Introduction
Prostate cancer (PCa) is a common malignant tumor in
the male genitourinary system, and its incidence rate is the second
highest among all types of tumor in men, following only lung
cancer. There are approximately 1,276,106 new cases and 358,989
deaths worldwide annually (1). With
the aging population, changes in dietary structure and widespread
implementation of prostatic-specific antigen screening, the
incidence rate of PCa in China has increased from 35.2/100,000 in
1998 to 5,300/100,000 in 2012 (2,3). For
early or localized PCa, close observation, radiotherapy and radical
prostatectomy (open prostatectomy, laparoscopic or robotic-assisted
laparoscopy) are effective treatment options. For advanced PCa,
androgen deprivation therapy (ADT) is one of the most commonly used
treatments. However, the average effective duration of ADT
treatment is 12–18 months. The majority of patients develop
resistance, progress to castration-resistant PCa (CRPC), and
succumb due to PCa (4,5). Furthermore, the median survival time of
patients with metastatic CRPC (mCRPC) is <2 years (6). Consequently, there is an urgent need
for more effective alternative or supplemental treatment options.
The development of biotechnology provides more opportunities for
investigating novel potential targets for gene-targeted PCa
treatment.
The GRB-associated binding protein 2 (GAB2) scaffold
protein is a member of the Gab family and is encoded by the
GAB2 (11q14.1) gene (7).
GAB2 serves a role in a number of physiological processes,
such as cell proliferation, differentiation, apoptosis and
migration, by binding to multiple receptors and participating in
numerous signal transduction pathways, such as the PI3K/AKT,
SHP2/ERK and JAK/STAT pathways (8,9).
Previous studies have reported that the GAB2 protein is abnormally
expressed in numerous types of malignant tumor, including glioma,
leukemia and melanoma, as well as ovarian, breast and lung cancer
(10–13). Abnormal GAB2 signaling is associated
with tumor progression, indicating that GAB2 may be a novel
oncogene (14). In the present
study, the Oncomine and Gene Expression Omnibus (GEO) databases
were searched for PCa gene expression level analyses; the
expression level of the GAB2 gene was higher in PCa tissues
than in benign prostatic hyperplasia tissues. Thus, it was
hypothesized that the GAB2 gene may possess a regulatory
effect on the progress of PCa, but its specific function and
mechanism have not yet been elucidated.
In the present study, the GAB2 gene was
knocked down in PCa cells using small interfering (si)RNA, and the
differentially expressed genes in the GAB2 knockdown cells were
screened via gene chip technology. The aim of the present study was
to investigate the downstream gene expression levels and signaling
associated with GAB2 and to provide theoretical and experimental
evidence for novel therapeutic targets for PCa.
Materials and methods
Data download and analysis
The GSE55945 chip data set was downloaded from the
GEO database (ncbi.nlm.nih.gov/geo/). The chip data set is based on
the GPL570 platform. In total, 21 samples were obtained including
13 cases of PCa and 8 cases of benign prostatic hyperplasia. The
GEO2R online analysis website (ncbi.nlm.nih.gov/geo/geo2r/) was used to process
GSE55945 gene chips in the GEO database to compare differences in
the expression levels of GAB2. The Oncomine database
(oncomine.org/) was used to analyze GAB2
expression levels in prostate adenocarcinoma and acinar prostate
adenocarcinoma.
Gene chip
The gene chip used in the present study was the
GeneChip™ PrimeView™ Human Gene Expression Array (cat. no. 901838;
Affymetrix; Thermo Fisher Scientific, Inc.) and was provided by
Shanghai Genechem Co., Ltd. This single GeneChip array can yield
data for >530,000 probes and 36,000 transcripts and variants,
which represent more than 20,000 genes mapped via UniGene or RefSeq
annotation. Sequences for the array design were selected from the
UniGene database 219 https://www.ncbi.nlm.nih.gov/unigene (constructed 30
March 2009), RefSeq 36th edition https://www.ncbi.nlm.nih.gov/refseq/ (13 July 2009)
and full-length human mRNA from GenBank™ http://www.ncbi.nlm.nih.gov/genbank (downloaded 12 May
2009).
