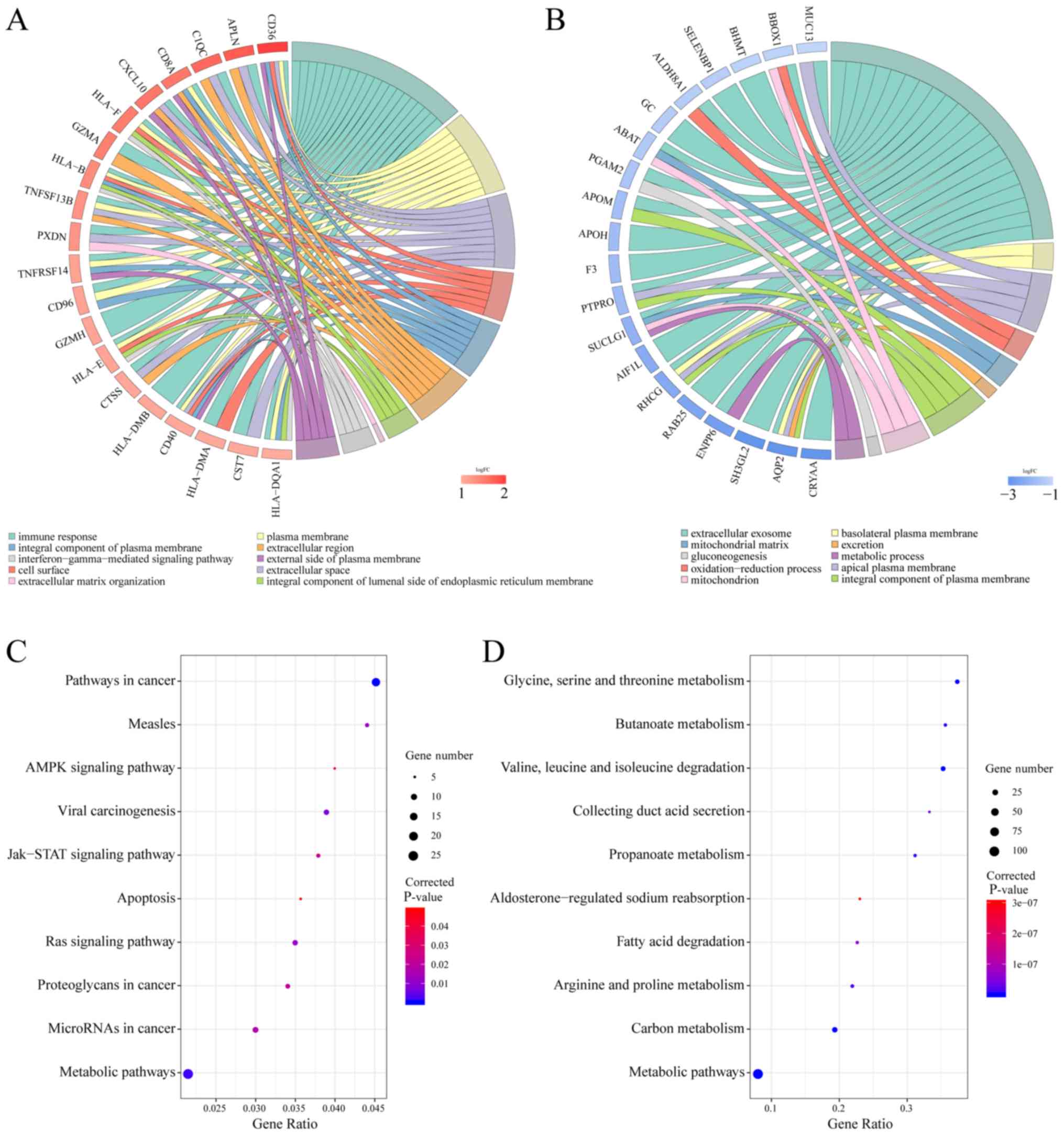
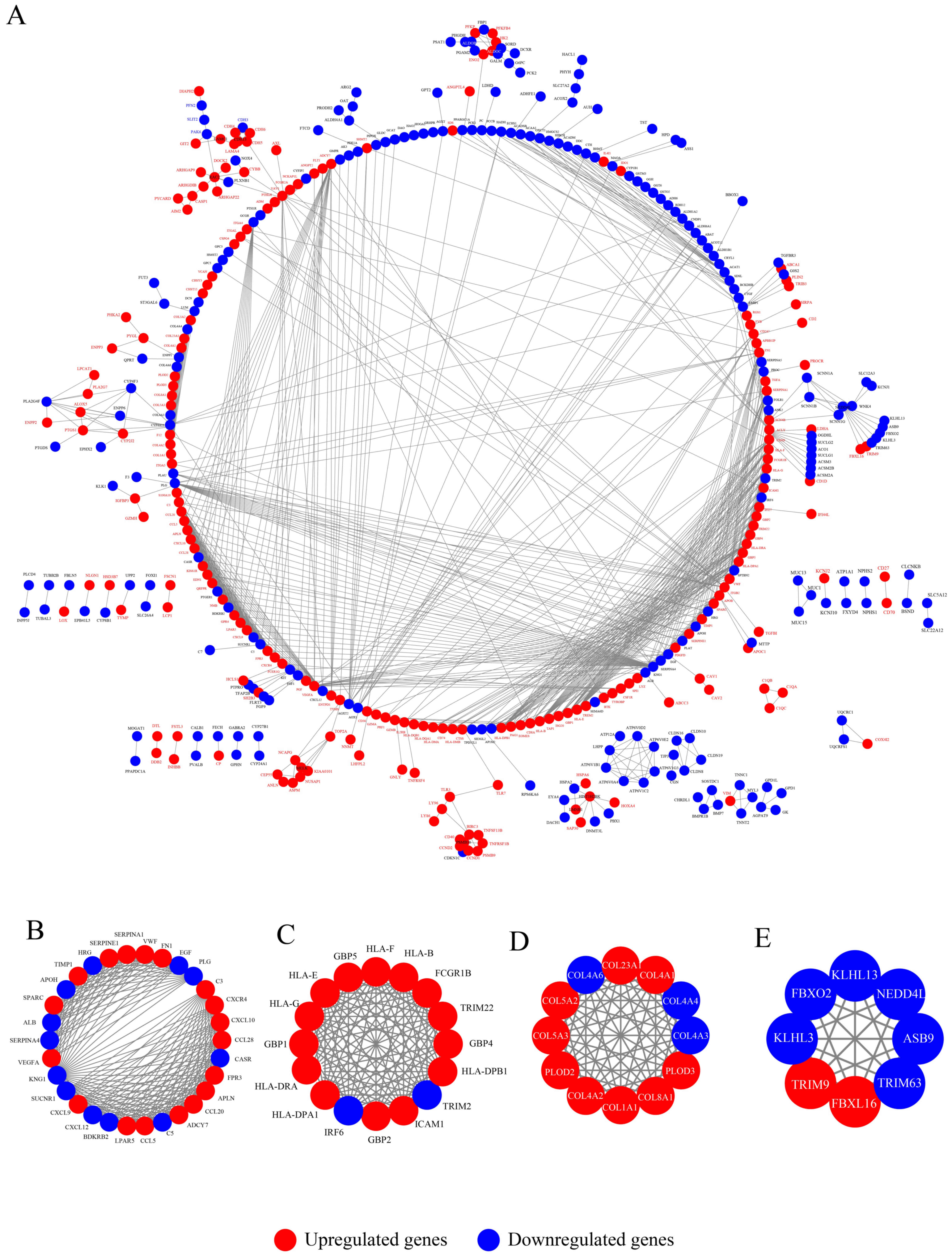
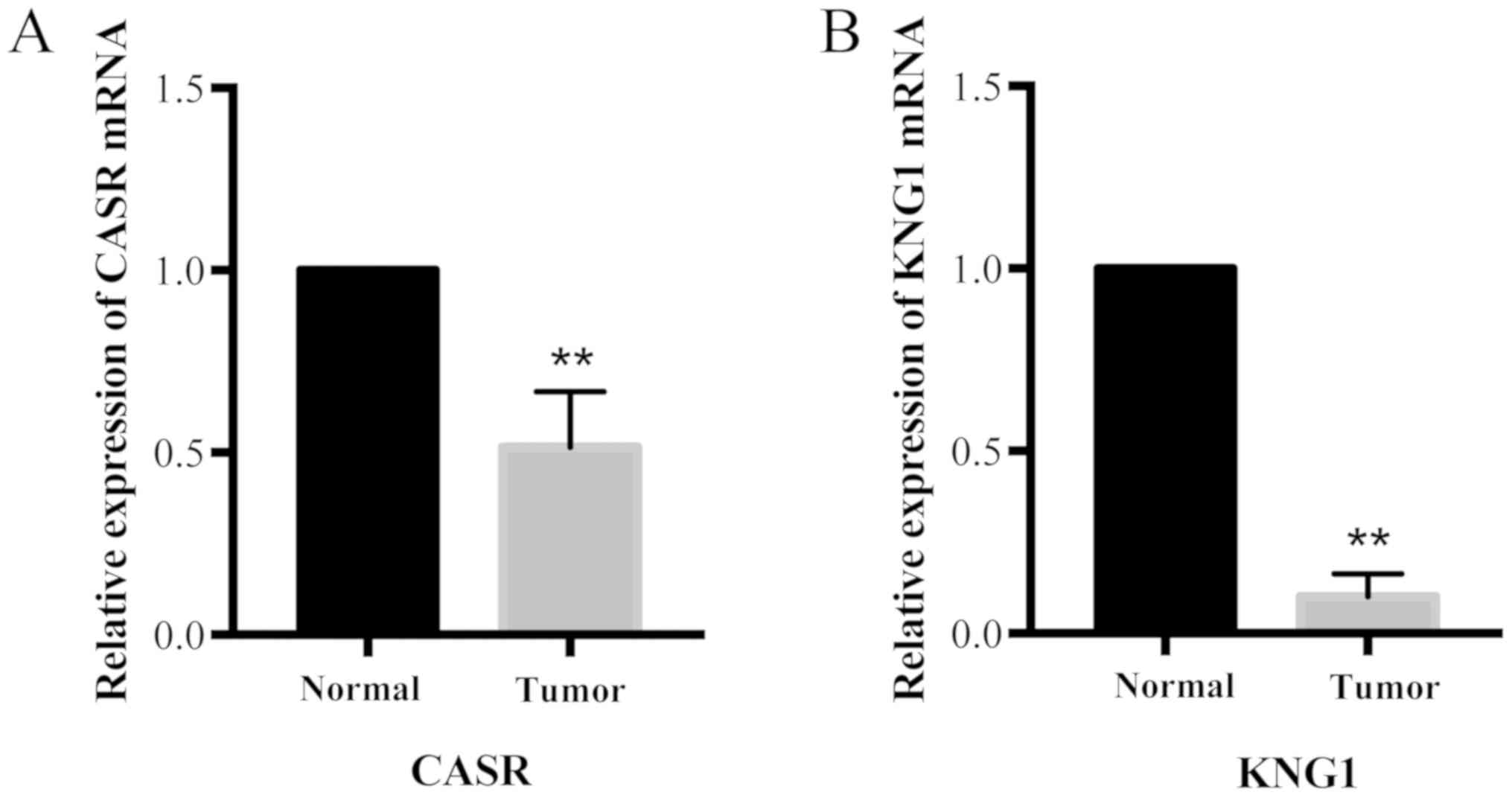
|
1
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ljungberg B, Bensalah K, Canfield S,
Dabestani S, Hofmann F, Hora M, Kuczyk MA, Lam T, Marconi L,
Merseburger AS, et al: EAU guidelines on renal cell carcinoma: 2014
update. Eur Urol. 67:913–924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tosco L, Van Poppel H, Frea B, Gregoraci G
and Joniau S: Survival and impact of clinical prognostic factors in
surgically treated metastatic renal cell carcinoma. Eur Urol.
