Introduction
With the third highest global incidence rate among
all malignancies (1), colorectal
cancer (CRC) continues to pose significant diagnostic, prognostic
and therapeutic challenges for clinicians. However, it can be
difficult to accurately predict metastasis in patients undergoing
curative surgery for localized CRC.
Currently, the dysregulation of functional long
non-coding RNAs (lncRNAs) and their involvement in human cancers
are receiving considerable attention (2–5); lncRNAs
are mRNA-like transcripts ranging from 200 nucleotides to ~100
kilobases in length that lack significant protein-coding ability
(6). Accumulating evidence suggests
that lncRNAs modulate various cellular functions, and regulate gene
expression in several ways, including epigenetic modification,
microRNA (miRNA/miR) sponging and mRNA stabilization (7). Long intergenic non-protein coding
RNA-467 (linc00467) is a recently identified lncRNA which has
rarely been investigated. The first study of linc00467 identified
its carcinogenic roles in neuroblastoma; following linc00467
knockdown, tumor cell proliferation was decreased while apoptosis
was enhanced, suggesting that linc00467 may act as a tumor
suppressor (8). However, the roles,
mechanisms, and potential targets of linc00467 in CRC have yet to
be fully elucidated (9). A previous
study revealed that miR-451a expression was significantly
downregulated in CRC tissues and cell lines, which was also
associated with a poor outcome in patients with CRC (10). It was therefore hypothesized that
miR-451a may be a potential target of linc00467.
To investigate its involvement in CRC, the
expression and potential role of linc00467 was assessed in CRC
tissues and cell lines. Bioinformatics analysis was conducted to
predict the target of linc00467, which was subsequently confirmed
in CRC samples and cell lines.
Materials and methods
Patients and tissue samples
Pairs of CRC and normal paracancerous tissues were
surgically resected from 31 patients with CRC in the Department of
Gastrointestinal Surgery, West China Hospital, Sichuan University
(Chengdu, China), between May 2010 and September 2011, and
immediately stored in liquid nitrogen. The paracancerous tissues
were ≥5 cm from the CRC tissues and contained no cancer cells, but
usually appeared inflamed and fibrotic. Patients were
pathologically diagnosed by two independent pathologists according
to the World Health Organization classification (11), and the tumors were staged according
to the Tumor-Node-Metastasis (TNM) Classification of Malignant
Tumors (12). None of the patients
underwent chemotherapy prior to surgical resection. The study
protocol was reviewed and approved by the Ethics Committee of
Sichuan University (approval no. K2016041), and written informed
consent was obtained from all patients.
Cell culture
A total of three CRC cell lines (HCT116, HT29 and
SW620) were obtained from the American Type Culture Collection. In
addition, a normal colonic epithelial cell line (NCM460) was
obtained from the Life Science Institute (http://www.sibs.cas.cn/sycs/) and used as a control.
HT29 cell line was characterized by Tsingke Biological Technology
(http://www.tsingke.net/shop/) using
short tandem repeat (STR) markers. The cells were cultured in
Dulbecco's modified Eagle's medium containing 10% foetal bovine
serum (Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100
µg/ml streptomycin, and maintained at 37°C in a humidified
incubator (5% CO2).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from patient tissues using
TRIzol® reagent (Takara Biotechnology Co., Ltd.),
according to the manufacturer's protocol. RNA concentration and
purity were determined using the Eppendorf BioPhotometer D30
(Eppendorf, Corporate), and RNA integrity was assessed with 1%
denaturing agarose gel electrophoresis. First-strand cDNA was
synthesized from 2 µg total RNA using 100 U Moloney murine
leukaemia virus reverse transcriptase (Invitrogen; Thermo Fisher
Scientific, Inc.), in a 10-µl reaction mixture that also contained
1 µl reverse transcription primers (10 mM each) and 10 U RNase
inhibitor. The reverse transcription conditions were as follows:
RNA samples and primers were mixed in a certain proportion on the
ice and reacted at 65°C for 5 min. The reverse transcription
primers were as follows: miR-451a forward,
5′-ACACTCCAGCTGGGAAACCGTTACCATTAC-3′ and reverse,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTTACAG-3′; U6 forward,
5′-CTCGCTTCGGCAGCAC-3′ and reverse, 5′-AACGCTTCACGAATTTGCG-3′;
linc00467 forward, 5′-GCGTAGGCCGGACATTTCTA-3′ and reverse,
5′-CCTGCCATGTTGGAAACTGC-3; and GAPDH forward,
5′-AGAAGGCTGGGGCTCATTTGC-3′ and reverse,
5′-ACAGTCTTCTGGGTGGCAGTG-3′. qPCR was conducted using the AceQ qPCR
SYBR Green Master Mix (Vazyme Biotech Co.) in a final volume of 20
µl, containing 10 µl master mix, 0.4 µl each of forward and reverse
primer (10 µM) and 2 µl cDNA. The thermocycling conditions were as
follows: An initial heating step at 95°C for 5 min, followed by 35
cycles of 10 sec at 95°C, and 60°C for 30 sec. miR-451a and
linc00467 expression were normalized to that of the internal U6 and
GAPDH controls, respectively. Each sample was run in triplicate and
the threshold cycle (Ct) numbers were averaged. Melting curve
analysis was performed using the CFX96 Touch™ qPCR System (Bio-Rad
Laboratories, Inc.) by increasing the temperature from 65°C to 95°C
in 0.1°C/sec increments for each fluorescence reading. Expression
level fold-changes were calculated using the relative
quantification (2−ΔΔCq) method (13).
