Introduction
Colorectal cancer (CRC) is a common malignant tumor
of the gastrointestinal tract. The incidence and mortality of the
digestive system malignancies rank second only to gastric,
esophageal and primary liver cancers (1). Moreover, CRC is prone to invade and
metastasize, which is the main cause of death. Surgical treatment
is still the first choice for the radical treatment of CRC.
However, despite the rapid development of surgical techniques, the
surgical cure rate and the 5-year survival rate of CRC patients
have been hovering around 50% (2).
The recurrence of cancer in liver, lung or peritoneal is one of the
causes of CRC treatment failure (3).
Therefore, early diagnosis and timely treatment is the key to
improving the prognosis of CRC patients.
MicroRNAs (miRNAs) are small, non-coding RNAs that
regulate gene expression. It has been reported that miRNAs have
important functions in cell-cell communication and have the
potential to detect and monitor diseases, such as cancer (4). A number of miRNAs have been found to
act as tumor suppressors or promoters in a number of cancers,
including CRC. For example, miR-141-3p has been shown to restrain
the cell growth and metastasis by targeting TRAF5 in CRC (5). Furthermore, miR-182 has been reported
to promote cell proliferation, invasion and tumor growth in CRC via
regulating DAB2IP (6). Recently, the
abnormal expression of miR-758 was found in human cancers, except
CRC. In particular, the overexpression of miR-758 was demonstrated
to inhibit cell proliferation, migration, invasion and promote
apoptosis in non-small cell lung cancer by negatively regulating
HMGB (7). Meng et al also
showed that miR-758 was downregulated in cervical cancer, and
miR-758 overexpression promoted cell invasion by regulating the
expression of MEPE (8). These
results suggest that miR-758 can be stably expressed in human
tissues. However, the specific role of miR-758 in CRC has not been
clarified yet.
TargetScan database (http://www.targetscan.org) predicts that paired box 6
(PAX6) is a target of miR-758. PAX6 has been found to be a highly
conserved transcription factor that plays a crucial role in the
development of human cancers (9).
For example, PAX6 overexpression has been shown to promote tumor
growth and inhibit apoptosis in human retinoblastoma (10). PAX-6 has also been shown to promote
breast cancer cell proliferation and tumorigenesis (11). In addition, Needhamsen et al
have demonstrated that PAX6 expression was regulated by miR-7
(12). However, the interaction
between miR-758 and PAX6 has not been reported in CRC. Furthermore,
the PI3K/AKT pathway is well known to be an important regulator in
human cancers. The PI3K/AKT pathway has been demonstrated to be
involved in tumor growth including CRC. The PI3K/AKT signaling
pathway is associated with dysregulation of miRNA and gene
expression in CRC (13). For
example, miR-532 has been shown to inhibit CRC progression by
directly targeting IGF-1R and suppressing the PI3K/AKT signaling
pathway (14). However, the way
miR-758 regulates the PI3K/AKT signaling pathway in CRC is
unclear.
In the present study, the molecular mechanism of
miR-758/PAX6/PI3K/AKT pathway was explored and the function of
miR-758 was investigated in CRC, aiming to open up a new avenue for
the diagnosis and treatment of CRC.
Materials and methods
Clinical tissues
The study was performed on 84 fresh CRC tissues
(colon and rectal cancers) confirmed by histopathology at the
Qingdao West Coast New Area Central Hospital (Qingdao, China) from
May 2017 to January 2019. All patients with CRC did not receive any
treatment before surgery. The study was approved by the
Institutional Ethics Committee of the Qingdao West Coast New Area
Central Hospital. Patients who participated in this research had
complete clinical data. Signed written informed consents were
obtained from the patients and/or guardians.
Cell culture
Normal human intestinal epithelial cells HIEC-6 and
CRC cell lines HCT-116 and SW620 were purchased from ATCC. The
cells were incubated in DMEM (10% FBS, 5% CO2, 37°C) for
subsequent experimentation.