63:646–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Grignon DJ and Che M: Clear cell renal
cell carcinoma. Clin Lab Med. 25:305–316. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim HS, Kim JH, Jang HJ, Han B and Zang
DY: Clinicopathologic significance of VHL gene alteration in
clear-cell renal cell carcinoma: An updated meta-analysis and
review. Int J Mol Sci. 19(pii): E25292018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song E, Song W, Ren M, Xing L, Ni W, Li Y,
Gong M, Zhao M, Ma X, Zhang X and An R: Identification of potential
crucial genes associated with carcinogenesis of clear cell renal
cell carcinoma. J Cell Biochem. 119:5163–5174. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yuan L, Zeng G, Chen L, Wang G and Wang X,
Cao X, Lu M, Liu X, Qian G, Xiao Y and Wang X: Identification of
key genes and pathways in human clear cell renal cell carcinoma
(ccRCC) by co-expression analysis. Int J Biol Sci. 14:266–279.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen L, Yuan L, Qian K, Qian G, Zhu Y, Wu
CL, Dan HC, Xiao Y and Wang X: Identification of biomarkers
associated with pathological stage and prognosis of clear cell
renal cell carcinoma by co-expression network analysis. Front
Physiol. 9:3992018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kolde R, Laur S, Adler P and Vilo J:
Robust rank aggregation for gene list integration and
meta-analysis. Bioinformatics. 28:573–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Wu J, Zhang D, Bing Z, Tian J, Ni
M, Zhang X, Meng Z and Liu S: Identification of potential key genes
associated with the pathogenesis and prognosis of gastric cancer
based on integrated bioinformatics analysis. Front Genet.
9:2652018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao X, Chen Y, Chen M, Wang S, Wen X and
Zhang S: Identification of key candidate genes and biological
pathways in bladder cancer. PeerJ. 6:e60362018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ni M, Liu X, Wu J, Zhang D, Tian J, Wang
T, Liu S, Meng Z, Wang K, Duan X, et al: Identification of
candidate biomarkers correlated with the pathogenesis and prognosis
of non-small cell lung cancer via integrated bioinformatics
analysis. Front Genet. 9:4692018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang X, Zhu S, Li L, Zhang L, Xian S, Wang
Y and Cheng Y: Identification of differentially expressed genes and
signaling pathways in ovarian cancer by integrated bioinformatics
analysis. Onco Targets Ther. 11:1457–1474. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu H, Brannon AR, Reddy AR, Alexe G,
Seiler MW, Arreola A, Oza JH, Yao M, Juan D, Liou LS, et al:
Identifying mRNA targets of microRNA dysregulated in cancer: With
application to clear cell renal cell carcinoma. BMC Syst Biol.
4:512010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peña-Llopis S, Vega-Rubin-de-Celis S, Liao
A, Leng N, Pavía-Jiménez A, Wang S, Yamasaki T, Zhrebker L,
Sivanand S, Spence P, et al: BAP1 loss defines a new class of renal
cell carcinoma. Nat Genet. 44:751–759. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wozniak MB, Le Calvez-Kelm F,
Abedi-Ardekani B, Byrnes G, Durand G, Carreira C, Michelon J,
Janout V, Holcatova I, Foretova L, et al: Integrative genome-wide
gene expression profiling of clear cell renal cell carcinoma in
Czech Republic and in the United States. PLoS One. 8:e578862013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Eckel-Passow JE, Serie DJ, Bot BM, Joseph
RW, Cheville JC and Parker AS: ANKS1B is a smoking-related
molecular alteration in clear cell renal cell carcinoma. BMC Urol.
14:142014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wotschofsky Z, Gummlich L, Liep J, Stephan
C, Kilic E, Jung K, Billaud JN and Meyer HA: Integrated microRNA
and mRNA signature associated with the transition from the locally
confined to the metastasized clear cell renal cell carcinoma
exemplified by miR-146-5p. PLoS One. 11:e01487462016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takahashi M, Tsukamoto Y, Kai T, Tokunaga
A, Nakada C, Hijiya N, Uchida T, Daa T, Nomura T, Sato F, et al:
Downregulation of WDR20 due to loss of 14q is involved in the
malignant transformation of clear cell renal cell carcinoma. Cancer
Sci. 107:417–423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
23
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:(Web Server Issue). W316–W322. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Greene FL, Gospodarowicz MK and Wittekend
C: American Joint Committee On Cancer (AJCC) cancer staging manual.