Cell culture and small interfering
(si)RNA knockdown experiments
PC-3 human PCa cells from the Cell Bank of Xi'an
Jiaotong University were cultured in medium containing 10% fetal
bovine serum (Hyclone Laboratories; GE Healthcare Life Sciences),
1% streptomycin mixture and ~90% RPMI-1640 complete medium (Gibco;
Thermo Fisher Scientific Inc.). The cells were cultured at a
constant temperature of 37°C with 95% O2, and 5%
CO2 in an incubator (relative humidity, ~95%). Adherent
cells were transfected with GAB2 siRNA as follows: The
commercial LV3-GAB2 lentivirus vector (containing green
fluorescent protein and the puromycin sequence) and GAB2
siRNA sequences were constructed by Shanghai GenePharma Co., Ltd.
The sequence of GAB2 siRNA was 5′-GGGACCTCCTGGTAGACAATA-3′,
and the sequence of scrambled fluorescein-labeled negative control
siRNA was: 5′-TTCTCCGAACGTGTCACGT-3′. When cells reached 40–50%
confluence in the presence of 8 µg/ml polypropylene, lentiviral
vectors were used to infect PC-3 cells at 50 multiplicity of
infection for 72 h. The stable GAB2 low expression level
subclones were maintained via 2–3 µg/ml puromycin-resistant
culturing (puromycin, Sigma-Aldrich; Merck KGaA). PC-3 human PCa
cells transfected with negative control (NC) siRNA were used as the
NC group, and cells transfected with GAB2 siRNA were used as
the knockdown (KD) group. Three samples in each group were
subjected to gene chip analysis.
Total RNA isolation and RT-qPCR
Total RNA was isolated via TRIzol extraction (cat.
no. 3101–100; Shanghai Pufei Biotechnology Co., Ltd.) according to
the manufacturer's instructions. First strand cDNA was synthesized
using M-MLV reverse transcriptase (cat. no. M1701; Promega
Corporation) and analyzed via qPCR using SYBR Ex Taq™ II (Takara
Bio, Inc.). Gene-specific primers were designed and synthesized by
Shanghai Genechem Co., Ltd. and their sequences were as follows:
GAB2 (forward primer: 5′-AGACCGCCAATCAGTGAAAAT-3′, reverse
primer: 5′-GGTGAAGTCGGCTGTTGTC-3′, 91bp) and GAPDH (forward primer:
5′-TGACTTCAACAGCGACACCCA-3′, reverse primer:
5′-CACCCTGTTGCTGTAGCCAAA-3′, 121bp). Real-time chain reactions were
performed using an iQ5TM Multicolor Real-Time PCR and Detection
System (Bio-Rad Laboratories, Inc.). Amplification conditions used
were 95°C for 5 min followed by 30 cycles of 94°C for 30 sec, 55°C
for 30 sec and 72°C for 35 sec, and finally 72°C for 3 min.
Relative quantitative analysis was performed according to the
following equation: F=2−∆∆Cq, where 2−ΔΔCq is
the expression level of the target gene for each KD group sample
(PC-3 human PCa cells with GAB2 gene knockdown) relative to
that of each NC group sample (PC-3 human PCa cells) (15).
Chip hybridization
Chip hybridization was performed using an Affymetrix
(Thermo Fisher Scientific, Inc.) expression profile chip and
GeneChip Hybridization Wash and Stain kit (cat. no. 900720;
Affymetrix; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Following hybridization at 45°C for 16
h at 20 × g in a GeneChip Hybridization Oven 645 (Affymetrix;
Thermo Fisher Scientific, Inc.), the hybridized chips were eluted
and stained using a GeneChip Fluidics Station 450 (Affymetrix;
Thermo Fisher Scientific, Inc.).
