Introduction
In 1999, Jacquemin et al (1) first reported the identification and
characterization of human one cut domain family member 2 (ONECUT2),
also known as OC-2. The other two members of the ONECUT
transcription factor family are hepatocyte nuclear factor 6 and
ONECUT3 (1). The ONECUT2 gene is
located on chromosome 18 of the human genome, consisting of two
exons and one intron (Fig. 1)
(1). ONECUT2 protein is
characterized by one cut domain and an atypical homologous domain;
the protein binds to specific DNA sequences to stimulate the
expression of target genes (1).
ONECUT2 is mainly distributed in the gallbladder, duodenum, liver
and small intestines in human tissues, and has low expression
levels in the brain, pancreas, stomach and testes (1).
As a transcription factor, ONECUT2 can widely
regulate the protein expression associated with cell proliferation,
migration, adhesion and differentiation, thus being involved in the
regulation of the development of an organism (2–9).
Emerging role of ONECUT2 in tumors
The role of ONECUT2 in tumorigenesis has been
investigated. Table I summarizes the
expression levels of ONECUT2 in different types of tumor, the
effect of ONECUT2 on tumor development and the potential mechanisms
of its regulation. The levels of ONECUT2 protein are elevated in
prostate cancer (10,11), colorectal cancer (12,13),
hepatocellular carcinoma (14),
ovarian cancer (15) and lung cancer
(16). Increased ONECUT2 expression
is associated with regulation of tumor cell proliferation (17), metastasis (15) and differentiation (18). However, the levels of ONECUT2 are
decreased in breast cancer and this is associated with inducing
tumor stemness (19). Additionally,
increased levels of ONECUT2 gene methylation in urine may be a
promising predictor of bladder cancer (20–22).
 | Table I.Level and effect of ONECUT2 in
different tumors. |
Table I.
Level and effect of ONECUT2 in
different tumors.
| Cancer type | ONECUT2 level | Effect of
ONECUT2 | Upstream | Downstream
targets | Pathway | Inhibitor |
|---|
| Prostate
cancer | ↑ | Promotes
proliferation; regulates neuroendocrine differentiation | REST | AR, FOXA1,
PEG10 | SMAD3 pathway | CSRM617, TH302 |
| Colorectal
cancer | ↑ | Promotes
proliferation, EMT and lymph node metastasis | miR-429 |
|
|
|
| Hepatocellular
carcinoma | ↑ | Predicts poor
survival time |
|
|
|
|
| Ovarian cancer | ↑ | Promotes
proliferation and EMT |
| VEGF | AKT/ERK
pathway |
|
| Lung cancer | ↑ | Promotes tumor
stemness | miR-543 |
|
|
|
| Breast cancer | ↓ | Induces tumor
stemness | miR-9-5p,
miR-195-5p, miR-203a-3p |
| EV-miRNA ONECUT2
axis |
|
| Bladder cancer | ↑ (methylation
level) | Predicts and
diagnoses disease |
|
|
|
|
ONECUT2 drives prostate cancer progression
to castration-resistant prostate cancer (CRPC)
Prostate cancer (PCa) is the second most commonly
diagnosed cancer, with an incidence rate of 13.5%, and the fifth
most lethal cause of cancer-associated death, with a mortality rate
of 6.7% among men worldwide, as reported in 2018 (23). Studies have shown increased
expression of ONECUT2 in prostate cancer (10,11).
Immunocoprecipitation experiements show that ONECUT2 and androgen
receptor (AR) coexist, indicating that they may interact with each
other (24). Dihydrotestosterone
stimulation can increase the binding of ONECUT2 and AR to
rs7463708, a single nucleotide polymorphism associated with
prostate cancer risk (25).
rs7463708 then interacts with the Prostate Cancer Associated
Transcript 1 (PCAT1) promoter and upregulates the expression of
PCAT1, which is an androgen late-response gene (24). PCAT1 combines with AR and
lysine-specific demethylase 1 in an androgen-dependent manner
(24). Through interaction with
chromatin, PCAT1 upregulates the expression of GNMT and DHCR24,
thus promoting tumor cell proliferation and tumor growth (24). This process may be a way to promote
the occurrence of prostate cancer (Fig.
2).
