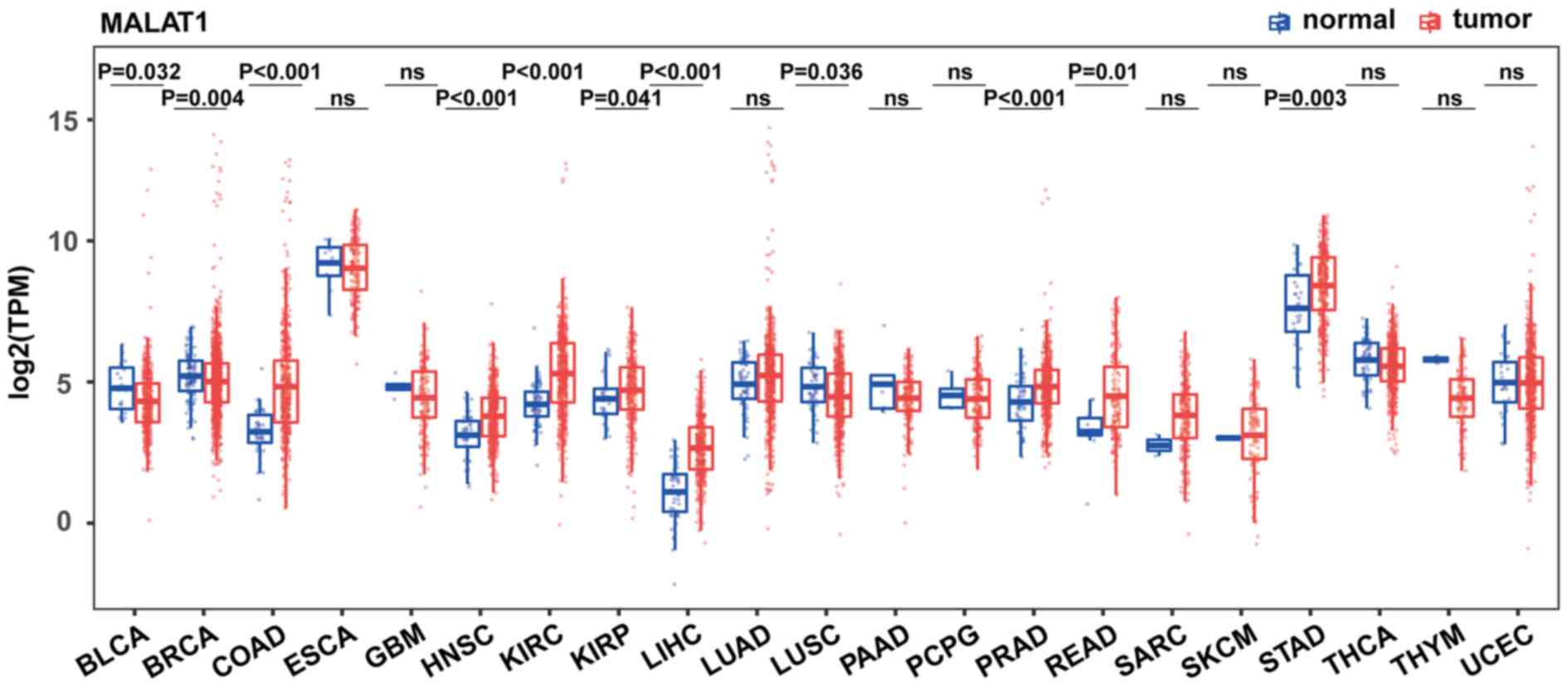
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Source, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lopez-Beltran A, Carrasco JC, Cheng L,
Scarpelli M, Kirkali Z and Montironi R: 2009 update on the
classification of renal epithelial tumours in adults. Int J Urol.
16:432–443. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bedke J, Gauler T, Grünwald V, Hegele A,
Herrmann E, Hinz S, Janssen J, Schmitz S, Schostak M, Tesch H, et
al: Systemic therapy in metastatic renal cell carcinoma. World J
Urol. 35:179–188. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brown C: Targeted therapy: An elusive
cancer target. Nature. 537:S106–S108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ulitsky I and Bartel DP: LincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang L, Lin C, Jin C, Yang JC, Tanasa B,
Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al:
LncRNA-dependent mechanisms of androgen receptor-regulated gene
activation programs. Nature. 500:598–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wilusz JE: Long noncoding RNAs: Re-writing
dogmas of RNA processing and stability. Biochim Biophys Acta.
1859:128–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gutschner T, Hämmerle M and Diederichs S:
MALAT1-a paradigm for long noncoding RNA function in cancer. J Mol
Med (Berl). 91:791–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y
and Yang K: Up-regulation of long noncoding RNA MALAT1 contributes
to proliferation and metastasis in esophageal squamous cell
carcinoma. J Exp Clin Cancer Res. 34:72015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Okugawa Y, Toiyama Y, Hur K, Toden S,
Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR and
Goel A: Metastasis-associated long non-coding RNA drives gastric
cancer development and promotes peritoneal metastasis.