Chip scanning and detection
analysis
Chip hybridization results were scanned using a
GeneChip Scanner 3000 (cat. no. 00-0210; Affymetrix; Thermo Fisher
Scientific, Inc.), and the raw data were read using Command Console
software (version 4.0; Affymetrix; Thermo Fisher Scientific, Inc.).
The quality control data were analyzed using R software (version
3.6.0) (16). The package was
normalized, and the algorithm used was MAS 5.0 (Affymetrix; Thermo
Fisher Scientific, Inc.).
Chip data bioinformatics analysis
The association between GAB2 gene expression
levels and PCa occurrence was found by mining the gene literature
(17,18). The IPA online integration analysis
software was used to analyze differentially expressed genes in PCa
cells before and after GAB2 gene knockdown. IPA is an
all-in-one online integrated analysis software (www.ingenuity.com) which helps to understand data from
gene expression data, microRNA data and small-scale experimental
data. IPA establishes a visual experimental system to understand
the properties of various molecules in genes, proteins, chemicals,
drugs and their interaction networks.
Statistical analysis
Data are expressed as the mean ± SD of three
replicate samples. Except for the IPA, all statistical analyses
were performed using SPSS software (version 22.0; IBM Corp.).
Between-group differences were compared using an unpaired t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Significant differences in gene
expression levels in PCa cells following GAB2 knockdown
The Oncomine and GEO databases were searched for PCa
gene expression level data, which demonstrated that the expression
level of the GAB2 gene was higher in PCa tissues than in
benign prostatic hyperplasia tissues (Fig. 1A and B). These results indicate that
GAB2 may be an oncogene of PCa and possess regulatory
effects on tumor development. The GAB2 gene was knocked down
in PC-3 human PCa cells and gene chip technology was used to
compare gene expression levels. GAB2 gene knockdown
efficiency in the KD group was 60.7%, compared with GAB2
gene expression levels in the NC group (Fig. 1C). The scatter plot demonstrates the
distribution of signal values between the experimental and control
groups on the plane of the rectangular coordinate system. The
ordinate and abscissa values of each point represent the expression
level value of a probe in the experimental and control groups,
respectively. The upper part of the green line indicates KD is
downregulated relative to NC; the lower part of the green line
indicates KD is upregulated relative to NC (Fig. 1D). The red dots on the volcano map
indicate genes with fold difference >1 and significance level
<0.05 (Fig. 2A). The points to
the left of X=−1 and to the right of X=1 are genes with differences
>2-fold. The majority of gene expression level changes are
notably different and differ >2-fold (Fig. 2A). The heat map shows the expression
levels of genes in each sample (Fig.
2B). Green indicates a relatively decreased signal value; black
indicates a moderate signal value; gray indicates that the signal
value was not detected. In the tree structure, two adjacent samples
or genes have higher similarity. The expression profiles of the KD
and NC groups are notably different (Fig. 2B). The results demonstrated
significant differences in 1,242 genes; 665 genes in the KD group
were upregulated, and 577 were downregulated compared with the NC
group.