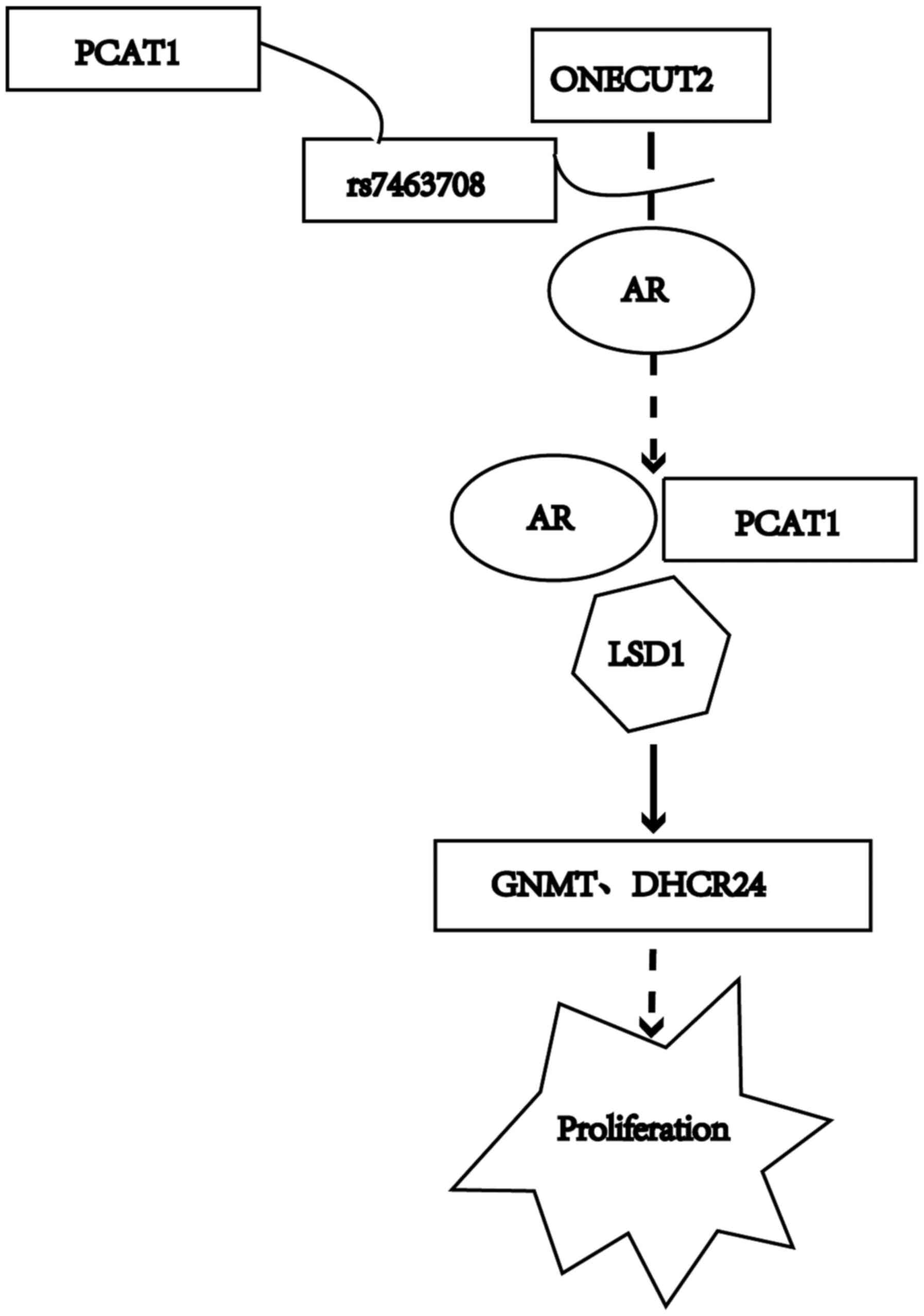 | Figure 2.Pathways of ONECUT2 promoting
prostate cancer cell proliferation. AR, androgen receptor. ONECUT2
binds with AR to rs7463708, which then interacts with PCAT1
promoter and upregulates PCAT1 expression; PCAT1 combines with AR
and LSD1, and then PCAT1 upregulates GNMT and DHCR24 expression,
thus promoting tumor cell proliferation. Different shapes indicate
different proteins or molecules. Continuous and curved lines
indicate direct interaction, while dashed lines indicate indirect
interaction. ONECUT2, One cut domain family member 2; GNMT, glycine
N-methyltransferase; PCAT1, Prostate Cancer Associated Transcript
1; AR, androgen receptor; LSD1, lysine-specific demethylase 1. |
Androgen deprivation therapy is the core therapy for
advanced and metastatic prostate cancer, but through a variety of
mechanisms, it eventually develops into resistance to androgen
therapy, namely CRPC (26,27). Nearly one-fifth of metastatic CRPC
(mCRPC) cases exhibit small cell neuroendocrine (NE) pathological
characteristics after treatment with effective AR pathway
inhibitors (28). One of the
mechanisms of resistance is the differentiation into NE prostate
cancer (NEPC). NEPC is a lethal invasive subtype of prostate
cancer, characterized by the loss of AR signal during NE
trans-differentiation (29).
