Introduction
Hepatocellular carcinoma (HCC) is the most common
human liver malignancy. Worldwide, the estimated number of
mortalities caused by HCC is ~810,000 per year, while the estimated
number of new cases of HCC is ~854,000 per year (1). As a nation with a large population and
high prevalence of hepatitis B virus (HBV) and HCV, China accounts
for ~422,100/year of total HCC mortalities worldwide (2). Thus, the mortality and severity of HCC
represent an urgent health problem in China.
Current management of HCC includes potentially
curative surgical resection and non-surgical therapeutics, such as
radiofrequency ablation, percutaneous ethanol injection and
trans-arterial embolization (3).
Moreover, combined chemotherapy and recently developed
immunotherapies constitute the treatment of patients with advanced
HCC (3). However, it remains
difficult to predict overall survival (OS) of patients with HCC for
treatment selection. Tumor classification systems, such as TNM,
Okuda, Cancer of the Liver Italian Program and Barcelona Clinic
Liver Cancer, have been widely used for prognostic assessment
(4,5). However, due to the heterogeneity of
malignancies, there is still no consensus on the most appropriate
staging system for predicting the survival of patients with HCC
(6,7). Previous studies have been conducted to
identify potential biomarkers. It has been revealed that frequently
used biomarkers, such as α-fetoprotein (AFP) (8) and carbohydrate antigen 19-9 (CA19-9)
(9), have a relatively poor
performance as predictors of long-term outcome.
Recently, there have been studies on the mechanism
of 5′-nucleotidase domain containing 2 (NT5DC2) in some
malignancies, including gliomas (10) and liver cancer types (11). NT5DC2 has been reported to be
associated with certain types of mental disorders, including
schizophrenia, attention deficit hyperactivity disorder (12,13) and
borderline personality disorders (14). NT5DC2 promotes tumorigenesis
of glioma stem-like cells by inducing the expression of FYN
proto-oncogene (Fyn) (10), as well
as increases tumor cell proliferation in HCC by stabilizing
epidermal growth factor receptor (11). Currently, to the best of our
knowledge, there is no research focusing on the value of
NT5DC2 in the prognosis of HCC.
The present study aimed to investigate the
suitability of NT5DC2 as a novel prognostic predictor for
HCC.
Materials and methods
Data source
The latest liver cancer (LIHC) project data,
including Level 3 RNA sequencing (RNA-seq) data and clinical data,
were obtained using the R package, TCGAbiolinks v2.16.0 (15), as an independent validation cohort.
There were 377 cases available including 255 males and 122 females.
The age range was between 16 to 90, with a median of 61. mRNA gene
expression levels are presented as a normalized value of level 3
RNA-seq data. Then, NT5DC2 mRNA expression was compared
between cancerous and paracancerous tissues. In HCC samples,
patients were assigned into two equal groups (NT5DC2-High
and NT5DC2-Low) based on the median normalized value of
NT5DC2. The reciprocal The Cancer Genome Atlas (TCGA)
clinical data (updated on 2019/08/08), comprising age, sex, weight,
Child-Pugh score (16), TNM stage
(17), residual tumor, Edmondson
grade (18), relapse-free survival
(RFS) and OS, were collected for further statistical analysis.
The Sequence Read Archive (SRA) dataset, SRP174991,
including paired HCC sample RNA-seq raw data (19), were downloaded using the Linux
package, prefetch (included in the Sra-tools version 2.10.6,
http://github.com/ncbi/sra-tools).
Sample and clinical data
collection
Patients diagnosed with HCC, receiving surgical
resection between January 2008 and December 2015, in the Department
of Hepatology and the Department of General Surgery, Peking Union
Medical College Hospital (PUMCH), were retrospectively enrolled in
this study. There were 134 cases available including 114 males and
20 females. The age range was between 32 to 82 years, with a median
of 56 years. The reciprocal clinical data including age, sex,
surface antigen of the hepatitis B virus (HBsAg), Anti-hepatitis C
(HCV), aspartate transaminase, alanine aminotransferase (ALT),
γ-glutamyl transpeptidase (GGT), alkaline phosphatase, AFP, CA19-9,
TNM stage and Edmonson grade, were obtained via the Hospital
Information System in PUMCH. The OS of enrolled patients was
assessed via phone calls and routine clinical follow-up. This
project was approved by the Ethic Committee, Peking Union Medical
College Hospital (approval no. JS-1569) and had been performed in
accordance with Declaration of Helsinki and its later amendments.
Written informed consent was provided by all patients before
surgery.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Paired HCC samples, which were preserved frozen
samples obtained during surgery, used in RT-qPCR were randomly
chosen from the PUMCH cohort. Samples from patients with HCC were
preserved in liquid nitrogen (−196°C) immediately after
resection.
Total RNA was extracted from liquid
nitrogen-preserving tissues using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) with
Phasemaker™ tubes (Invitrogen; Thermo Fisher Scientific,
Inc.), according to manufacturer's protocol. RT was performed using
the GoScript™ RT system (Promega Corporation) with oligo-dT
(15) primer at 42°C for 1 h,
according to the manufacturer's instructions. RT-qPCR was performed
with 2X GoTaq Master Mix (cat. no. A6001; Promega Corporation)
using an Applied Biosystems™ 7500 Fast Dx Real-Time PCR System
(Thermo Fisher Scientific, Inc.). Thermocycling conditions: 95°C
for 2 min, 40 cycles: 95°C for 10 sec, 60°C for 30 sec.
NT5DC2 mRNA expression was normalized to GAPDH mRNA
expression. The gene expression was calculated via
2−ΔΔCq method (20). The
primers validated for amplification efficiency and selected for
RT-qPCR were as follows: NT5DC2 forward (F),
5′-GCAGCCATCTACGCCAACA-3′ and reverse (R),
5′-TCACGGGCGGTACTGAAGA-3′; and GAPDH F,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and R,
5′-GGCTGTTGTCATACTTCTCATGG-3′.