Classical pathway and upstream
regulation analysis
IPA online integration analysis software was used to
analyze the differential gene list and the enrichment of the gene
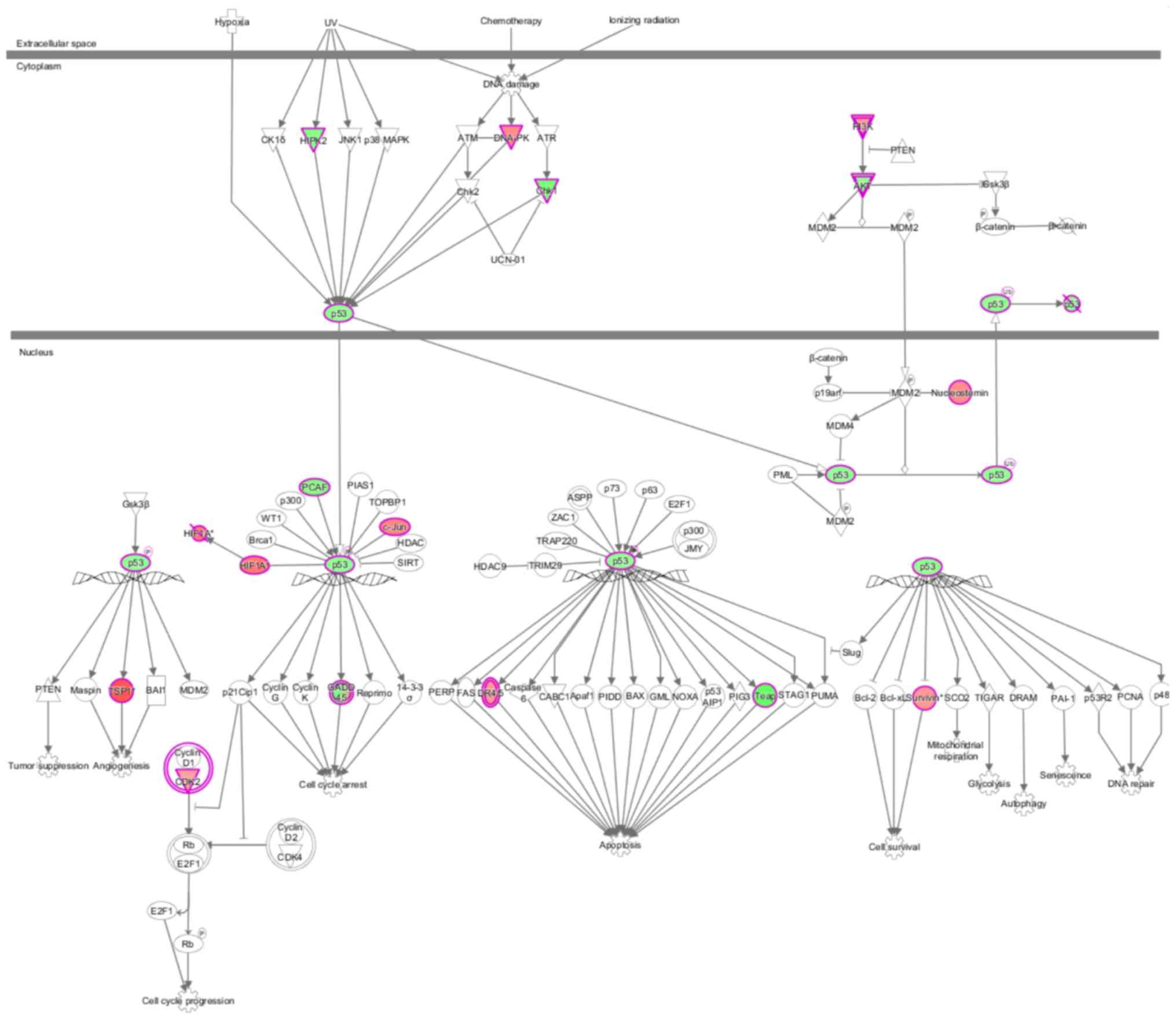
sets contained in each pathway. p53 signaling was inhibited after
knocking down GAB2 (Z-score=−0.535), and GAB2 was
involved in regulating the superpathway of cholesterol biosynthesis
(Fig. 3). The behavior of each
molecule in the p53 signaling pathway in the present study is shown
in Fig. 4. Green represents a
decrease; red represents an increase; color intensity is associated
with the change magnification. Magenta indicates molecules on the
differential gene list. Fig. 5 shows
the performance of each gene when the pathway is activated,
according to the analysis of IPA commercial software (www.ingenuity.com). In the present study,
DNA-dependent protein kinase (DNA-PK), checkpoint kinase 1 (Chk1),
AKT, thrombospondin 1 (TSP1) and death receptor (DR) 4/5 were
abnormally expressed compared with previously reported results,
indicating that these molecules may participate in tumorigenesis
via novel regulatory pathways (Figs.