The expression of the AR gene is negatively
correlated with the cell proliferation signal in mCRPC, and the
decrease in AR activity is consistent with disease progression
(30), which strongly implies that
an alternative transcription pathway is in action. Rotinen et
al (18) hypothesized that
ONECUT2 is a targetable master regulator of CRPC. Computational
modeling was applied to 38 transcriptome datasets from 2,115 PC
cases, including 260 samples of mCRPC. The modeling predicted that
ONECUT2 activity is comparable to that of histone-lysine
N-methyltransferase EZH2 (EZH2), a known CRPC driver, and that
ONECUT2 forms a network with other key transcription factors,
including POU domain class 5 transcription factor 1, paired box
protein Pax-5, AR and EZH2. In mCRPC, the expression levels and
activity of ONECUT2 are negatively correlated with those of AR, and
chromatin immunoprecipitation sequencing revealed ONECUT2 binding
to the AR promoter; therefore, ONECUT2 may act as a direct
repressor of AR gene expression (18). After ONECUT2 expression is enhanced,
the expression of AR and the AR regulatory genes KLK3, KLK2 and EHF
is downregulated, and the expression of the prostate-specific
antigen (PSA) gene and the PSA enhancer activity of the classical
regulatory target of AR are also inhibited (18). ONECUT2 can activate the transcription
program of mCRPC, and endogenous ONECUT2 can combine with the
forkhead box A1 (FOXA1) promoter and gene to inhibit the expression
of FOXA1 mRNA and protein. FOXA1, an inhibitor of NE
differentiation, disappears with the development of mCRPC (31). These results indicate that ONECUT2
suppresses the AR axis to promote the emergence of NEPC.
RE1-silencing transcription factor (REST) is a
master repressor of neuronal differentiation (32) and REST loss is implicated in NE-CRPC
(29). Chromatin
immunoprecipitation-quantitative PCR demonstrates the direct
binding of REST at the ONECUT2 promoter, and there is an inverse
association between ONECUT2 and REST expression in NEPC, suggesting
that REST is a disease-related expression inhibitor of ONECUT2
(29). Paternally expressed gene 10
(PEG10) has been identified as a driving factor for the transition
from adenocarcinoma (ADC) to NEPC in the LTL331 model, a
patient-derived xenograft model of NEPC trans-differentiation
(33). During the transition,
ONECUT2 combines with the PEG10 promoter and increases the PEG10
mRNA level, suggesting that ONECUT2 is the direct upstream
activator of PEG10 transcription. It can be concluded that ONECUT2
is located between REST and PEG10 during the transformation of
prostate ADC into NEPC (18).
Using structure-based drug design, Rotinen et
al (18) constructed a
three-dimensional model of the ONECUT2-HOX domain and screened out
CXRM617 as a small molecular inhibitor of ONECUT2. In vitro,
CXRM617 can inhibit the proliferation and induce the apoptosis of
prostate cancer cell lines with high expression of ONECUT2. CXRM617
significantly reduces the tumor volume and weight, and reduces the
occurrence and growth of diffuse metastasis in vivo. CXRM617
can be used as the main candidate drug for the development of mCRPC
therapy (34).
Guo et al (24) proposed that ONECUT2 drives tumor
aggressiveness partially through synergizing with hypoxia.
Specifically, ONECUT2 activates SMAD3, which regulates the hypoxia
signal by regulating HIF1α chromatin binding, resulting in a higher
degree of hypoxia in NEPC than in prostate ADC. The use of TH-302,
a hypoxia-activated prodrug, can significantly reduce the growth of
NEPC.
Previous research notes that ONECUT2 is a novel
target for the treatment of CRPC (18,29)
(Fig. 3). Further preclinical
studies are required to determine the prostate cancer types most
likely to respond to ONECUT2 inhibition, as well as to understand
ONECUT2-driven castration resistance mechanisms, identify
predictive biomarkers, and understand the role of ONECUT2 in tumor
heterogeneity and tumor evolution.
 | Figure 3.Pathways of ONECUT2 driving
neuroendocrine differentiation. ONECUT2 suppresses the AR axis and
inhibits FOXA1 expression, an inhibitor of neuroendocrine
differentiation, to promote the emergence of NEPC. ONECUT2 is
located between REST and PEG10 during the transformation of
prostate adenocarcinoma into NEPC. ONECUT2 regulates hypoxia
signaling by activating SMAD3 to drive NEPC, and the use of TH-302,
a hypoxia-activated prodrug, can significantly reduce the growth of
NEPC. Different shapes indicate different proteins or molecules.