Haemotoxylin-eosin (H&E) and
immunohistochemical (IHC) staining of tissue microarrays
(TMAs)
TMAs were constructed from formalin-fixed, paraffin
embedded tissue blocks stored at the Department of Pathology,
PUMCH. All specimens were previously fixed in 10% neutral formalin
solution at room temperature for 12 h before paraffin embedded.
Tissue blocks of paired cancer, paracancerous tissue and randomly
selected healthy tissue, as previously established by pathologists,
were selected and TMAs were prepared. Tissue cores were re-embedded
in paraffin, resected into 8-µm thick slides. HE staining was
performed using Gemini AS Automated Slide Stainer (Thermo Fisher
Scientific, Inc.) with a normal H&E staining program in room
temperature. IHC staining was performed with HIER pH 6 retrieval.
Slides were blocked by normal goat serum working solution (cat. no.
SP-9001; OriGene Technologies, Inc.) at room temperature for 15
min, then added 100 µl Anti-NT5DC2 rabbit polyclonal
antibody (1:50; cat. no. NBP2-13679; Novus Biologicals, LLC), and
incubated at 37°C for 60 min. Then, 100 µl biotin-labeled goat
anti-rabbit polyclonal antibody solution (ready to use; cat. no.
SP-9001; OriGene Technologies, Inc.) was added to slides and
incubated at room temperature for 15 min. The reactions were
visualized by diaminobenzidine (DAB) for 30 sec at room temperature
and counterstained with hematoxylin for 10 sec at room temperature.
Normal goat serum working solution, biotin-labeled goat anti-rabbit
polyclonal antibody solution and streptavidin-HRP reagent were
included in the SPlink Biotin-Streptavidin-horseradish peroxidase
(HRP) Detection kit (cat. no. SP-9001; OriGene Technologies, Inc.).
1X DAB was diluted from the 20X DAB kit (cat. no. ZLI-9017; OriGene
Technologies, Inc.). Sample results were evaluated by pathologists
under a light microscope at the PUMCH. IHC-TMA slides were scanned
using Panaroma SCAN II (3DHISTech Ltd.). In total, three random
×400 fields of view of the IHC chips were captured for mean optical
density (MOD) analysis via ImageJ 1.52 (National Institutes of
Health). The MOD was calculated as follows: MOD=Integrated OD/Sum
area of samples. MODs were compared between cancerous tissues,
para-HCC tissues and distal healthy liver tissues in IHC TMAs, and
reciprocal receiver operating characteristic (ROC) curves were
generated.
Statistical analysis
All experiments were repeated three times. P<0.05
was considered to indicate a statistically significant difference.
Quantitative analyses were performed using a Kolmogorov-Smirnov
test for normality of the distribution and an F-test for equality
of variances. Samples with normal distribution were represented as
mean ± SD, while samples with non-normal distribution were
represented as median (interquartile range). Categorical samples
were expressed as n% (n). Moreover, two groups with normal
distribution were compared via Student's t-test; two paired groups
with normal distribution were compared using paired t-test.
Multiple groups with normal distribution were compared using
one-way ANOVA test followed by Tukey's post hoc test. Groups with
non-normal distribution were compared using Mann-Whitney U test.
Fisher's exact test and χ2 test were used for comparison
of categorical data. Kaplan-Meier survival analyses and Rényi tests
were performed between NT5DC2-High and NT5DC2-Low
groups. ROC curve was used to assess the diagnostic value of
NT5DC2 in IHC TMAs. The median cut-off value was selected
according to ROC curve.
Univariate Cox proportional hazards regression and
multivariate Cox proportional hazards regression models were
applied to analyze the impact of potentially confounding factors on
OS. Potential confounding factors in both groups were evaluated via
a univariate Cox regression model. Potentially significant factors
(P<0.1) in the univariate Cox regression model were included in
a multivariate Cox regression model. The hazard ratio (HR), 95% CI
and statistical significance are presented in the tables. All
statistical analyses were performed via R software 3.5.3 (21).
Bioinformatic analyses
To investigate NT5DC2 function in patients
with HCC, an integrative analysis of NT5DC2 was performed.
Patients of the LIHC dataset were categorized into two groups
according to NT5DC2 expression: NT5DC2-Low and
NT5DC2-High. Gene Set Enrichment Analysis (GSEA) was
performed using GSEA version 4.0.1 with MSigDB 7.0 (22,23).
Gene sets with significant enrichment were listed, and RNA-seq data
from the SRA dataset were mapped to the reference genome using
hisat2 v2.1.0 (24). The reads were
processed with featureCounts v1.6.0 (25) and DESeq2 v1.24.0 (26). The references consisted of the human
reference genome (GRCh38) and the Ensembl annotated human
transcriptome (GRCh38.v98). Normalized values were compared between
the two groups and NT5DC2 expression levels were analyzed
using the R package, ggplot2 v3.2.1 (https://ggplot2.tidyverse.org/). A Kaplan-Meier
survival curve was generated using the R packages, survival
v2.44-1.1 (https://CRAN.R-project.org/package=survival) and
survminer v0.4.6 (https://rpkgs.datanovia.com/survminer/index.html).