4 and 5). The shape and color of
the molecule, and the interaction relationship between the
molecules are shown in Figures
S1-4. Next, activation or inhibition of upstream regulators,
including transcription factors, cytokines, small RNAs, receptors,
kinases, chemical molecules and drugs, was predicted for all
differential genes. Results of the upstream regulatory factor
analysis are presented in Table SI.
In the present study, insulin-induced gene 1 (INSIG1) was predicted
to be suppressed following GAB2 knockdown, and there were 18
genes that were consistently suppressed and are related to this
gene. The interaction between INSIG1 and its directly associated
downstream molecules is shown in Fig.
6. Only the expression level states of INSIG1 and C-X-C motif
chemokine ligand 6 (CXCL6) were inconsistent with previously
reported results. INSIG1 can inhibit CXCL6 at the mRNA level; in
the present study, however, CXCL6 was significantly downregulated
when INSIG1 was inhibited.
 | Figure 6.Upstream regulatory factor network
map. Orange line indicates consistent activated expression level
state between the upstream regulator and the gene; yellow line
indicates inconsistent expression level state; gray line indicates
there is no predictive information associated with the expression
level state in the data set. INSIG1, insulin induced gene 1; ACACA,
acetyl-CoA carboxylase α; ACLY, ATP citrate lyase; ACSS2, acyl-CoA
synthetase short chain family member 2; CXCL6, C-X-C motif
chemokine ligand 6; CYP51A1, cytochrome P450 family 51 subfamily A
member 1; DHCR24, 24-dehydrocholesterol reductase; DHCR7,
7-dehydrocholesterol reductase; ELOVL6, ELOVL fatty acid elongase
6; FADS1, fatty acid desaturase 1; FDFT1, farnesyl-diphosphate
farnesyltransferase 1; HMGCS1, 3-hydroxy-3-methylglutaryl-CoA
synthase 1; IDI1, isopentenyl-diphosphate Δ isomerase 1; LDLR, low
density lipoprotein receptor; LIPA, lipase A; LPCAT3,
lysophosphatidylcholine acyltransferase 3; LPIN1, lipin 1; PMVK,
phosphomevalonate kinase; SCARB1, scavenger receptor class B member
1; SQLE, squalene epoxidase; UCP2, uncoupling protein 2. |
Disease function and regulation effect
analysis
Up- and downregulation of differential gene
expression levels are associated with activation or inhibition of
functions and diseases. IPA software was used to analyze the
changes of 500 functions (Table
SII). Z-score >2 means that the function is significantly
activated, and Z-score <-2 means that the function is
significantly inhibited. A total of 24 functions changed
significantly, including ‘viral infection’ (Z-score=3.176) and
‘infection of tumor cell lines’ (Z-score=2.934). The functions with
significant inhibition were ‘morbidity or mortality’
(Z-score=−2.906) and ‘organismal death’ (Z-score=−2.879) (Fig. 7). The regulatory effect network map
shows the interaction between genes and regulators and functions in
the data set. In the present study, mouse double minute 2 homolog,
methyl-CpG-binding protein 2 and SAM pointed domain containing ETS
transcription factor possessed an activating effect on advanced
malignant tumor, melanoma cell line viability and muscle cell
proliferation, as well as an inhibitory effect on cancer cell death
via β-actin, survivin, dipeptidyl peptidase 4, hypoxia inducible
factor 1α, homeodomain-interacting protein kinase 2, immunoglobulin
heavy constant µ, interleukin-1 receptor-associated kinase 1,
lysine acetyltransferase 2B, prostate specific antigen, protein
tyrosine phosphatase receptor type F, syndecan 1, SMAD4,
thrombospondin 1, TP53, vascular endothelial growth factor A,
vimentin and other genes (Fig.
8).
Interaction network analysis
The IPA network generation algorithm was used to
split the network map between molecules into multiple networks and
to score each network. All networks were sorted by score value.