Expression vector construction and
transfection
A hsa-linc00467-containing region was amplified from
human genomic DNA and inserted into pHBcas9n+puro (Hanbio
Biotechnology Co., Ltd., Shanghai, China); the linc00467 region was
obtained using Zhang Feng's CRISPR gRNA design tool (http://crispr.mit.edu/). The primer sequences were as
follows: Forward, 5′-CACCGGGTCTTCGAGAAGACCT-3′ and reverse,
5′-AAACAGGTCTTCTCGAAGACCC-3′. Position 113 and 577 of the
hsa-linc00467-containing regions were amplified from human genomic
DNA and inserted into pSicheck 2.0 (Sinasun Company, Beijing,
China; http://sinasun.biomart.cn/), and the
corresponding primer sequences were as follows: 113 Forward,
5′-CCGCTCGAGGCGCTGTGACGTTC-3′ and reverse,
5′-ATTTGCGGCCGCCCTGTTTGGTCCG-3′; and 577 forward,
5′-CCGCTCGAGAGAGGGACTGAAACTGGG-3′ and reverse,
5′-ATTTGCGGCCGCCTCCGCATCCTTCTTTGG-3′. A has-miR451a-containing
region was also amplified from human genomic DNA and inserted into
pCDNA-3.0 (Hanbio Biotechnology Co., Ltd.), and the primer
sequences were as follows: Forward,
5′-CGCGGATCCAGCCTGACAAGGACAGG-3′ and reverse,
5′-CCGCTCGAGCCCACCCCTGCCTTGTTTG-3′. Transfection was performed
using Lipofectamine® 3000 reagent according to the
manufacturer's protocol (Invitrogen; Thermo Fisher Scientific,
Inc.). Plasmid concentration and purity were determined using the
Eppendorf BioPhotometer D30 (Eppendorf, Corporate). In cellular
proliferation assay, each well was transfected with 100 ng plasmid
and subsequent experiments were performed 24 h post-transfection.
In flow cytometric detection of apoptosis, 2,500 ng plasmid was
added to each well and subsequent experiments were performed 48 h
post-transfection. In the dual-luciferase assay, 400 ng plasmid was
added to each well and subsequent experiments were performed 48 h
post-transfection.
Cellular proliferation assay
Cellular proliferation was assessed using a 3-(4,
5-dimethylthiazole-2-yl)-2, 5-diphenyltetrazolium bromide (MTT)
assay. Lentivirus-infected cells were seeded into 96-well plates at
a density of 2×103 cells per well, and cultured for 24,
48 or 72 h. Next, 20 µl MTT solution was added to each well and the
cells were incubated for a further 4 h until a purple precipitate
was visible. For each well, 150 µl dimethyl sulfoxide was added to
dissolve the purple precipitate and oscillated away from light for
5 min. The spectrophotometric absorbance of each well (at 490 nm)
was measured at different time points using a microplate reader
absorbance test plate (Molecular Devices). The experimental results
were calculated based on the following formula: Inhibition
ratio=(1-ApHBcas9n-linc
00467/ApHBcas9n-NC).