Cell transfection
miR-758 mimics, miR-758 inhibitor, and PAX6
overexpression plasmid were purchased from Shanghai GenePharma Co.,
Ltd. HCT-116 cells were transfected using Lipofectamine®
2000 (Invitrogen, Thermo Fisher Scientifc, Inc.), respectively.
HCT-116 cells with human homologous sequences were used as control
(miR-NC). According to the manufacturer's protocol, the cells were
plated in 6-well plates (at 70–90% confluence) and were transfected
with 100 pmol miR-758 mimics, inhibitor and their control or 2.5 µg
PAX6 overexpression plasmid. After 6 h of incubation at 37°C with
5% CO2, the transfection efficiency was detected and
subsequent assays were performed.
RNA isolation and RT-qPCR
Total RNA isolation was performed using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The cDNA solution was then obtained using a PrimeScript
Reverse Transcription kit (Qiagen China Co., Ltd). The temperature
conditions for reverse transcription were as follows: 37°C for 15
min and 85°C for 5 sec. RT-qPCR was performed using SYBR Green I
(Takara Biotechnology Co., Ltd.) following the manufacturer's
instructions. The thermocycling conditions for PCR amplification
were as follows: 5 min at 95°C, followed by 40 cycles of 95°C for
30 sec and 60°C for 45 sec. U6 and GAPDH were used as the controls
for miR-758 and PAX6, respectively. The expression of miR-758 and
PAX6 was quantified using the 2−ΔΔCq method (15). Primers were designed as follows:
miR-758 forward, 5′-ACACTCCAGCTGGGTTTGTGACCTGGTCCA-3′ and reverse,
5′-TGGTGTCGTGGAGTCG-3′; U6 forward, 5′-CGCTTCGGCAGCACATATACTA-3′
and reverse, 5′-GCGAGCACAGAATTAATACGAC-3′; PAX6 forward,
5′-TCTTTGCTTGGGAAATCCG-3′ and reverse, 5′-CTGCCCGTTCAACATCCTTAG-3′;
and GAPDH forward, 5′-GGCACTGAGAAGCGGGGCCG-3′ and reverse,
5′-GAAGATGGTGATGGGATTTC-3′.
Transwell assay
An 8.0-µm pore polycarbonate membrane insert
(Corning, Inc.) was used to determine the invasion and migration of
CRC cells. The upper chamber was coated with Matrigel (BD
Biosciences) for invasion assay. Next, the transfected HCT-116
cells (3×103 cells/well) were placed in the upper
chamber. DMEM with 10% FBS was added to the lower chamber. After 24
h, the cells were fixed and stained with 0.5% crystal violet at
room temperature for 30 min. For cell migration assay, Matrigel was
not required and the procedure followed was identical to that of
the cell invasion assay. Observation and image capturing were
performed using a light microscope (magnification, ×200).
CCK-8 assay
The prepared HCT-116 cells were incubated in a
96-well plate for 24 h (at 37°C, 5% CO2). HCT-116 cells
(4×103/well) were then incubated for 24, 48, 72 and 96
h. Next, the cells were incubated for 4 h with 10 ml of CCK-8
(Dojindo Molecular Technologies, Inc.) solution. The absorbance at
450 nm was observed with a microplate reader (Molecular Devices,
LLC).
