akhurst T: Staging of non-small-cell lung
cancer. PET Clin. 13:1–10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Blandin Knight S, Crosbie PA, Balata H,
Chudziak J, Hussell T and Dive C: Progress and prospects of early
detection in lung cancer. Open Biol. 7(pii): 1700702017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Woodard GA, Jones KD and Jablons DM: Lung
cancer staging and prognosis. Cancer Treat Res. 170:47–75. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu DM, Liu T, Deng SH, Han R and Xu Y:
SLC39A4 expression is associated with enhanced cell migration,
cisplatin resistance, and poor survival in non-small cell lung
cancer. Sci Rep. 7:72112017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xue J, Yang J, Luo M, Cho WC and Liu X:
MicroRNA-targeted therapeutics for lung cancer treatment. Expert
Opin Drug Discov. 12:141–157. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang D, Sun W, Zhou Y, Li P, Chen F, Chen
H, Xia D, Xu E, Lai M, Wu Y and Zhang H: Mutations of key driver
genes in colorectal cancer progression and metastasis. Cancer
Metastasis Rev. 37:173–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu J, Zhan Y, Feng J, Luo J and Fan S:
MicroRNAs associated with therapy of non-small cell lung cancer.
Int J Biol Sci. 14:390–397. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hwang DW, Kim HY, Li F, Park JY, Kim D,
Park JH, Han HS, Byun JW, Lee YS, Jeong JM, et al: In vivo
visualization of endogenous miR-21 using hyaluronic acid-coated
graphene oxide for targeted cancer therapy. Biomaterials.
121:144–154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, Duo Y, Bi J, Zeng X, Mei L, Bao S,
He L, Shan A, Zhang Y and Yu X: Targeted delivery of anti-miR-155
by functionalized mesoporous silica nanoparticles for colorectal
cancer therapy. Int J Nanomedicine. 13:1241–1256. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu J, Zhang X, Huang Y, Zhang Q, Zhou J,
Zhang X and Wang X: miR-200b and miR-200c co-contribute to the
cisplatin sensitivity of ovarian cancer cells by targeting DNA
methyltransferases. Oncol Lett. 17:1453–1460. 2019.PubMed/NCBI
|
|
12
|
An JC, Shi HB, Hao WB, Zhu K and Ma B:
miR-944 inhibits lung adenocarcinoma tumorigenesis by targeting
STAT1 interaction. Oncol Lett. 17:3790–3798. 2019.PubMed/NCBI
|
|
13
|
Dong M, Xie Y and Xu Y: miR-7-5p regulates
the proliferation and migration of colorectal cancer cells by
negatively regulating the expression of Krüppel-like factor 4.
Oncol Lett. 17:3241–3246. 2019.PubMed/NCBI
|
|
14
|
Yang F, Chen L and Wang ZJ: MicroRNA-32
inhibits the proliferation, migration and invasion of human colon
cancer cell lines by targeting E2F transcription factor 5. Eur Rev
Med Pharmacol Sci. 23:4156–4163. 2019.PubMed/NCBI
|
|
15
|
Qiao G, Li J, Wang J, Wang Z and Bian W:
miR-381 functions as a tumor suppressor by targeting ETS1 in
pancreatic cancer. Int J Mol Med. 44:593–607. 2019.PubMed/NCBI
|
|
16
|
Li J, Yu M, Liu Z and Liu B: Clinical
significance of serum miR-25 in non-small-cell lung cancer. Br J
Biomed Sci. 76:111–116. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jin X, Guan Y, Sheng H and Liu Y:
Crosstalk in competing endogenous RNA network reveals the complex
molecular mechanism underlying lung cancer. Oncotarget.
8:91270–91280. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li C, Qin F, Hu F, Xu H, Sun G, Han G,
Wang T and Guo M: Characterization and selective incorporation of
small non-coding RNAs in non-small cell lung cancer extracellular
vesicles. Cell Biosci. 8:22018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu C, Huang Q and Zhu H: miR-383
inhibited the cell cycle progression of gastric cancer cells via
targeting cyclin E2. DNA Cell Biol. 38:849–856. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao W, Zhang Y, Niu M, Bo Y, Li H, Xue X,
Lu Y, Zheng X, Tang Y, Cui J, et al: Identification of
miR-145-5p-centered competing endogenous RNA network in laryngeal
squamous cell carcinoma. Proteomics. 19:e19000202019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tao L, Wu YQ and Zhang SP: MiR-21-5p
enhances the progression and paclitaxel resistance in
drug-resistant breast cancer cell lines by targeting PDCD4.