Flow cytometric detection of
apoptosis
Following transfection with pHBcas9n-linc00467 or
pHBcas9n-NC for 48 h, ~1×106 cells were collected for
apoptosis analysis. Annexin V-FITC solution (5 µl; Vazyme Biotech
Co., Ltd.) was added to each sample and gently mixed, and the cells
were incubated at room temperature for 10 min in the dark; 10 min
before analysis, 5 µl propidium iodide staining solution was added,
followed by 4005 µl binding buffer, and the cells were flow
cytometrically analyzed (DxFLEX; Beckman Coulter, Inc.). Data for
early and late apoptosis are all included.
Dual-luciferase assay
293T cells were seeded into a 24-well plate
(5×104 per well) and transfected with 200 ng
psiCheck2-NC(Sinasun Company, Beijing, China)+pCDNA-miR451a (Hanbio
Biotechnology Co., Ltd.), psiCheck2-113+pCDNA-miR451a or
psiCheck2-577+pCDNA-miR451a using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.). The 3′UTR region of
linc00467 was amplified and inserted into the plasmid with the
following primers: 113 Forward 5′-CCGCTCGAGGCGCTGTGACGTTC-3′ and
reverse, 5′-ATTTGCGGCCGCCCTGTTTGGTCCG-3′. 577 forward
5′-CCGCTCGAGAGAGGGACTGAAACTGGG-3′ and reverse,
5′-ATTTGCGGCCGCCTCCGCATCCTTCTTTGG-3′. After a 48-h incubation
period, cell lysates were collected to assess Firefly and
Renilla luciferase activities using the Dual-Luciferase
reporter system (Promega Corporation).
Bioinformatics analysis
The potential binding sites between linc00467 and
miR-451a were predicted by uploading their sequences to the online
software RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid?id=rnahybrid_view_submission).
A total of 10 potential binding sites were predicted, and the top 2
sites were selected for further investigation.
Statistical analysis
All data are expressed as the mean ± standard
deviation. Relative gene expression was analysed using the Livak
and Schmittgen method (13).
Differences between groups were determined using the Student's
t-test or analysis of variance, followed by Tukey's post hoc test.
The non-parametric Wilcoxon-Mann-Whitney or Kruskal-Wallis tests
were used to analyse the association between the linc00467
expression level and various clinicopathological characteristics.
P<0.05 was considered to indicate a statistically significant
difference.
Results
The expression of linc00467 is
upregulated and is associated with TNM stage and serum
carcinoembryonic antigen (CEA) level in CRC
Of the 31 patients included in the present study, 15
were males and 16 were females, and all were aged between 30 and 90
years. As shown in Table I and
Fig. 1, compared with paracancerous
tissues from the same patients, linc00467 was overexpressed in
>60% of the 31 CRC specimens, with an average increase of 1.75
times the expression level in paracancerous tissues. Furthermore,
linc00467 expression was significantly associated with metastasis
(P<0.05) and an advanced clinical stage of CRC (P<0.05). In
cases with lymph node metastasis, linc00467 was overexpressed in
92% (11/12) of patients. However, linc00467 was overexpressed in
only 32% (6/19) of patients without lymph node metastasis.
Furthermore, linc00467 was overexpressed in only 40% (8/20) of
patients with serum CEA ≤3.4 ng/ml, but in 82% (9/11) of patients
with a CEA level >3.4 ng/ml.
 | Table I.Association between linc00467
expression and pathological features in patients with CRC. |
Table I.
Association between linc00467
expression and pathological features in patients with CRC.