Western blot analysis
Protein samples were obtained using RIPA lysis
buffer (Beyotime Institute of Biotechnology). Total protein was
quantified using a bicinchoninic acid protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.) and 40 µg protein/lane were
separated via SDS-PAGE on a 10% gel. The separated proteins were
transferred onto polyvinylidene fluoride membranes (Thermo Fisher
Scientific, Inc.) and blocked for 3 h at room temperature with 5%
non-fat milk in PBS (Thermo Fisher Scientific, Inc.) containing
0.1% Tween-20 (Sigma-Aldrich; Merck KGaA). The membranes were then
incubated with E-cadherin (rabbit monoclonal; dilution, 1:1,000;
cat. no. ab1416; Abcam), N-cadherin (rabbit polyclonal; dilution,
1:1,000; cat. no. ab18203; Abcam), Bcl-2 (rabbit monoclonal;
dilution, 1:1,000; cat. no. ab185002; Abcam), Bax (mouse
monoclonal; dilution, 1:1,000; cat. no. ab77566; Abcam), PI3K
(rabbit monoclonal; dilution, 1:1,000; cat. no. ab32089; Abcam),
p-PI3K (rabbit monoclonal; dilution, 1:1,000; cat. no. ab154598;
Abcam), AKT (rabbit polyclonal; dilution, 1:1,000; cat. no. ab8805;
Abcam), p-AKT (rabbit monoclonal; dilution, 1:1,000; cat. no.
ab81283; Abcam) and GAPDH (rabbit monoclonal; dilution, 1:1,000;
cat. no. ab181602; Abcam) primary antibodies overnight at 4°C.
After washing, the membranes were incubated with horseradish
peroxidase-conjugated secondary antibodies (dilution, 1:5,000; cat.
no. ab7090; Abcam) for 1 h at room temperature. Protein bands were
visualized using ECL kit (Beyotime Institute of Biotechnology).
Dual-luciferase reporter assay
WT-PAX6-3′UTR or MUT-PAX6-3′UTR was inserted into
the pmirGLO luciferase reporter vector (Promega Corporation).
HCT-116 cells were transfected with the luciferase vector and
miR-758 mimics using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientifc, Inc.) according to the manufacturer's
protocol. After 24 h, the Renilla and firefly luciferase
activity were detected using a dual-luciferase reporter assay
system (Promega Corporation). Firefly luciferase activity was
normalized to Renilla luciferase activity.
Statistical analysis
Data were analyzed using SPSS 17.0 (IBM Corp.) or
GraphPad Prism 6 (GraphPad Software, Inc.) software and were
expressed as the mean ± SD. The differences were analyzed using
Student's t-test or one-way ANOVA followed by Tukey's post hoc
test. The univariate Kaplan-Meier method with log-rank test and the
Chi-square test were used to analyze the association between
miR-758 and patient survival or clinical features. Spearman's rank
correlation analysis was performed to analyze the correlation
between miR-758 and PAX6 expression levels. P<0.05 was
considered to indicate a statistically significant difference.
Results
Downregulation of miR-758 is
associated with aggressive behavior and poor prognosis in CRC
RT-qPCR was performed to assess the expression of
miR-758 in CRC. miR-758 expression was decreased in CRC tissues
compared with that in normal tissues (P<0.01, Fig. 1A). The downregulation of miR-758 was
also detected in HCT-116 and SW620 cells in comparison with miR-758
expression in HIEC-6 cells (P<0.01, Fig. 1B). Additionally, the low expression
of miR-758 was found to be associated with differentiation, TNM
stage and lymph-node metastasis in CRC patients (P<0.05,
Table I). Moreover, the poor
prognosis in CRC patients was found to be associated with the
downregulation of miR-758 (P<0.05, Fig. 1C). These results suggest that miR-758
may be involved in CRC progression. Since the difference of miR-758
expression between the HCT-116 and HIEC-6 cells was more
significant than that between SW620 and HIEC-6 cells, HCT-116 cells
were selected for the following experiments.
 | Table I.Relationship between miR-758
expression and clinicopathological characteristics of CRC
patients. |
Table I.