Fig. 9 shows the genetic interaction
network map: KCNMA1 (potassium calcium-activated channel subfamily
M α 1), cell division cycle 73, nardilysin convertase, insulin-like
growth factor 1 and dynein cytoplasmic 1 light intermediate chain 1
are the primary genetic interaction nodes. The interaction network
primarily affects ‘cellular assembly and organization’, ‘cellular
function and maintenance’ and ‘connective tissue disorders’.
Discussion
The progression of PCa is complex; abnormal
expression levels of a number of tumor-associated genes or the
inactivation of numerous tumor suppressor genes may lead to the
occurrence and progression of PCa (19). Therefore, by studying the genes and
genomic changes related to PCa, new ideas for clinical prevention
and treatment of PCa may be found. Analysis of the Oncomine and GEO
databases demonstrated that the expression level of the GAB2
gene was higher in PCa tissues than in benign prostatic hyperplasia
tissues, indicating that the GAB2 gene may possess a
regulatory effect on the progression of PCa. Gene chip technology
allows rapid, efficient and parallel detection of large-scale gene
expression levels, which facilitates improved understanding of the
mechanism of the GAB2 gene in the development and
progression of PCa (20). The
present study detected significant differences in the performance
of 1,242 genes between the KD and NC groups; 665 genes were
upregulated, and 577 were downregulated. These results indicated
that the GAB2 gene serves a regulatory role in PCa that is
mediated via a number of pathways acting on numerous genes.
After determining the differential gene expression
levels in the KD and NC groups, the differential gene results of
the chip analysis, including probe number and fold-change were
introduced into the IPA online integration analysis tool. The
software further analyzed the molecular pathways activated or
inhibited by GAB2 gene knockdown to predict upstream
regulatory molecules, analyze the association between disease and
function, and determine the network of gene interactions in order
to provide novel insights into the progression of PCa. Previous
findings have demonstrated that activation of the p53 signaling
pathway leads to cell cycle inhibition and apoptosis in PCa cells
(19,20). Singh et al (21), reported that targeting the p53
pathway can inhibit PCa cell proliferation and metastasis (22,23). The
present study demonstrated that GAB2 gene expression levels
are upregulated in PCa cells, but the association between
GAB2 and the p53 signaling pathway has not yet been
reported. In the present study, p53 signaling in PCa cells was
suppressed following GAB2 knockdown, but DNA-PK was
abnormally expressed in the p53 signaling pathway. By contrast,
Dylgjeri et al (24)
demonstrated that pharmacological targeting of DNA-PK inhibits
tumor growth both in vitro and in vivo. This
difference may be because the complex molecular networks within
tumor cells have not yet been elucidated. In addition to DNA-PK,
Chk1, AKT, TSP1, and DR4/5 were also abnormally expressed following
GAB2 knockdown; the involvement of these molecules in PCa
should be further investigated.
Yang et al (22) reported that INSIG1 serves a key role
in cholesterol homeostasis. Similarly, in the present classical
pathway analysis, GAB2 was demonstrated to be involved in
regulating the superpathway of cholesterol biosynthesis, and INSIG1
was predicted to be strongly suppressed (determined via upstream
regulatory analysis). Previous studies have shown that PCa risk
factors include chronic inflammation of the prostate (22,25,26) and
sexually transmitted infections (27–32). The
present study demonstrated that viral infection pathways are
significantly activated. These results indicated that PCa may be
caused by viral infections that increase the expression level of
GAB2. Regulatory effects and interaction network analyses
have indicated that numerous genes affect tumorigenesis and
progression; a number of pathways are involved, such as ‘advanced
malignant tumors’, ‘melanoma cell line viability’, ‘muscle cell
proliferation’, ‘cancer cell death’, ‘cellular assembly and
organization’, ‘cellular function and maintenance’ and ‘connective
tissue disorders’.