Results
NT5DC2 mRNA is upregulated in
cancerous tissue compared with paracancerous tissue in multiple
datasets
In the LIHC dataset of TCGA, 377 cases were
available, but eight cases were excluded due to the lack of RNA-seq
data. Moreover, 10 cases of non-HCC liver cancer were excluded. One
case was excluded due to the lack of OS data. Finally, 358
patients, for whom survival data were available, were included in
the analysis. A total of 358 HCC tissues and 40 para-HCC tissues
were examined. Normalized NT5DC2 expression was significantly
upregulated in 358 HCC tissues compared with the para-HCC tissues
(P<0.0001; Fig. 1A). The
available paired samples were selected (n=50) in TCGA LIHC dataset,
and a ROC curve analysis was performed. The area under the curve
(AUC) of paired HCC samples was 0.942 (Fig. 1B).
 | Figure 1.NT5DC2 is upregulated in
cancerous tissue compared with paracancerous tissue in multiple
datasets. (A) Violin plot of the difference in NT5DC2
expression value between HCC and paired para-HCC samples in TCGA
LIHC cohort. (B) ROC curve of NT5DC2 expression in
distinguishing between HCC and paired para-HCC tissues in TCGA LIHC
cohort. The AUC was calculated and illustrated. (C) Violin plot of
the difference in NT5DC2 expression value between HCC and
paired para-HCC samples in SRP174991. (D) ROC curve of
NT5DC2 expression for distinguishing between HCC and paired
para-HCC tissues in SRP174991. AUC was calculated and illustrated.
(E) Violin plot of the difference in NT5DC2 expression
between HCC and paired para-HCC samples in selected HCC and paired
para-HCC samples using reverse transcription-quantitative PCR. (F)
ROC curve of NT5DC2 expression for distinguishing between
HCC and paired para-HCC tissues in SRP174991. AUC was calculated
and illustrated. (G) Selected tissue specimens in TMA of HCC,
para-HCC and healthy liver samples stained using H&E or IHC.
(H) Violin plot demonstrating the difference in NT5DC2
expression between HCC and paired para-HCC samples in a TMA of the
PUMCH cohort. Expression between groups were compared using one-way
ANOVA followed by Tukey's test. (I) ROC curve of NT5DC2
expression for distinguishing between HCC and paired para-HCC
tissues in IHC-TMA of HCC, para-HCC and healthy liver samples. AUC
was calculated and illustrated. *P<0.05, ****P<0.0001. ROC,
receiver operating characteristic; AUC, area under the curve;
NT5DC2, 5′-nucleotidase domain containing 2; HCC,
hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; LIHC,
liver cancer; TMA, tissue microarray; H&E, hematoxylin and
eosin; IHC, immunohistochemistry; PUMCH, Peking Union Medical
College Hospital; CA, cancer; PARA, paracancerous. |
In the SRP174991 dataset, NT5DC2 expression
was compared in 35 pairs of HCC and para-HCC samples from patients
with HBV-related HCC. The results demonstrated a significant
upregulation of NT5DC2 in HCC tissues (P<0.0001; Fig. 1C). In the 35 paired HCC SRP174991
samples, the AUC of paired HCC samples was 0.810 (Fig. 1D).
NT5DC2 mRNA expression was examined using
RT-qPCR in 44 HCC tissues and 44 para-HCC tissues randomly selected
from the PUMCH cohort. The mRNA expression of NT5DC2 was
significantly higher in cancerous compared with paracancerous
tissues (P<0.05; Fig. 1E). ROC
curve analysis was performed and the AUC was 0.637 (Fig. 1F).
IHC was conducted to analyze a TMA comprising 38
randomly selected non-HCC liver tissues, 32 paracancerous liver
tissues and 32 HCC liver tissues (Fig.
1G). IHC staining identified a higher expression of
NT5DC2 in HCC tissues compared with both para-HCC tissues
(P<0.05) and distal healthy liver tissues (P<0.0001; Fig. 1H). Furthermore, ROC curves of
NT5DC2 expression in HCC and para-HCC tissues were
generated, and the AUC of NT5DC2 was 0.734 (Fig. 1I).
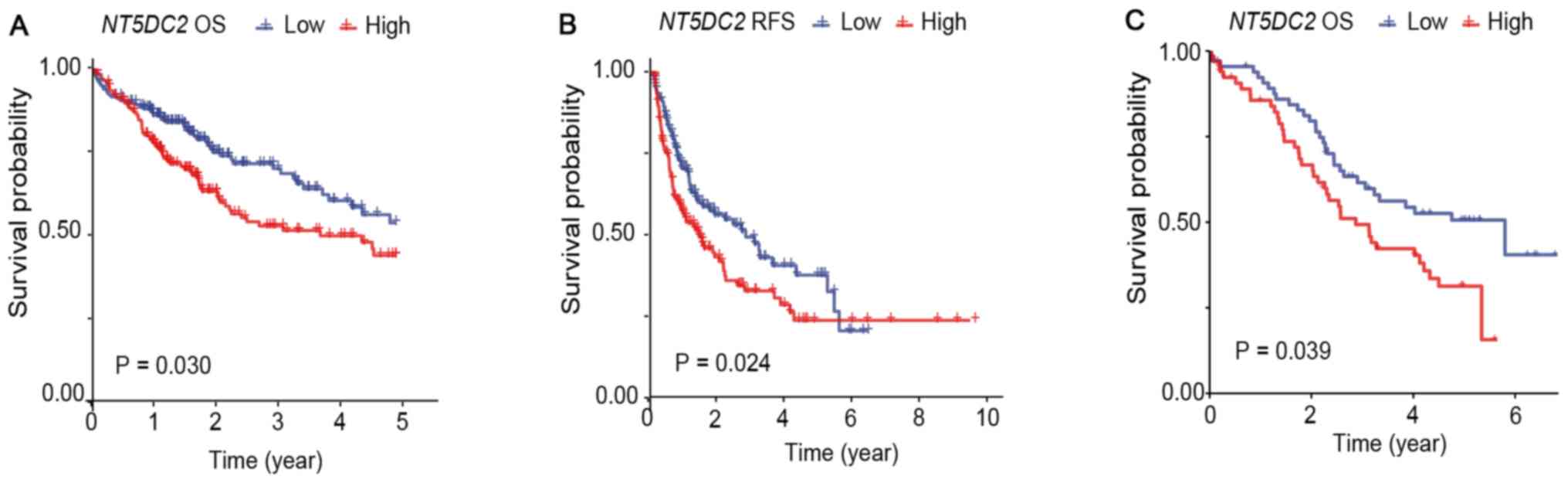
NT5DC2 gene upregulation reduces OS
and RFS in patients with HCC
In the LIHC cohort of TCGA, 358 patients with
available survival data were included in the survival analysis.