Relationship between miR-758
expression and clinicopathological characteristics of CRC
patients.
|
|
| miR-758 |
|
|---|
|
|
|
|
|
|---|
| Characteristics | Cases | High | Low | P-value |
|---|
| Age (years) |
|
|
| 0.71 |
| ≥60 | 34 | 13 | 21 |
|
|
<60 | 50 | 14 | 36 |
|
| Sex |
|
|
| 0.53 |
|
Male | 32 | 14 | 18 |
|
|
Female | 52 | 13 | 39 |
|
|
Differentiation |
|
|
| 0.01a |
|
Well | 58 | 16 | 42 |
|
|
Moderate-poor | 26 | 11 | 15 |
|
| TNM stage |
|
|
| 0.012a |
|
I–II | 64 | 18 | 46 |
|
|
III–IV | 20 | 9 | 11 |
|
| Lymph node
metastasis |
|
|
| 0.02a |
| No | 65 | 17 | 48 |
|
|
Yes | 19 | 10 | 9 |
|
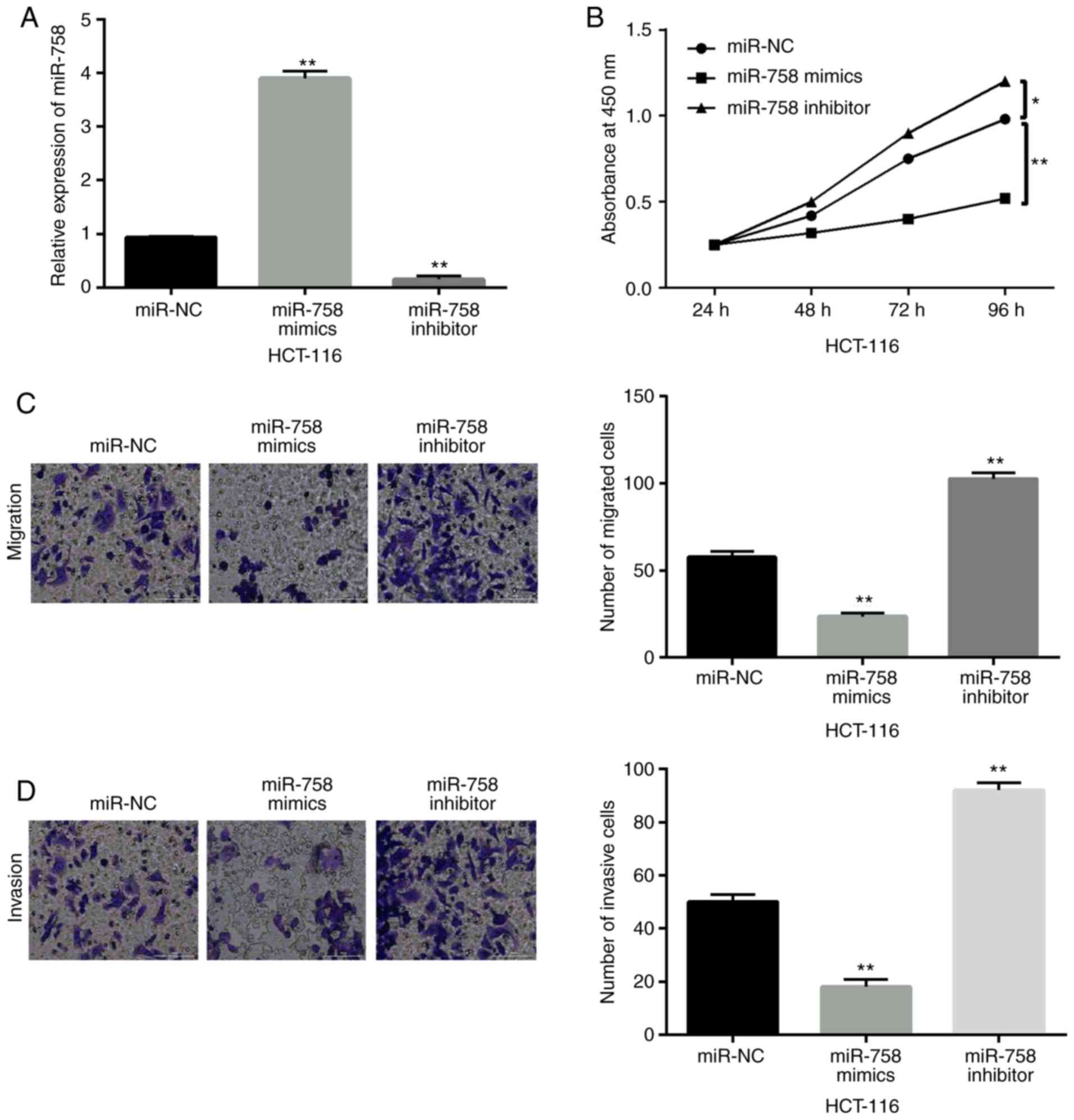
miR-758 restrains cell viability and
metastasis in CRC
To investigate the function of miR-758 in CRC, a
gain-loss function experiment was performed in HCT-116 cells with
miR-758 mimics or inhibitor. The expression of miR-758 was
increased by its mimics and decreased by its inhibitor (P<0.01,
Fig. 2A). Functionally, the
overexpression of miR-758 restrained cell proliferation
(P<0.01), whereas the downregulation of miR-758 accelerated the
proliferation of HCT-116 cells (P<0.05) (Fig. 2B). In addition, miR-758 mimics
inhibited cell migration, whereas miR-758 inhibitor promoted cell
migration in HCT-116 cells (P<0.01, Fig. 2C). Consistently, miR-758 mimics also
inhibited cell invasion, whereas miR-758 inhibitor promoted HCT-116
cell invasion (P<0.01, Fig. 2D).
Based on these results, miR-758 restrained CRC progression by
inhibiting cell viability and metastasis.
miR-758 directly targets PAX6
For the prediction of the downstream target of
miR-758, the TargetScan database was used (http://www.targetscan.org). It was predicted that
miR-758 has a site that binds to the 3′-UTR of PAX6 (Fig. 3A). A luciferase reporter assay was
then designed to confirm this prediction. miR-758 mimics were
identified to reduce the luciferase activity of WT-PAX6 (P<0.01,
Fig. 3B). However, miR-758 mimics
had no effect on MUT-PAX6 luciferase activity. To further confirm