In the present study, the GAB2 gene was
knocked down in PCa cells. Gene chip technology and bioinformatics
analyses were then used to investigate the signaling pathways,
disease and function, and gene interaction networks associated with
GAB2. The aforementioned experimental and bioinformatic
analysis results demonstrated that GAB2 regulates pathways
such as the superpathway of cholesterol biosynthesis and p53
signaling in cells and serves a role in diseases and functions,
such as ‘non-melanoma solid tumors’, ‘viral infections’ and
‘morbidity or mortality’.
The present study has some limitations, such as the
lack of animal experiments and human studies. Therefore, it is not
clear whether the GAB2 gene has the same function in vivo
and whether it can be used as a therapeutic target for patients
with PCa. The purpose of the present study was to analyze and
summarize the changes of associated pathways following knockdown of
the GAB2 gene, and to provide a novel perspective for the
prevention and treatment of PCa by identifying potential research
directions and therapeutic targets.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
Natural Science Foundation of Shaanxi Province China (grant nos.
2020JM-370 and 2020JM-390).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YD and XQ conceived and designed the experiments.
YD, XQ, XZ, LM and JT performed the experiments. XQ analyzed the
data and wrote the manuscript. All authors have read and approved
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang B, Liao GQ, Wen XF, Chen WH, Cheng S,
Stolzenburg JU, Ganzer R and Neuhaus J: Nuclear magnetic resonance
spectroscopy as a new approach for improvement of early diagnosis
and risk stratification of prostate cancer. J Zhejiang Univ Sci B.
18:921–933. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ha Chung B, Horie S and Chiong E: The
incidence, mortality, and risk factors of prostate cancer in Asian
men. Prostate Int. 7:1–8. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Heidenreich A, Aus G, Bolla M, Joniau S,
Matveev VB, Schmid HP and Zattoni F; European Association of
Urology, : EAU guidelines on prostate cancer. Eur Urol. 53:68–80.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miyamoto H, Messing EM and Chang C:
Androgen deprivation therapy for prostate cancer: Current status
and future prospects. Prostate. 61:332–353. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kirby M, Hirst C and Crawford ED:
Characterising the castration-resistant prostate cancer population:
A systematic review. Int J Clin Pract. 65:1180–1192. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Souza AG, Bastos VAF, Silva IBB, Marangoni
K and Goulart VA: Different gene therapy strategies: A overview for
prostate cancer. Curr Gene Ther. 16:287–291. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vaughan TY, Verma S and Bunting KD:
Grb2-associated binding (Gab) proteins in hematopoietic and immune
cell biology. Am J Blood Res. 1:130–134. 2011.PubMed/NCBI
|
|
9
|
Nyga R, Pecquet C, Harir N, Gu H,
Dhennin-Duthille I, Régnier A, Gouilleux-Gruart V, Lassoued K and
Gouilleux F: Activated STAT5 proteins induce activation of the PI
3-kinase/Akt and Ras/MAPK pathways via the Gab2 scaffolding
adapter. Biochem J. 390:359–366. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brown LA, Kalloger SE, Miller MA, Shih
IeM, McKinney SE, Santos JL, Swenerton K, Spellman PT, Gray J,
Gilks CB and Huntsman DG: Amplification of 11q13 in ovarian
carcinoma. Genes Chromosomes Cancer. 47:481–489. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zatkova A, Schoch C, Speleman F, Poppe B,
Mannhalter C, Fonatsch C and Wimmer K: GAB2 is a novel target of
11q amplification in AML/MDS. Genes Chromosomes Cancer. 45:798–807.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sármay G, Angyal A, Kertész A, Maus M and
Medgyesi D: The multiple function of Grb2 associated binder (Gab)
adaptor/scaffolding protein in immune cell signaling. Immunol Lett.
104:76–82. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu Y and Rohrschneider LR: The gift of
Gab. FEBS Lett. 515:1–7. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ke Y, Wu D, Princen F, Nguyen T, Pang Y,
Lesperance J, Muller WJ, Oshima RG and Feng GS: Role of Gab2 in
mammary tumorigenesis and metastasis. Oncogene. 26:4951–4960. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
R Core Team, . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing (version 3.6.0). (Vienna, Austria). 2019.URL
https://www.R-project.org/.
|
|
17
|
Felciano RM, Bavari S, Richards DR,
Billaud JN, Warren T, Panchal R and Krämer A: Predictive systems
biology approach to broad-spectrum, host-directed drug target
discovery in infectious diseases. Pac Symp Biocomput. 17–28.