These patients were categorized into two groups based on the median
value of NT5DC2 expression: NT5DC2-High (n=179) and
NT5DC2-Low (n=179). The baseline data of the two groups are
listed in Table I. Kaplan-Meier and
Rényi analysis results demonstrated significant differences in OS
(P=0.030; Fig. 2A) and RFS (P=0.024;
Fig. 2B).
 | Table I.Baseline characteristics of TCGA LIHC
dataset (n=358). |
Table I.
Baseline characteristics of TCGA LIHC
dataset (n=358).
| Characteristic |
NT5DC2-High |
NT5DC2-Low | P-value |
|---|
| Age, years | 57.78 (13.95) | 61.26 (12.33) | 0.013 |
| Sex |
|
| 0.164 |
|
Male | 64.3 (115) | 71.5 (128) |
|
|
Female | 35.7 (64) | 28.5 (51) |
|
| Weight, kg | 66.0 (20.0) | 73.0 (24.5) | 0.003 |
| Child-Pugh
score |
|
| 0.138 |
| Grade
A | 51.4 (92) | 67.0 (120) |
|
| Grade
B | 7.3 (13) | 4.4 (8) |
|
| Grade
C | 0 (0) | 0.6 (1) |
|
| TNM Stage |
|
| <0.001 |
| Stage
I | 35.8 (64) | 57.2 (103) |
|
| Stage
II | 26.8 (48) | 18.9 (34) |
|
| Stage
III | 30.2 (54) | 15.6 (28) |
|
| Stage
IV | 1.1 (2) | 1.1 (2) |
|
| Residual tumor |
|
| 0.650 |
| R0 | 88.3 (158) | 88.3 (159) |
|
| R1 | 3.4 (6) | 4.5 (8) |
|
| R2 | 0 (0) | 0.6 (1) |
|
| Rx | 6.7 (12) | 4.5 (8) |
|
| Edmondson
grade |
|
| 0.026 |
| G1 | 10.1 (18) | 19.6 (35) |
|
| G2 | 45.8 (82) | 49.7 (89) |
|
| G3 | 38.0 (68) | 28.5 (51) |
|
| G4 | 3.9 (7) | 2.2 (4) |
|
In the PUMCH cohort, a total of 134 patients were
diagnosed with HCC and received surgical therapy (Table II). These patients were
retrospectively enrolled in this study and categorized into two
groups according to the median IHC MOD: NT5DC2-High (n=67)
and NT5DC2-Low (n=67). Kaplan-Meier analysis was performed,
and the Rényi test indicated that patients in the
NT5DC2-High group had a significantly poorer OS (P=0.039)
compared with the NT5DC2-Low group (Fig. 2C).
 | Table II.Cox Proportional Hazards regression
models for overall survival in The Cancer Genome Atlas liver cancer
cohort (n=359). |
Table II.
Cox Proportional Hazards regression
models for overall survival in The Cancer Genome Atlas liver cancer
cohort (n=359).
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years | 1.012
(0.998-1.026) | 0.109 |
|
|
| Male | 0.841
(0.585-1.210) | 0.351 |
|
|
|
NT5DC2/1,000 | 2.227
(1.369-3.625) | 0.001 | 1.695
(1.016-1.727) | 0.043 |
| TNM stage | 1.512
(1.284-1.781) | <0.001 | 1.460
(1.235-1.727) | <0.001 |
| Edmondson
grade | 1.081
(0.852-1.372) | 0.522 |
|
|
NT5DC2 gene upregulation is a risk
factor associated with poor OS and RFS
In TCGA LIHC cohort, 358 patients with available
survival data were categorized into two groups based on the median
value of NT5DC2 expression: NT5DC2-High (n=179) and
NT5DC2-Low (n=179). It was found that the two groups were
different in age (P=0.013), weight (P=0.003), TNM stage
(P<0.001) and Edmondson grade (P=0.026; Table I).
To exclude potential confounding biases, clinically
significant variables, such as age, sex, NT5DC2/1,000 units
(NT5DC2/1,000), TNM stage and Edmondson grade were included
in univariate Cox proportional hazards regression models to
evaluate their influence on OS. The results suggested that both
NT5DC2/1,000 (P=0.001; HR=2.227; 95% CI=1.369-3.625) and TNM
stage (P<0.001; HR=1.512; 95% CI=1.284-1.781) were risk factors
associated with reduced OS (Table
II). These potential risk factors were included in a
multivariate Cox proportional hazards regression model, which
indicated that NT5DC2/1,000 (P=0.043; HR=1.695; 95%
CI=1.016-1.727) and TNM stage (P<0.001; HR=1.460; 95%
CI=1.235-1.727) were independent risk factors for reduced OS
(Table II).