the relationship between miR-758 and PAX6, the PAX6 expression was
observed in HCT-116 cells with miR-758 mimics or inhibitor. RT-qPCR
showed that the overexpression of miR-758 reduced the expression of
PAX6, whereas the downregulation of miR-758 promoted the expression
of PAX6 (P<0.01, Fig. 3C). In
addition, PAX6 was shown to be upregulated in CRC tissues compared
with normal tissues (P<0.01, Fig.
3D). Moreover, miR-758 was negatively correlated with PAX6
expression in CRC tissues (P<0.01, R2=0.7269;
Fig. 3E). The results suggest that
miR-758 directly targets PAX6 and negatively regulates PAX6
expression in CRC.
miR-758 inhibits CRC progression
through targeting PAX6
To explore the interaction between PAX6 and miR-758,
PAX6 overexpression vectors were transfected into HCT-116 cells
with miR-758 mimics. The results showed that the decreased
expression of PAX6 induced by miR-758 mimics was restored by PAX6
overexpression vector in HCT-116 cells (Fig. 4A). Functionally, the overexpression
of PAX6 attenuated the inhibitory effect of miR-758 on the cell
proliferation of HCT-116 cells (P<0.01, Fig. 4B). Consistently, miR-758-mediated
inhibition of cell migration and invasion was also weakened by PAX6
overexpression (P<0.01, Fig. 4C and
D). These results suggest that the upregulation of PAX6 impairs
the inhibitory effect of miR-758 on CRC.
miR-758 blocks EMT and regulates
PI3K/AKT pathway in CRC
To further illustrate the molecular mechanism of
miR-758 in CRC, the way miR-758 regulates EMT (E-cadherin,
N-cadherin), apoptosis (Bcl-2, Bax) and PI3K/AKT pathway (PI3K,
p-PI3K, AKT, p-AKT) in HCT-116 cells was investigated. Fig. 5 shows that the overexpression of
miR-758 promoted E-cadherin and Bax expression levels, whereas
miR-758 overexpression inhibited N-cadherin and Bcl-2 expression
levels in HCT-116 cells. In addition, miR-758 mimics inhibited the
expression of p-PI3K and p-AKT, whereas miR-758 inhibitor promoted
their expression in HCT-116 cells (Fig.