Neoplasma. 66:746–755. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang R, Li G, Zhuang G, Sun S and Song Z:
Overexpression of microRNA-423-3p indicates poor prognosis and
promotes cell proliferation, migration, and invasion of lung
cancer. Diagn Pathol. 14:532019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen S, Wang L, Yao B, Liu Q and Guoa C:
miR-1307-3p promotes tumor growth and metastasis of hepatocellular
carcinoma by repressing DAB2 interacting protein. Biomed
Pharmacother. 117:1090552019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu X, Lu Y, Xu Y, Hou S, Huang J, Wang B,
Zhao J, Xia S, Fan S, Yu X, et al: Exosomal transfer of miR-501
confers doxorubicin resistance and tumorigenesis via targeting of
BLID in gastric cancer. Cancer Lett. 459:122–134. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zou JG, Ma LF, Li X, Xu FL, Fei XZ, Liu Q,
Bai QL and Dong YL: Circulating microRNA array (miR-182, 200b and
205) for the early diagnosis and poor prognosis predictor of
non-small cell lung cancer. Eur Rev Med Pharmacol Sci.
23:1108–1115. 2019.PubMed/NCBI
|
|
27
|
Sun Y, Yang B, Lin M, Yu H, Chen H and
Zhang Z: Identification of serum miR-30a-5p as a diagnostic and
prognostic biomarker in colorectal cancer. Cancer Biomark.
24:299–305. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du H, Xu Q, Xiao S, Wu Z, Gong J, Liu C,
Ren G and Wu H: MicroRNA-424-5p acts as a potential biomarker and
inhibits proliferation and invasion in hepatocellular carcinoma by
targeting TRIM29. Life Sci. 224:1–11. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang SY, Li Y, Jiang YS and Li RZ:
Investigation of serum miR-411 as a diagnosis and prognosis
biomarker for non-small cell lung cancer. Eur Rev Med Pharmacol
Sci. 21:4092–4097. 2017.PubMed/NCBI
|
|
30
|
Liang Z, Li X, Liu S, Li C, Wang X and
Xing J: MiR-141-3p inhibits cell proliferation, migration and
invasion by targeting TRAF5 in colorectal cancer. Biochem Biophys
Res Commun. 514:699–705. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen Z, Gao Y, Gao S, Song D and Feng Y:
MiR-135b-5p promotes viability, proliferation, migration and
invasion of gastric cancer cells by targeting Krüppel-like factor 4
(KLF4). Arch Med Sci. 16:167–176. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Long X, Shi Y, Ye P, Guo J, Zhou Q and
Tang Y: MicroRNA-99a suppresses breast cancer progression by
targeting FGFR3. Front Oncol. 9:14732020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu Y, Yang C, Yang S, Cheng F, Rao J and
Wang X: miR-665 promotes hepatocellular carcinoma cell migration,
invasion, and proliferation by decreasing Hippo signaling through
targeting PTPRB. Cell Death Dis. 9:9542018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao XG, Hu JY, Tang J, Yi W, Zhang MY,
Deng R, Mai SJ, Weng NQ, Wang RQ, Liu J, et al: miR-665 expression
predicts poor survival and promotes tumor metastasis by targeting
NR4A3 in breast cancer. Cell Death Dis. 10:4792019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu J, Jiang Y, Wan Y, Zhou S, Thapa S and
Cheng W: MicroRNA-665 suppresses the growth and migration of
ovarian cancer cells by targeting HOXA10. Mol Med Rep.
18:2661–2668. 2018.PubMed/NCBI
|
|
36
|
Qi Y, Dai Y and Gui S: Protein tyrosine
phosphatase PTPRB regulates Src phosphorylation and tumour
progression in NSCLC. Clin Exp Pharmacol Physiol. 43:1004–1012.
2016. View Article : Google Scholar : PubMed/NCBI
|