Age, sex and TNM stage were also included in
univariate Cox proportional hazards regression models to evaluate
the impact of these factors on RFS. The results demonstrated that
NT5DC2/1,000 (P<0.001; HR=2.360; 95%
CI=1.547-3.599) and TNM stage (P<0.001; HR=1.515; 95%
CI=1.312-1.750) were risk factors for shorter RFS (Table III). These potential risk factors
were included in a multivariate Cox proportional hazards regression
model, which identified that the NT5DC2/1,000
(P=0.011; HR=1.780; 95% CI=1.143-2.772) and TNM stage
(P<0.001; HR=1.444; 95% CI=1.242-1.678) was independent risk
factors for reduced RFS (Table
III).
 | Table III.Cox Proportional Hazards regression
model for relapse-free survival in The Cancer Genome Atlas liver
cancer cohort (n=359). |
Table III.
Cox Proportional Hazards regression
model for relapse-free survival in The Cancer Genome Atlas liver
cancer cohort (n=359).
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years | 0.996
(0.984-1.008) | 0.481 |
|
|
| Male | 1.056
(0.761-1.466) | 0.744 |
|
|
|
NT5DC2/1000 | 2.360
(1.547-3.599) | <0.001 | 1.780
(1.143-2.772) | 0.011 |
| TNM stage | 1.515
(1.312-1.750) | <0.001 | 1.444
(1.242-1.678) | <0.001 |
| Edmondson
grade | 1.096
(0.892-1.348) | 0.382 |
|
|
In the PUMCH dataset, multiple factors, including
ALT, GGT and CA19-9, significantly differed between
NT5DC2-High and NT5DC2-Low groups (Table IV). These potential risk factors,
age, sex, NT5DC2-IHC MOD/10 units (NT5DC2/10), TNM
stage, AFP per 1,000 units (AFP/1,000) and HBsAg were individually
included in univariate Cox proportional hazards regression model.
Univariate Cox regression models suggested that NT5DC2/10
(P=0.002; HR=1.185; 95% CI=1.064-1.320), AFP/1,000 (P<0.001;
HR=1.017; 95% CI=1.007-1.026) and TNM stage (P<0.001; HR=1.703;
95% CI=1.338-2.168) were risk factors for poor OS (Table V). Potential risk factors were
included in a multivariate Cox proportional hazards regression
model, indicating that NT5DC2/10 (P<0.001; HR=1.219; 95%
CI=1.090-1.364) and TNM stage (P<0.001; HR=1.830; 95%
CI=1.409-2.377) were independent risk factors for poor OS (Table V).
 | Table IV.Baseline data of PUMCH dataset
(n=134). |
Table IV.
Baseline data of PUMCH dataset
(n=134).
| Characteristic |
NT5DC2-High |
NT5DC2-Low | P-value |
|---|
| Agea, years | 55.27±11.44 | 57.93±10.52 | 0.164 |
| Sex |
|
| 0.084 |
|
Male | 86.6 (58) | 73.1 (49) |
|
|
Female | 13.4 (9) | 26.9 (18) |
|
| HBsAg, %
positive | 79.1 (53) | 73.1 (49) | 0.544 |
| Anti-HCV, %
positive | 9.0 (6) | 9.0 (6) | 1.000 |
| AST, U/l | 37.0 (37.5) | 36.00 (25.5) | 0.140 |
| ALT, U/l | 39.0 (36.0) | 29.00 (29.2) | 0.010 |
| GGT, U/l | 69.0 (98.5) | 55.00 (59.5) | 0.015 |
| ALP, U/l | 76.0 (43.0) | 78.00 (49.5) | 0.478 |
| AFP, ng/ml | 176.5 (2019.0) | 30.00 (481.9) | 0.063 |
| CA19-9, U/ml | 24.2 (40.85) | 22.00 (23.65) | <0.001 |
| Edmondson
grade |
|
| 0.777 |
| G1 | 23.9 (17) | 26.8 (18) |
|
| G2 | 62.7 (42) | 61.2 (41) |
|
| G3 | 27.2 (6) | 7.5 (5) |
|
| G4 | 4.3 (1) | 0 (0) |
|
| TNM Stage |
|
| 0.732 |
| I | 59.7 (40) | 55.2 (37) |
|
| II | 13.4 (9) | 11.9 (8) |
|
|
III | 25.4 (17) | 28.4 (19) |
|
| IV | 1.5 (1) | 4.5 (3) |
|
 | Table V.Cox Proportional Hazards regression
model of overall survival in the Peking Union Medical College
Hospital cohort (n=134). |
Table V.
Cox Proportional Hazards regression
model of overall survival in the Peking Union Medical College
Hospital cohort (n=134).
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years | 0.995
(0.973-1.019) | 0.710 |
|
|
| Male | 1.221
(0.625-2.384) | 0.559 |
|
|
|
NT5DC2/10 | 1.185
(1.064-1.320) | 0.002 | 1.219
(1.090-1.364) | <0.001 |
| AFP/1000 | 1.017
(1.007-1.026) | <0.001 | 1.008
(0.998-1.018) | 0.136 |
| TNM stage | 1.703
(1.338-2.168) | <0.001 | 1.830
(1.409-2.377) | <0.001 |
| HBsAg | 1.245
(0.720-2.154) | 0.433 |
|
|
NT5DC2 may participate in
cancer-related biological processes in HCC
GSEA was performed in TCGA LIHC cohort. Gene
expression data of TCGA LIHC cohort were divided into two groups
based on normalized NT5DC2 gene expression:
NT5DC2-High and NT5DC2-Low. GSEA was performed based
on the phenotype of the group (Fig.
3B). In cancer-related phenotype, a liver cancer survival
related gene set, LEE_LIVER_CANCER_SURVIVAL_UP, and a
metastasis-related gene set, LIAO_METASTASIS, were enriched.
Moreover, a proliferation-related gene set was enriched,
CHIANG_LIVER_CANCER_SUBCLASS_PROLIFERATION_UP. It was suggested
that the bile acid may be downregulated, since the bile acid
metabolism-related gene set was also enriched,
HALLMARK_BILE_ACID_METABOLISM.