5). However, miR-758 mimics did not affect the expression of
PI3K and AKT (Fig. 5). Moreover,
downregulation of miR-758 showed opposite effect on the expression
of these genes (Fig. 5). Taken
together, miR-758 overexpression blocked EMT and the PI3K/AKT
pathway in CRC.
Discussion
It has been shown that the abnormal expression of
miRNAs is associated with CRC progression. For example, miR-769 has
been reported to be downregulated in CRC and restrain cancer
progression by targeting CDK1 (16).
In the present study, the expression of miR-758 was also shown to
be reduced in CRC. The downregulation of miR-758 was associated
with aggressive behavior and poor prognosis in CRC patients.
Functionally, miR-758 was presented to be able to inhibit cell
viability and metastasis in CRC. Importantly, miR-758 was shown to
block EMT and PI3K/AKT pathway in CRC, and induce apoptosis by
regulating Bcl-2/Bax expression in CRC. These results imply that
miR-758 serves as a tumor suppressor in CRC progression.
Previous studies have also reported the
downregulation of miR-758 in human cancers, such as ovarian and
gastric cancer (17,18). Also, miR-758 has been found to
inhibit cell migration, invasion and proliferation in bladder
cancer, hepatocellular carcinoma and glioblastoma (19–21).
These results are consistent with the results of the present study.
Downregulation of miR-758 has also been shown to predict poor
prognosis in patients with non-small cell lung cancer (22) and the same result was also found in
CRC patients. In addition, the present study showed that miR-758
induced apoptosis by suppressing Bcl-2 and enhancing Bax expression
in CRC cells. Different from previous studies, EMT and PI3K/AKT
pathway were also shown to be restrained by the overexpression of
miR-758 in CRC cells. Up to our knowledge, these findings have not
been reported by previous studies.
As previous studies have shown, miR-758 usually
regulates tumorigenesis by mediating some target genes, such as
SUCNR1 and CD36 (23,24). In the present study, miR-758 was
shown to directly target PAX6. Upregulation of PAX6 was detected in
CRC tissues. More importantly, upregulation of PAX6 attenuated the
inhibitory effect of miR-758 in CRC. In consistency with the
results of the present study, PAX6 has also been reported to be
upregulated in retinoblastoma and breast cancer (25,26). In
particular, as a target of miR-7, PAX6 was found to promote
proliferation and invasion of CRC cells (27). These findings indicate that PAX6 acts
as an oncogene in CRC. As a target gene, PAX6 has been regulated by
several miRNAs including miR-335 and miR-365 (28,29). In
this research, a negative correlation between miR-758 and PAX6 was
also found in CRC tissues. The interaction between miRNAs and PAX6
has also been investigated in other cancers. For example, miR-335
has been shown to attenuate the proliferation and invasion of
breast cancer cells by regulating PAX6 (30). Wang et al demonstrated that
miR-365b-3p blocked the cell cycle progression and promoted
apoptosis in human retinoblastoma cells by downregulating PAX6
(31). Similarly, miR-758 restrained
cell viability and metastasis in CRC by targeting PAX6, indicating
that miR-758 is an inhibitory miRNA in CRC.