| Variable | N (%) | Median linc00467
expression | P-value |
|---|
| Age, years |
|
| 0.64 |
| ≤58 | 15 (48.39) | 2.11 |
|
|
>58 | 16 (51.61) | 3.08 |
|
| Sex |
|
| 0.40 |
| Male | 16 (51.61) | 3.10 |
|
|
Female | 15 (48.39) | 2.08 |
|
| Differentiation |
|
| 0.44 |
| Poor | 4 (12.90) | 4.62 |
|
|
Moderate | 27 (87.10) | 2.31 |
|
| CEA, ng/ml |
|
| <0.01a |
| ≤3.4 | 20 (64.52) | 1.62 |
|
|
>3.4 | 11 (35.48) | 4.41 |
|
| Lymphatic
metastasis |
|
| <0.01a |
| Yes | 12 (38.71) | 5.65 |
|
| No | 19 (61.29) | 0.69 |
|
| T-stage |
|
| 0.16 |
| T2 | 8 (25.81) | 0.79 |
|
| T3 | 6 (19.35) | 2.59 |
|
| T4 | 17 (54.84) | 3.32 |
|
| Clinical stage |
|
| <0.01a |
| I+II | 18 (58.06) | 0.44 |
|
|
III+IV | 13 (41.94) | 5.01 |
|
Knocking out linc00467 inhibits the
proliferation of CRC cells
As shown in Fig. 2A,
the relative expression levels of linc00467 in HCT116, HT29 and
SW620 cells were 3.26-, 5.94- and 2.24-fold, respectively, compared
with that in NCM460 cells. HCT116 and HT29 cells were transfected
with pHBcas9n-lin00467 or pHBcas9n-NC plasmids for 24 h using
Lipofectamine® 3000 reagent. As shown in Fig. 2B, linc00467-knockout decreased the
proliferation of HCT116 and HT29 cells by 90.87 and 71.84%,
respectively. The inhibitory effects on proliferation were evident
at 24 h, but most apparent at 72 h post-transfection (Fig. 2C). The PHBcas9n-lin00467 plasmid was
completely sequenced following construction; a peak in the
linc00467 insertion sequence fragment indicates that construction
of the target gene knockout plasmid was successful (Fig. 2D and E).
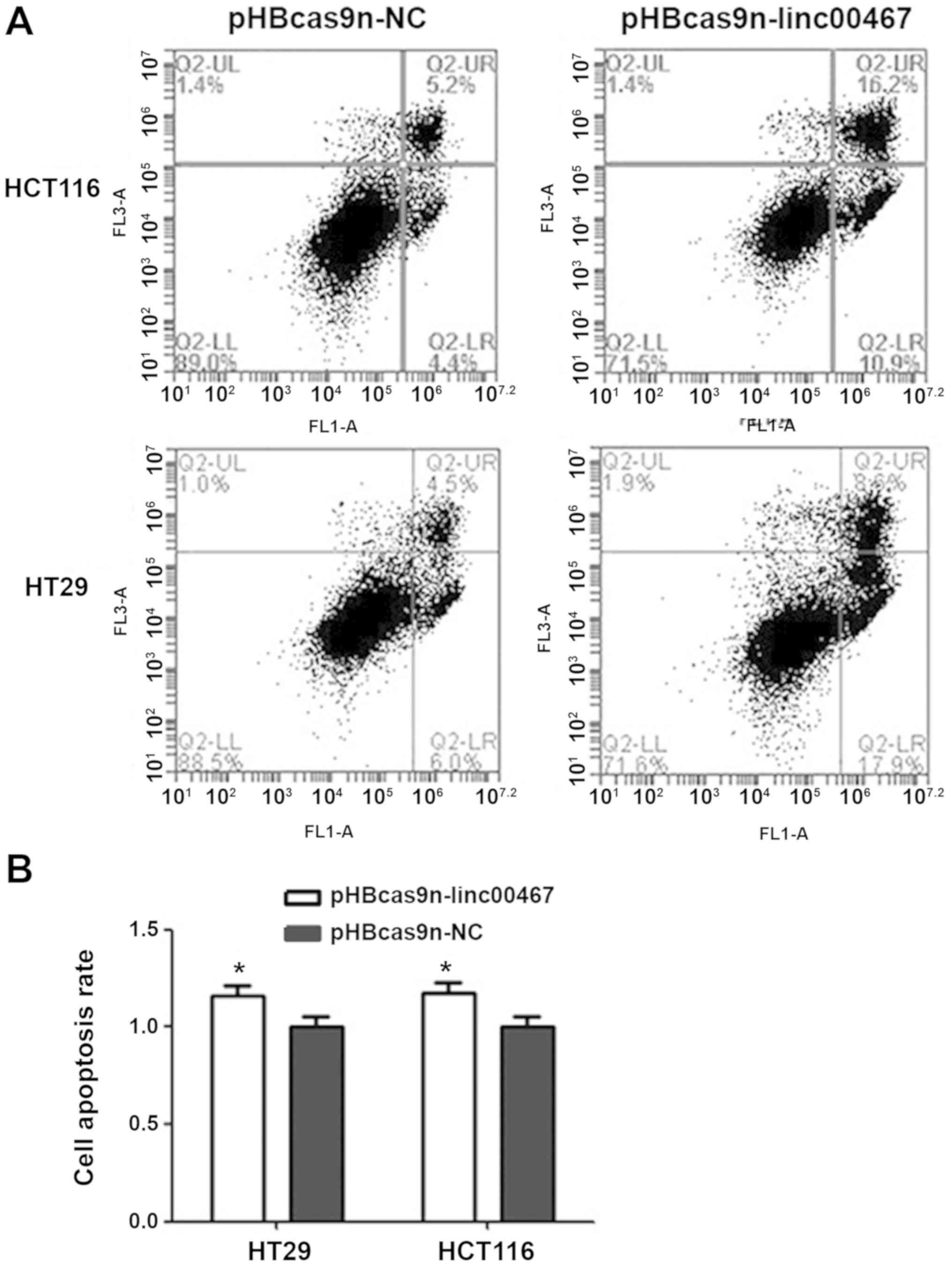
Knocking out linc00467 induces
apoptosis in CRC cells
Following linc00467-knockout, HCT116 cell apoptosis
was increased by 17.5% compared with the pHBcas9n-NC plasmid
control group (Fig. 3A and B). The
number of cells undergoing early apoptosis was increased by 6.5%,
while the number of those in late apoptosis increased by 11%. HT29
cell apoptosis was increased by 16% following linc00467-knockout.
The number of early and late apoptotic cells increased by 11.9 and
4.1%, respectively, compared with those in the pHBcas9n-NC
group.
linc00467 binds miR-451a as an
endogenous competitor
Bioinformatics analysis predicted that miR-451a was
a potential target of linc00467, with several possible binding
sites. Positions 113 and 577 were the most likely binding sites