Carcinogenesis. 35:2731–2739. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Park JY, Lee JE, Park JB, Yoo H, Lee SH
and Kim JH: Roles of long non-coding RNAs on tumourigenesis and
glioma development. Brain Tumour Res Treat. 2:1–6. 2014. View Article : Google Scholar
|
|
17
|
Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng
J, Xiao W, Yu G, Yao W, Zhou H, et al: LncRNA MALAT1 functions as a
competing endogenous RNA to regulate ZEB2 expression by sponging
miR-200s in clear cell kidney carcinoma. Oncotarget. 6:38005–38015.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen R, Liu Y, Zhuang H, Yang B, Hei K,
Xiao M, Hou C, Gao H, Zhang X, Jia C, et al: Quantitative
proteomics reveals that long non-coding RNA MALAT1 interacts with
DBC1 to regulate p53 acetylation. Nucleic Acids Res. 45:9947–9959.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Engreitz JM, Sirokman K, McDonel P,
Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M
and Lander ES: RNA-RNA interactions enable specific targeting of
noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell.
159:188–199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dawe HR, Minamide LS, Bamburg JR and
Cramer LP: ADF/cofilin controls cell polarity during fibroblast
migration. Curr Biol. 13:252–257. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang W, Eddy R and Condeelis J: The
cofilin pathway in breast cancer invasion and metastasis. Nat Rev
Cancer. 7:429–440. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bravo-Cordero JJ, Magalhaes MA, Eddy RJ,
Hodgson L and Condeelis J: Functions of cofilin in cell locomotion
and invasion. Nat Rev Mol Cell Biol. 14:405–415. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Warren AY and Harrison D: WHO/ISUP
classification, grading and pathological staging of renal cell
carcinoma: Standards and controversies. World J Urol. 36:1913–1926.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/Bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res. 44:e712016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
R Core Team (2012), . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing. (Vienna, Austria). ISBN 3-900051-07-0, URL
http://www.R-project.org/.
|
|
28
|
RStudio Team (2015), . RStudio: Integrated
Development for R. RStudio, Inc. (Boston, MA). URL
http://www.rstudio.com/.
|
|
29
|
Rooney MS, Shukla SA, Wu CJ, Getz G and
Hacohen N: Molecular and genetic properties of tumours associated
with local immune cytolytic activity. Cell. 160:48–61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang HM, Yang FQ, Chen SJ, Che J and
Zheng JH: Upregulation of long non-coding RNA MALAT1 correlates
with tumour progression and poor prognosis in clear cell renal cell
carcinoma. Tumour Biol. 36:2947–2955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
interacts with miR-205. Cancer Res. 75:1322–1331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Han Y, Liu Y, Zhang H, Wang T, Diao R,
Jiang Z, Gui Y and Cai Z: Hsa-miR-125b suppresses bladder cancer
development by down-regulating oncogene SIRT7 and oncogenic long
non-coding RNA MALAT1. FEBS Lett. 587:3875–3882. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Malakar P, Shilo A, Mogilevsky A, Stein I,
Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV
and Karni R: Long noncoding RNA MALAT1 promotes hepatocellular
carcinoma development by SRSF1 upregulation and mTOR activation.
Cancer Res. 77:1155–1167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tripathi V, Shen Z, Chakraborty A, Giri S,
Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A and
Prasanth KV: Long noncoding RNA MALAT1 controls cell cycle
progression by regulating the expression of oncogenic transcription
factor B-MYB. PLoS Genet. 9:e10033682013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang
Y, Xiao Z, Siverly AN, Lawhon SE, Ton BN, et al: Long noncoding RNA
MALAT1 suppresses breast cancer metastasis. Nat Genet.
50:1705–1715. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li C, Miao R, Liu S, Wan Y, Zhang S, Deng
Y, Bi J, Qu K, Zhang J and Liu C: Down-regulation of miR-146b-5p by
long noncoding RNA MALAT1 in hepatocellular carcinoma promotes
cancer growth and metastasis. Oncotarget. 8:28683–28695. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang L, Lin C, Liu W, Zhang J, Ohgi KA,
Grinstein JD, Dorrestein PC and Rosenfeld MG: ncRNA- and Pc2
methylation-dependent gene relocation between nuclear structures
mediates gene activation programs. Cell. 147:773–788. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun Z, Ou C, Liu J, Chen C, Zhou Q, Yang
S, Li G, Wang G, Song J, Li Z, et al: YAP1-induced MALAT1 promotes
epithelial-mesenchymal transition and angiogenesis by sponging
miR-126-5p in colorectal cancer. Oncogene. 38:2627–2644. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Audibert A, Weil D and Dautry F: In vivo
kinetics of mRNA splicing and transport in mammalian cells. Mol
Cell Biol. 22:6706–6718. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen Q and Pollard TD: Actin filament
severing by cofilin is more important for assembly than
constriction of the cytokinetic contractile ring. J Cell Biol.
195:485–498. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kanellos G, Zhou J, Patel H, Ridgway RA,
Huels D, Gurniak CB, Sandilands E, Carragher NO, Sansom OJ, Witke
W, et al: ADF and cofilin1 control actin stress fibres, nuclear
integrity, and cell survival. Cell Rep. 13:1949–1964. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pang D, Yang C, Li C, Zou Y, Feng B, Li L,
Liu W, Luo Q, Chen Z and Huang C: Polyphyllin II inhibits liver
cancer cell proliferation, migration and invasion through
downregulated cofilin activity and the AKT/NF-κB pathway. Biol
Open. 9:bio0468542020. View Article : Google Scholar : PubMed/NCBI
|