In conclusion, miR-758 was downregulated in CRC,
which was associated with aggressive behavior and poor prognosis in
CRC patients. Moreover, miR-758 restrained cell viability and
metastasis in CRC via targeting PAX6 and was presented to block EMT
and inactivate the PI3K/AKT pathway in CRC. The limitation of this
study is that the role of miR-758 was only investigated in colon
cancer cells HCT-116. The function of miR-758 in other subtypes of
CRC cells is unclear. Further investigation of the specific
regulatory mechanism of miR-758 in CRC will be the aim of our
future research.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ was involved in the conception and design of the
study. JX performed PCR, Transwell and CCK-8 assays. HZ was
responsible for western blot analysis and dual-luciferase reporter
assay. JS analyzed and interpreted the patient data. NL and XH
assisted with statistical analysis. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Institutional Ethics
Committee of Qingdao West Coast New Area Central Hospital (Qingdao,
China). Patients who participated in this research had complete
clinical data. Signed written informed consents were obtained from
the patients and/or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Primavesi F, Stattner S, Jager T, Gobel G,
Presl J, Tomanova K, Buchner S, Maglione M, Resch T, Hutter J, et
al: Progressive oncological surgery is associated with increased
curative resection rates and improved survival in metastatic
colorectal cancer. Cancers (Basel). 11:E2182019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kopetz S, Chang GJ, Overman MJ, Eng C,
Sargent DJ, Larson DW, Grothey A, Vauthey JN, Nagorney DM and
McWilliams RR: Improved survival in metastatic colorectal cancer is
associated with adoption of hepatic resection and improved
chemotherapy. J Clin Oncol. 27:3677–3683. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liang Z, Li X, Liu S, Li C, Wang X and
Xing J: MiR-141-3p inhibits cell proliferation, migration and
invasion by targeting TRAF5 in colorectal cancer. Biochem Biophys
Res Commun. 514:699–705. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li X, Zhang X, Zhang Q and Lin R: miR-182
contributes to cell proliferation, invasion and tumor growth in
colorectal cancer by targeting DAB2IP. Int J Biochem Cell Biol.
111:27–36. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou GH, Lu YY, Xie JL, Gao ZK, Wu XB, Yao
WS and Gu WG: Overexpression of miR-758 inhibited proliferation,
migration, invasion, and promoted apoptosis of non-small cell lung
cancer cells by negatively regulating HMGB. Biosci Rep.
39:BSR201808552019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meng X, Zhao Y, Wang J, Gao Z, Geng Q and
Liu X: Regulatory roles of miRNA-758 and matrix extracellular
phosphoglycoprotein in cervical cancer. Exp Ther Med. 14:2789–2794.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Elso C, Lu X, Weisner PA, Thompson HL,
Skinner A, Carver E and Stubbs L: A reciprocal translocation
dissects roles of Pax6 alternative promoters and upstream
regulatory elements in the development of pancreas, brain, and eye.
Genesis. 51:630–646. 2013.PubMed/NCBI
|
|
10
|
Li L, Li B, Zhang H, Bai S, Wang Y, Zhao B
and Jonas JB: Lentiviral vector-mediated PAX6 overexpression
promotes growth and inhibits apoptosis of human retinoblastoma
cells. Invest Ophthalmol Vis Sci. 52:8393–8400. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zong X, Yang H, Yu Y, Zou D, Ling Z, He X
and Meng X: Possible role of Pax-6 in promoting breast cancer cell
proliferation and tumorigenesis. BMB Rep. 44:595–600. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Needhamsen M, White RB, Giles KM, Dunlop
SA and Thomas MG: Regulation of Human PAX6 Expression by miR-7.
Evol Bioinform Online. 10:107–113. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Slattery ML, Mullany LE, Sakoda LC, Wolff
RK, Stevens JR, Samowitz WS and Herrick JS: The PI3K/AKT signaling
pathway: Associations of miRNAs with dysregulated gene expression
in colorectal cancer. Mol Carcinog. 57:243–261. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song Y, Zhao Y, Ding X and Wang X:
microRNA-532 suppresses the PI3K/Akt signaling pathway to inhibit
colorectal cancer progression by directly targeting IGF-1R. Am J
Cancer Res. 8:435–449. 2018.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Xu M, Lu P and Zhou F:
microRNA-769 is downregulated in colorectal cancer and inhibits
cancer progression by directly targeting cyclin-dependent kinase 1.