since they generated the highest predictive scores (RNAhybrid
online software). Compared with the pSicheck2-NC+pCDNA-miR451a
group, the fluorescence intensity of the
psiCheck2-577+pCDNA-miR451a group was only decreased to 75.83%,
while that of the psiCheck2-113+pCDNA-miR451a group was decreased
to 31.62% (Fig. 4A; P<0.01).
HT29 and HCT116 cells were transfected with
pHBcas9n-lin00467 for 48 h, and miR-451a expression was quantified
by RT-qPCR. In HT29 cells, the expression level of miR-451a was
1.45-fold higher in the pHBcas9n-lin00467 group (P<0.05) than in
the pHBcas9n-NC group. In HCT116 cells, miR-451a expression was
1.88-fold higher in the pHBcas9n-lin00467 group compared with the
pHBcas9n-NC group (Fig. 4B).
HT29 and HCT116 cells were transfected with
pCDNA-miR451a for 48 h, and the expression level of linc00467 was
subsequently determined. In HT29 cells, linc00467 expression was
reduced by 68% in the pCDNA-miR451a group, compared with the
pCDNA-NC group. In HCT116 cells, linc00467 expression in the
pCDNA-miR451a group was decreased by 76% compared with the pCDNA-NC
group. These findings indicate that miR451a overexpression
decreases the level of linc00467 in CRC cells (Fig. 4C).
Discussion
The results of the present study suggest that
linc00467 may be a suitable predictor of CRC metastasis, as its
expression was revealed to be associated with lymph node metastasis
in patients with CRC. In addition, linc00467 expression in CRC was
revealed to be associated with the serum CEA level, and its
expression in CRC cell lines was significantly higher than that in
normal colorectal epithelial cells, which was consistent with the
results in human CRC specimens.
To the best of our knowledge, our previous study was
the first to illustrate that miR-451a expression was decreased in
CRC tissues, and that it may act as a tumor suppressor (10). It was also indicated that miR-451a
and linc00467 were expressed to differing degrees in CRC, which was
consistent with the characteristics of the competing endogenous
(ceRNA) theory (12,14). The ceRNA theory hypothesizes that the
combination of miRNA and mRNA can inactivate a target gene and
decrease the expression of miRNA (15,16). The
results of the present study confirm that linc00467 overexpression
in CRC leads to the downregulation and antagonism of miR-451a. As a
tumor-suppressor, the function of miR-451a was abolished by the
overexpression of linc00467.