Mechanically, two Cyclin-Rb-E2F pathway related gene
sets, REN_BOUND_BY_E2F and LEE_LIVER_CANCER_MYC_E2F1_DN, were
significantly enriched. In addition,
SARRIO_EPITHELIAL_MESENCHYMAL_TRANSITION_UP, as an
epithelial-mesenchymal-transition (EMT)-related gene set, was
upregulated. Gene set YAMASHITA_LIVER_CANCER_STEM_CELL_DN, which
was related to liver cancer stem-like cells, was also enriched. The
plot of GSEA enrichment is presented in Fig. 3.
Discussion
Reliable biomarkers may aid in the correct clinical
decisions by surgeons when in dilemma over diagnosis (27). Most traditional markers of HCC are
suitable for early diagnosis. However, commonly used biomarkers,
such as AFP (28), carcinoembryonic
antigen (29) and CA19-9 (30), have shown poor performance in HCC
prognosis before surgery. The present study demonstrated that the
evaluation of NT5DC2 expression via IHC staining and
RNA-seq, but not RT-qPCR, was capable of distinguishing HCC tissues
from paired para-HCC tissues. In the current study, RT-qPCR
exhibited the lowest AUC, suggesting its unsuitability for clinical
use. RNA-seq of HCC and para-HCC tissues resulted in a higher AUC
compared with IHC staining. However, IHC staining is cheaper and
simpler compared with RNA-seq, and the analysis is easily
performed, especially in bioptic samples (31).
The present results suggested that NT5DC2 was
a potential predictor of OS, although in TCGA LIHC cohort, the
Rényi P-value was initially not significant. Notably, in this
cohort, OS referred to all-cause mortality. It was identified that,
in this cohort, the average age of enrolled patients at diagnosis
was ~60 years. Therefore, long-term follow-up could exceed the life
expectancy of enrolled patients. HCC has a comparatively low 5-year
survival rate (32); therefore, an
endpoint of follow-up was set up as the 5th year. Then, the Rényi
P-value became significant. In the multivariate Cox regression
model, NT5DC2 was an independent risk factor for reduced OS
and RFS, indicating that it was a potential prognostic factor for
HCC. This Cox regression model also demonstrated that the TNM stage
was associated with both OS and RFS, in accordance with previous
studies (33–35).
In the current study it was identified that whether
IHC or RNA-sequencing were used, NT5DC2 could be a
beneficial indicator of the prognosis of HCC. TNM stage and
NT5DC2 were found to be independent risk factors; hence,
NT5DC2 can increase the prognostic value of the TNM stage.
As a common method of pathological diagnosis, the accessibility of
IHC is not inferior to serum biomarkers such as AFP (27). However, the present results indicated
that AFP was a risk factor dependent on TNM stage. A previous study
has shown that the increase in serum AFP levels was related to the
tumor volume of HCC (36). Since TNM
stage already contains the tumor volume, the prognostic value of
AFP may be limited, especially when used simultaneously with the
TNM stage (37). Therefore,
NT5DC2 is more suitable as a supplement to TNM stage
compared with AFP.
In the present study, GSEA analysis in
NT5DC2-High and NT5DC2-Low groups, and it was found
that NT5DC2 upregulation was associated with metastasis and
survival, which was in accordance with the Kaplan-Meier analyses
results. While the detailed mechanism of how NT5DC2
upregulation affects OS and RFS in patients with HCC remains
elusive, the GSEA result may provide some suggestions. For example,
the enriched gene set included REN_BOUND_BY_E2F, a gene set of E2F
bound targets, which was a result of Anti-E2F chromatin
immunoprecipitation and sequencing in mammal cell-lines (38). Thus, NT5DC2 upregulation could
reflect Cyclin-retinoblastoma tumor suppressor (Rb)-E2F pathway
activation (39). Elevated E2F or
E2F target expression is associated with poor HCC prognosis
(40,41). In a previous study of HCC,
overexpression of NT5DC2 significantly promoted the change
of cell cycle from G1 to S phase and the expression of
cyclins was upregulated (11). The
Rb-E2F pathway is an important G1/S phase checkpoint,
and the G1/S ratio change that occurs during
NT5DC2 overexpression is likely due to the Cyclin-Rb-E2F
pathway, but this requires further verification (39).
An enrichment in EMT-related genes was also
identified in the current study, which was consistent with a
previous report (42). EMT is a
strong clinical indicator for HCC, and is crucial for HCC
metastasis (42). It has been shown
that transforming growth factor-β-induced EMT may also induce a
stemness phenotype in HCC cell-lines (43). In glioma stem-like cells, NT5DC2 was
revealed to be highly expressed and induce the upregulation of Fyn,
resulting in an aggressive phenotype (10). Moreover, in the current study, the
cancer stem cell-related dataset,
YAMASHITA_LIVER_CANCER_STEM_CELL_DN_96, was enriched in association
with NT5DC2 expression, as assessed using GSEA. However, the
molecular mechanism via which NT5DC2 influences HCC stemness
requires further investigation.
A strength of the current study was that the role of
NT5DC2 as a risk factor for OS was demonstrated in two
independent HCC cohorts. However, this study has some limitations.
First, in the PUMCH cohort, >70% of patients with HCC were HBV-
or HCV-positive. Thus, the results of IHC-stained TMA may not apply
to patients from Western countries, where HBV/HCV infection rates
are much lower (44). Second, this
study did not demonstrate NT5DC2 suitability as a serum
biomarker, possibly representing a limitation to its clinical use.
However, since previous IHC analysis of multiple biomarkers,
including human epidermal growth factor receptor 2F (45) and programmed death-ligand 1 (46), was highly consistent between surgical
resection samples and bioptic samples, IHC staining may still be a
suitable method for the analysis of bioptic samples.