(7th). Philadelphia. (Springer). 2009.PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ge S, Xia X, Ding C, Zhen B, Zhou Q, Feng
J, Yuan J, Chen R, Li Y, Ge Z, et al: A proteomic landscape of
diffuse-type gastric cancer. Nat Commun. 9:10122018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Leek JT and Storey JD: Capturing
heterogeneity in gene expression studies by surrogate variable
analysis. PLoS Genet. 3:1724–1735. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu F, Wu S and Gou X: Identification of
biomarkers and potential molecular mechanisms of clear cell renal
cell carcinoma. Neoplasma. 65:242–252. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang Y, Lu Q, Shao X, Mo B, Nie X, Liu W,
Chen X, Tang Y, Deng Y and Yan J: Development of A three-gene
prognostic signature for hepatitis B virus associated
hepatocellular carcinoma based on integrated transcriptomic
analysis. J Cancer. 11:989–2002. 2018.PubMed/NCBI
|
|
34
|
Zhou Y, Cao HB, Li WJ and Zhao L: The
CXCL12 (SDF-1)/CXCR4 chemokine axis: Oncogenic properties,
molecular targeting, and synthetic and natural product CXCR4
inhibitors for cancer therapy. Chin J Nat Med. 16:801–810.
2018.PubMed/NCBI
|
|
35
|
Wang M, Yang X, Wei M and Wang Z: The role
of CXCL12 axis in lung metastasis of colorectal cancer. J Cancer.
9:3898–3903. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wei S, Wang Y, Xu H and Kuang Y: Screening
of potential biomarkers for chemoresistant ovarian carcinoma with
miRNA expression profiling data by bioinformatics approach. Oncol
Lett. 10:2427–2431. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang D, Luo L and Guo J: miR-129-1-3p
inhibits cell migration by targeting BDKRB2 in gastric cancer. Med
Oncol. 31:982014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li C, Xie J, Lu Z, Chen C, Yin Y, Zhan R,
Fang Y, Hu X and Zhang CC: ADCY7 supports development of acute
myeloid leukemia. Biochem Biophys Res Commun. 465:47–52. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Frees S, Breuksch I, Haber T, Bauer HK,
Chavez-Munoz C, Raven P, Moskalev I, D Costa N, Tan Z, Daugaard M,
et al: Calcium-sensing receptor (CaSR) promotes development of bone
metastasis in renal cell carcinoma. Oncotarget. 9:15766–15779.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu J, Fang J, Cheng Z, Fan L, Hu W, Zhou F
and Shen H: Overexpression of the Kininogen-1 inhibits
proliferation and induces apoptosis of glioma cells. J Exp Clin
Cancer Res. 37:1802018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tang J, Kong D, Cui Q, Wang K, Zhang D,
Yuan Q, Liao X, Gong Y and Wu G: Bioinformatic analysis and
identification of potential prognostic microRNAs and mRNAs in
thyroid cancer. PeerJ. 6:e46742018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu J, Huang Y, Lin C, Li X, Fang X, Zhong
C, Yuan Y and Zheng S: Identification of kininogen 1 as a serum
protein marker of colorectal adenoma in patients with a family
history of colorectal cancer. J Cancer. 9:540–547. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang H, Liu J, Fu X and Yang A:
Identification of key genes and pathways in tongue squamous cell
carcinoma using bioinformatics analysis. Med Sci Monit.
23:5924–5932. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yap LF, Velapasamy S, Lee HM, Thavaraj S,
Rajadurai P, Wei W, Vrzalikova K, Ibrahim MH, Khoo AS, Tsao SW, et
al: Down-regulation of LPA receptor 5 contributes to aberrant LPA
signalling in EBV-associated nasopharyngeal carcinoma. J Pathol.
235:456–465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Znaor A, Lortet-Tieulent J, Laversanne M,
Jemal A and Bray F: International variations and trends in renal
cell carcinoma incidence and mortality. Eur Urol. 67:519–530. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lopez R, Wang R and Seelig G: A molecular
multi-gene classifier for disease diagnostics. Nat Chem.
10:746–754. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen X, Xie D, Zhao Q and You ZH:
MicroRNAs and complex diseases: From experimental results to
computational models. Brief Bioinform. 20:515–539. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen X, Wang L, Qu J, Guan NN and Li JQ:
Predicting miRNA-disease association based on inductive matrix
completion. Bioinformatics. 34:4256–4265. 2018.PubMed/NCBI
|
|
49
|
Chen X, Yin J, Qu J and Huang L: MDHGI:
Matrix decomposition and heterogeneous Graph inference for
miRNA-disease association prediction. PLoS Comput Biol.
14:e10064182018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen X and Yan GY: Novel human
lncRNA-disease association inference based on lncRNA expression
profiles. Bioinformatics. 29:2617–2624. 2013. View Article : Google Scholar : PubMed/NCBI
|