The potential targets of miR-451a and linc00467 were
evaluated with RNAhybrid using the miRNA response element (MRE) of
linc00467 (17,18). Following bioinformatics prediction,
two binding sites with the highest scores among the potential MREs
were selected, namely positions 113 and 577. A luciferase assay
directly demonstrated that linc00467 possesses a binding site for
miR451a, which is a target gene directly regulated via linc00467
locus 113.
The present findings are also supported by other
studies. For example, in CRC, H19 can promote
epithelial-mesenchymal transition by functioning as an miR-138 and
miR200a sponge (19). Furthermore,
lncRNA-ROR promotes proliferation, invasiveness and metastasis by
associating with miR-145 (20). The
current results suggest that linc00467 is upregulated in CRC
tissues and as an endogenous competitor, downregulates miR-451a
expression, which subsequently regulates a variety of downstream
oncogenes to promote CRC metastasis.
In conclusion, the present study demonstrates that
linc00467 is upregulated in CRC, and that knocking out linc00467
can inhibit the proliferation of CRC cells and induce apoptosis.
Additionally, as a novel tumor suppressor in CRC, miR-451a was
found to be downregulated and associated with TNM stage and distant
metastasis in CRC. As an endogenous competitive RNA molecule,
linc00467 binds to miR-451a and subsequently regulates the
expression of tumor suppressive factors in CRC.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
YB and HW designed and conducted the study and wrote
the manuscript. BH, KX and YQL conducted the study and helped to
write the manuscript. YL and SM helped to analyze the data. YZ and
LZ conceived and designed the study. All authors reviewed and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The study protocol was reviewed and approved by the
Ethics Committee of Sichuan University (no. K2016041).
Patient consent for publication
Written informed consent was obtained from patients
prior to sampling.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin C and Yang L: Long noncoding RNA in
cancer: Wiring signaling circuitry. Trends Cell Biol. 28:287–301.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lipovich L, Johnson R and Lin CY: MacroRNA
underdogs in a microRNA world: Evolutionary, regulatory, and
biomedical significance of mammalian long non-protein-coding RNA.
Biochim Biophys Acta. 1799:597–615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Atmadibrata B, Liu PY, Sokolowski N, Zhang
L, Wong M, Tee AE, Marshall GM and Liu T: The novel long noncoding
RNA linc00467 promotes cell survival but is down-regulated by
N-Myc. PLoS One. 9:e881122014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He X, Li S, Yu B, Kuang G, Wu Y, Zhang M,
He Y, Ou C and Cao P: Up-regulation of LINC00467 promotes the
tumorigenesis in colorectal cancer. J Cancer. 10:6405–6413. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu K, Zhang YY, Han B, Bai Y, Xiong Y,
Song Y and Zhou LM: Suppression subtractive hybridization
identified differentially expressed genes in colorectal cancer:
microRNA-451a as a novel colorectal cancer-related gene. Tumor
Biol. 39:10104283177055042017. View Article : Google Scholar
|
|
11
|
Jernman J, Välimäki MJ, Louhimo J, Haglund
C and Arola J: The novel WHO 2010 classification for
gastrointestinal neuroendocrine tumours correlates well with the
metastatic potential of rectal neuroendocrine tumours.
Neuroendocrinology. 4:317–324. 2012. View Article : Google Scholar
|
|
12
|
Hu H, Krasinskas A and Willis J:
Perspectives on current tumor-node-metastasis (TNM) staging of
cancers of the colon and rectum. Semin Oncol. 4:500–510. 2011.
View Article : Google Scholar
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wapinski O and Chang HY: Corrigendum: Long
noncoding RNAs and human disease. Trends Cell Biol. 21:5612011.
View Article : Google Scholar
|
|
16
|
Crick FH, Barnett L, Brenner S and
Watts-Tobin RJ: General nature of the genetic code for proteins.
Nature. 192:1227–1232. 1961. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, et al: The lncRNA H19 promotes
epithelial to mesenchymal transition by functioning as miRNA
sponges in colorectal cancer. Oncotarget. 6:22513–22525. 2015.
View Article : Google Scholar : PubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBI
|
|
20
|
Zhou P and Sun L, Liu D, Liu C and Sun L:
Long non-coding RNA lincRNA-ROR promotes the progression of colon
cancer and holds prognostic value by associating with miR-145.
Pathol Oncolo Res. 22:733–740. 2016. View Article : Google Scholar
|