2013.PubMed/NCBI
|
|
18
|
Calvano SE, Xiao W, Richards DR, Felciano
RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK,
et al: A network-based analysis of systemic inflammation in humans.
Nature. 437:1032–1037. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Singal R, Ferdinand L, Reis IM and
Schlesselman JJ: Methylation of multiple genes in prostate cancer
and the relationship with clinicopathological features of disease.
Oncol Rep. 12:631–637. 2004.PubMed/NCBI
|
|
20
|
Xin-Hong G, Xiao-Cheng J and Liang-Bi C:
Gene chip technology and the studies of gene expression profiles. J
Biol. 2001.
|
|
21
|
Singh SK, Banerjee S, Acosta EP, Lillard
JW and Singh R: Resveratrol induces cell cycle arrest and apoptosis
with docetaxel in prostate cancer cells via a p53/p21WAF1/CIP1 and
p27KIP1 pathway. Oncotarget. 8:17216–17228. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang T, Espenshade PJ, Wright ME, Yabe D,
Gong Y, Aebersold R, Goldstein JL and Brown MS: Crucial step in
cholesterol homeostasis: Sterols promote binding of SCAP to
INSIG-1, a membrane protein that facilitates retention of SREBPs in
ER. Cell. 110:489–500. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dennis LK, Lynch CF and Torner JC:
Epidemiologic association between prostatitis and prostate cancer.
Urology. 60:78–83. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dylgjeri E, McNair C, Goodwin JF, Raymon
HK, McCue PA, Shafi AA, Leiby BE, de Leeuw R, Kothari V, McCann JJ,
et al: Pleiotropic impact of DNA-PK in cancer and implications for
therapeutic strategies. Clin Cancer Res. 25:5623–5637. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Daniels NA, Ewing SK, Zmuda JM, Wilt TJ
and Bauer DC; Osteoporotic Fractures in Men (MrOS) research group,
: Correlates and prevalence of prostatitis in a large
community-based cohort of older men. Urology. 66:964–970. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang J, Li J, Yunxia Z, Zhu H, Liu J and
Pumill C: The role of prostatitis in prostate cancer:
Meta-analysis. PLoS One. 8:e851792013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dillner J, Knekt P, Boman J, Lehtinen M,
Af Geijersstam V, Sapp M, Schiller J, Maatela J and Aromaa A:
Sero-epidemiological association between human-papillomavirus
infection and risk of prostate cancer. Int J Cancer. 75:564–567.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nelson WG, De Marzo AM and Isaacs WB:
Prostate cancer. N Engl J Med. 349:366–381. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Taylor ML, Mainous AG III and Wells BJ:
Prostate cancer and sexually transmitted diseases: A meta-analysis.
Fam Med. 37:506–512. 2005.PubMed/NCBI
|
|
30
|
Sutcliffe S, Giovannucci E, De Marzo AM,
Leitzmann MF, Willett WC and Platz EA: Gonorrhea, syphilis,
clinical prostatitis, and the risk of prostate cancer. Cancer
Epidemiol Biomarkers Prev. 15:2160–2166. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wagenlehner FM, Elkahwaji JE, Algaba F,
Bjerklund-Johansen T, Naber KG, Hartung R and Weidner W: The role
of inflammation and infection in the pathogenesis of prostate
carcinoma. BJU Int. 100:733–737. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng I, Witte JS, Jacobsen SJ, Haque R,
Quinn VP, Quesenberry CP, Caan BJ and Van Den Eeden SK:
Prostatitis, sexually transmitted diseases, and prostate cancer:
The California Men's health study. PLoS One. 5:e87362010.
View Article : Google Scholar : PubMed/NCBI
|