Overall, the present study demonstrated the value of
NT5DC2 as an prognostic biomarker in multiple HCC cohorts. In the
multivariate Cox regression model, NT5DC2 upregulation was an
independent risk factor of poor OS in both cohorts and poor RFS in
LIHC cohort. GSEA indicated the enrichment of a series of survival-
and metastasis-related gene-sets, such as
LEE_LIVER_CANCER_SURVIVAL_UP and LIAO_METASTASIS.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81372578), Chinese Academy
of Medical Sciences Innovation Fund (grant no. 2017-I2M-4-003).
Availability of data and materials
The datasets used or analyzed during the present
study are available from the corresponding author upon reasonable
request. The bioinformatics datasets generated and/or analyzed
during the current study are available in The Cancer Genome Atlas
repository, https://www.cancer.gov/tcga.
Authors' contributions
JMC and XDH was responsible for research design and
provided study material. JMC provided the pathological sections and
performed the immunochemical scoring together with JZC. PHW
collected and assembled the clinical and follow-up data. JMC, JZC
and PHW analyzed and interpreted the data. JMC and XDH drafted and
finalized the manuscript. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
This project was approved by the Ethic Committee,
Peking Union Medical College Hospital (approval no. JS-1569) and
had been performed in accordance with Declaration of Helsinki and
its later amendments. All persons gave their written informed
consent prior to their inclusion in the study. Any details that may
disclose the identity of the subjects under study were not included
in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Global Burden of Disease Cancer
Collaboration, . Fitzmaurice C, Allen C, Barber RM, Barregard L,
Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al:
Global, regional, and national cancer incidence, mortality, years
of life lost, years lived with disability, and disability-adjusted
life-years for 32 cancer groups, 1990 to 2015: A systematic
analysis for the global burden of disease study. JAMA Oncol.
3:524–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
NCCN, . NCCN Clinical Practice Guidelines
in Oncology (NCCN Guidelines®): Hepatobiliary Cancers
(Version 3.2018). 2018.
|
|
4
|
Zhang JF, Shu ZJ, Xie CY, Li Q, Jin XH, Gu
W, Jiang FJ and Ling CQ: Prognosis of unresectable hepatocellular
carcinoma: Comparison of seven staging systems (TNM, Okuda, BCLC,
CLIP, CUPI, JIS, CIS) in a Chinese cohort. PLoS One. 9:e881822014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen ZH, Hong YF, Lin J, Li X, Wu DH, Wen
JY, Chen J, Ruan DY, Lin Q, Dong M, et al: Validation and ranking
of seven staging systems of hepatocellular carcinoma. Oncol Lett.
14:705–714. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu PH, Hsu CY, Hsia CY, Lee YH, Su CW,
Huang YH, Lee FY, Lin HC and Huo TI: Prognosis of hepatocellular
carcinoma: Assessment of eleven staging systems. J Hepatol.
64:601–608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sherman M: Staging for hepatocellular
carcinoma: Complex and confusing. Gastroenterology. 146:1599–1602.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ahmed Mohammed HF and Roberts LR: Should
AFP (or any biomarkers) be used for HCC surveillance? Curr Hepatol
Rep. 16:137–145. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou L, Rui JA, Wang SB, Chen SG and Qu Q:
Carbohydrate antigen 19-9 increases the predictive efficiency of
α-fetoprotein for prognosis of resected hepatocellular carcinoma.
Am Surg. 84:80–85. 2018.PubMed/NCBI
|
|
10
|
Guo S, Ran H, Xiao D, Huang H, Mi L, Wang
X, Chen L, Li D, Zhang S, Han Q, et al: NT5DC2 promotes
tumorigenicity of glioma stem-like cells by upregulating fyn.
Cancer Lett. 454:98–107. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li KS, Zhu XD, Liu HD, Zhang SZ, Li XL,
Xiao N, Liu XF, Xu B, Lei M, Zhang YY, et al: NT5DC2 promotes tumor
cell proliferation by stabilizing EGFR in hepatocellular carcinoma.
Cell Death Dis. 11:3352020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
van Hulzen KJE, Scholz CJ, Franke B, Ripke
S, Klein M, McQuillin A, Sonuga-Barke EJ; PGC ADHD Working Group, ;
Kelsoe JR, Landén M, et al: Genetic overlap between
attention-deficit/hyperactivity disorder and bipolar disorder:
evidence from genome-wide association study meta-analysis. Biol
Psychiatry. 82:634–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zayats T, Jacobsen KK, Kleppe R, Jacob CP,
Kittel-Schneider S, Ribasés M, Ramos-Quiroga JA, Richarte V, Casas
M, Mota NR, et al: Exome chip analyses in adult attention deficit
hyperactivity disorder. Transl Psychiatry. 6:e9232016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Prados J, Stenz L, Courtet P, Prada P,
Nicastro R, Adouan W, Guillaume S, Olié E, Aubry JM, Dayer A and
Perroud N: Borderline personality disorder and childhood
maltreatment: A genome-wide methylation analysis. Genes Brain
Behav. 14:177–188. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/Bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res. 44:e712016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Amin M, Edge S, Greene FL, Schilsky RL,
Byrd DR, Gaspar LE, Washington MK, Gershenwald JE, Compton CC and
Hess KR: AJCC cancer staging manual. 8th. Springer; New York, NY:
2017, View Article : Google Scholar
|
|
18
|
Zhou L, Rui JA, Zhou WX, Wang SB, Chen SG
and Qu Q: Edmondson-Steiner grade: A crucial predictor of
recurrence and survival in hepatocellular carcinoma without
microvascular invasio. Pathol Res Pract. 213:824–830. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang Y, Sun A, Zhao Y, Ying W, Sun H,
Yang X, Xing B, Sun W, Ren L, Hu B, et al: Proteomics identifies
new therapeutic targets of early-stage hepatocellular carcinoma.