Onco Targets Ther. 11:9013–9025. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu X, Li Y, Kong D, Hu L, Liu D and Wu J:
Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p
to facilitate the malignancy of ovarian cancer. J Cell Physiol.
234:10800–10808. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo J, Zhang Z, Pan L and Zhou Y:
Identification of miR-758-3p as potential modulator of CBX5
expression in gastric cancer. Technol Cancer Res Treat.
17:15330338188160612018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu X, Chen B, Shi H, Zhou J, Zhou F, Cao J
and Sun X: miR-758-3p suppresses human bladder cancer cell
proliferation, migration and invasion by targeting NOTCH2. Exp Ther
Med. 17:4273–4278. 2019.PubMed/NCBI
|
|
20
|
Jiang D, Cho W, Li Z, Xu X, Qu Y, Jiang Z,
Guo L and Xu G: MiR-758-3p suppresses proliferation, migration and
invasion of hepatocellular carcinoma cells via targeting MDM2 and
mTOR. Biomed Pharmacother. 96:535–544. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu J, Jiang J, Hui X, Wang W, Fang D and
Ding L: MiR-758-5p suppresses glioblastoma proliferation, migration
and invasion by targeting ZBTB20. Cell Physiol Biochem.
48:2074–2083. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang S and Jiang M: The long non-coding
RNA-DANCR exerts oncogenic functions in non-small cell lung cancer
via miR-758-3p. Biomed Pharmacother. 103:94–100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang H, Zheng J, Lin J, Chen J, Yu Z,
Chen C and Liu T: miR-758 mediates oxLDL-dependent vascular
endothelial cell damage by suppressing the succinate receptor
SUCNR1. Gene. 663:1–8. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li BR, Xia LQ, Liu J, Liao LL, Zhang Y,
Deng M, Zhong HJ, Feng TT, He PP and Ouyang XP: miR-758-5p
regulates cholesterol uptake via targeting the CD36 3′UTR. Biochem
Biophys Res Commun. 494:384–389. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li X, Yang L, Shuai T, Piao T and Wang R:
MiR-433 inhibits retinoblastoma malignancy by suppressing Notch1
and PAX6 expression. Biomed Pharmacother. 82:247–255. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zou Q, Yi W, Huang J, Fu F, Chen G and
Zhong D: MicroRNA-375 targets PAX6 and inhibits the viability,
migration and invasion of human breast cancer MCF-7 cells. Exp Ther
Med. 14:1198–1204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Y, Li Y, Liu Y, Xie P, Li F and Li G:
PAX6, a novel target of microRNA-7, promotes cellular proliferation
and invasion in human colorectal cancer cells. Dig Dis Sci.
59:598–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheng Q, Cao H, Chen Z, Ma Z, Wan X, Peng
R and Jiang B: PAX6, a novel target of miR-335, inhibits cell
proliferation and invasion in glioma cells. Mol Med Rep.
10:399–404. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan F, Liu J, Pang H, Tian Y, Yuan K, Li
Y, Wang J, Bian S, Zheng Y, Dong D, et al: MicroRNA-365 suppressed
cell proliferation and migration via targeting PAX6 in
glioblastoma. Am J Transl Res. 11:361–369. 2019.PubMed/NCBI
|
|
30
|
Meng Y, Zou Q, Liu T, Cai X, Huang Y and
Pan J: microRNA-335 inhibits proliferation, cell-cycle progression,
colony formation, and invasion via targeting PAX6 in breast cancer
cells. Mol Med Rep. 11:379–385. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang J, Wang X, Wu G, Hou D and Hu Q:
MiR-365b-3p, down-regulated in retinoblastoma, regulates cell cycle
progression and apoptosis of human retinoblastoma cells by
targeting PAX6. FEBS Lett. 587:1779–1186. 2013. View Article : Google Scholar : PubMed/NCBI
|