Nature. 567:257–261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
R Core Team, . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: 2018
|
|
22
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liao Y, Smyth GK and Shi W: featureCounts:
An efficient general purpose program for assigning sequence reads
to genomic features. Bioinformatics. 30:923–930. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sia D, Villanueva A, Friedman SL and
Llovet JM: Liver cancer cell of origin, molecular class, and
effects on patient prognosis. Gastroenterology. 152:745–761. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang SL, Liu LP, Yang S, Liu L, Ren JW,
Fang X, Chen GG and Lai PB: Preoperative serum α-fetoprotein and
prognosis after hepatectomy for hepatocellular carcinoma. Br J
Surg. 103:716–724. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu J, Xia Y, Shi L, Li X, Wu L and Yan Z:
Elevated serum carcinoembryonic antigen is associated with a worse
survival outcome of patients after liver resection for
hepatocellular carcinoma: A propensity score matching analysis. J
Gastrointest Surg. 20:2063–2073. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hsu CC, Goyal A, Iuga A, Krishnamoorthy S,
Lee V, Verna EC, Wang S, Chen FN, Rodriguez R, Emond J, et al:
Elevated CA19-9 is associated with increased mortality in a
prospective cohort of hepatocellular carcinoma patients. Clin
Transl Gastroenterol. 6:e742015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Suryavanshi M, Mehta A, Jaipuria J, Kumar
D, Vishwakarma G, Panigrahi MK, Verma H, Saifi M, Sharma S, Tandon
S, et al: Clinical utility of RT-PCR in assessing HER 2 gene
expression versus traditional IHC and FISH in breast cancer
patients. Breast Cancer. 25:416–430. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zeng H, Zheng R, Guo Y, Zhang S, Zou X,
Wang N, Zhang L, Tang J, Chen J, Wei K, et al: Cancer survival in
China, 2003–2005: A population-based study. Int J Cancer.
136:1921–1930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tabrizian P, Jibara G, Shrager B, Schwartz
M and Roayaie S: Recurrence of hepatocellular cancer after
resection: Patterns, treatments, and prognosis. Ann Surg.
261:947–955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma X, Gu J, Wang K, Zhang X, Bai J, Zhang
J, Liu C, Qiu Q and Qu K: Identification of a molecular subtyping
system associated with the prognosis of Asian hepatocellular
carcinoma patients receiving liver resection. Sci Rep. 9:70732019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cao J, Wang P, Chen J and He X: PIGU
overexpression adds value to TNM staging in the prognostic
stratification of patients with hepatocellular carcinoma. Hum
Pathol. 83:90–99. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu C, Xiao GQ, Yan LN, Li B, Jiang L, Wen
TF, Wang WT, Xu MQ and Yang JY: Value of α-fetoprotein in
association with clinicopathological features of hepatocellular
carcinoma. World J Gastroenterol. 19:1811–1819. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kamarajah SK, Frankel TL, Sonnenday C, Cho
CS and Nathan H: Critical evaluation of the American joint
commission on cancer (AJCC) 8th edition staging system for patients
with hepatocellular carcinoma (HCC): A surveillance, epidemiology,
end results (SEER) analysis. J Surg Oncol. 117:644–650. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ren B, Cam H, Takahashi Y, Volkert T,
Terragni J, Young RA and Dynlacht BD: E2F integrates cell cycle
progression with DNA repair, replication, and G(2)/M checkpoints.
Genes Dev. 16:245–256. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kent LN and Leone G: The broken cycle: E2F
dysfunction in cancer. Nat Rev Cancer. 19:326–338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kent LN, Rakijas JB, Pandit SK, Westendorp
B, Chen HZ, Huntington JT, Tang X, Bae S, Srivastava A, Senapati S,
et al: E2f8 mediates tumor suppression in postnatal liver
development. J Clin Invest. 126:2955–2969. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kent LN, Bae S, Tsai SY, Tang X,
Srivastava A, Koivisto C, Martin CK, Ridolfi E, Miller GC, Zorko
SM, et al: Dosage-dependent copy number gains in E2f1 and E2f3
drive hepatocellular carcinoma. J Clin Invest. 127:830–842. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Giannelli G, Koudelkova P, Dituri F and
Mikulits W: Role of epithelial to mesenchymal transition in
hepatocellular carcinoma. J Hepatol. 65:798–808. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Malfettone A, Soukupova J, Bertran E,
Crosas-Molist E, Lastra R, Fernando J, Koudelkova P, Rani B, Fabra
Á, Serrano T, et al: Transforming growth factor-β-induced
plasticity causes a migratory stemness phenotype in hepatocellular
carcinoma. Cancer Lett. 392:39–50. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
de Martel C, Maucort-Boulch D, Plummer M
and Franceschi S: World-wide relative contribution of hepatitis B
and C viruses in hepatocellular carcinoma. Hepatology.
62:1190–1200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang T, Hsieh ET, Henry P, Hanna W,
Streutker CJ and Grin A: Matched biopsy and resection specimens of
gastric and gastroesophageal adenocarcinoma show high concordance
in HER2 status. Hum Pathol. 45:970–975. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang H, Agulnik J, Kasymjanova G, Wang A,
Jiménez P, Cohen V, Small D, Pepe C, Sakr L, Fiset PO, et al:
Cytology cell blocks are suitable for immunohistochemical testing
for PD-L1 in lung cancer. Ann Oncol. 29:1417–1422. 2018. View Article : Google Scholar : PubMed/NCBI
|