Continuous lines indicate direct interaction, while dashed lines
indicate indirect interaction. REST, RE1-silencing transcription
factor; AR, androgen receptor; ONECUT2, one cut domain family
member 2; PEG10, paternally expressed gene 10; NEPC, neuroendocrine
prostate cancer; FOXA1, forkhead box A1. |
ONECUT2 affects colorectal cancer (CRC)
progression by regulating epithelial-mesenchymal transition
(EMT)
CRC is one of the most common gastrointestinal
malignancies, with a 5-year survival rate of ~55%. Upregulated
ONECUT2 expression in CRC (12,13) is
associated with lymph node metastasis, and a low level of ONECUT2
is associated with a good prognosis (13), but the specific mechanism involved
has not been clarified.
EMT is one of the well-defined processes involved
during the invasion and distant metastasis of primary epithelial
tumors (35). MicroRNAs
(miRNAs/miRs) of the miR-200 family have been experimentally shown
to regulate EMT (12,36). miR-429, as a member of the miR-200
family, can inhibit the tumorigenicity of CRC cells in vivo;
this miRNA can reverse the change in EMT induced by TGF-β and
inhibit EMT inducible factors (such as ZEB1, ZEB2, SNAI and SLUG)
(12). Sun et al (12) confirmed that ONECUT2 is the target
gene of miR-429. High expression of ONECUT2 can reverse the
inhibition of EMT inducible factors and the inhibition of migration
and invasion mediated by miR-429. Ranković et al (13) also showed that downregulation of the
miR-200 family is observed at the invasive front rather than in the
central region of tumors in CRC, and that the expression of ONECUT2
was correlated with nodal metastases (13). These results suggest that the miR-200
family and their target gene ONECUT2 contribute to the transition
from adenoma to CRC and to the development of metastases.
Hui et al (37) constructed lncRNA-miRNA-mRNA
expression networks using the Gene Expression Omnibus database and
found that ONECUT2 associates with multiple miRNAs in the competing
endogenous RNA network, including has-miR139-5p in CRC.
Has-miR139-5p expression levels are significantly decreased within
colon tumors and are correlated with tumor grade. Has-miR-139-5p,
by directly targeting the BCL2 pathway, can reduce tumor metastasis
and drug sensitivity in CRC. Has-miR-139-5p interacts with ONECUT2
and the expression levels are negatively correlated, suggesting
that has-miR-139-5p and ONETCUT2 may have a similar regulatory
mechanism to that between has-miR-139-5p and BCL2.
ONECUT2 may be involved in the inhibitory
pathways of hepatocellular carcinoma (HCC)
The prognosis of HCC remains poor due to tumor
progression and a high tumor recurrence rate (38). miR-9 dysregulation is implicated in a
number of human malignancies. miR-9 inhibits ovarian cancer cell
growth (39), regulates gastric
cancer cell growth and exhibits tumor suppressive activity in human
gastric cancer pathogenesis (40),
promotes tumor metastasis in esophageal squamous cell carcinoma
(41) and increases cervical cancer
cell motility (42). A previous
in vitro study revealed that miR-9 restoration retards HCC
cell proliferation and migration (14). Zhang et al (14) confirmed that ONECUT2, IGF2BP1 and
ANXA2 are miR-9 targets, and that they are aberrantly upregulated
in HCC. The upregulation of ONECUT2, IGF2BP1 and IL-6 is
significantly correlated with poor post-surgery survival rate. It
is suggested that miR-9 acts as a tumor suppressor by inhibiting a
series of target genes, including those of the
miR-9/IGF2BP1/AKT/ERK axis.
ONECUT2 promotes ovarian cancer cell
proliferation and migration
Ovarian cancer is one of the deadliest of all the
gynecological malignancies. ONECUT2 expression is significantly
increased in malignant ovarian ADC cell lines and in most malignant
ovarian cancer tissues; however, it is significantly decreased in
less malignant cancer cell lines, suggesting that the abnormal
expression of ONECUT2 is closely associated with the degree of
deterioration.
Silencing ONECUT2 expression using siRNA was shown
to significantly decrease the tumor inhibition rate by ~73% in an
ovarian cancer xenograft mouse model (15). Silencing ONECUT2 could inhibit the
phosphorylation of AKT and ERK, implying that ONECUT2 may promote
the proliferation of ovarian cancer cells through the AKT/ERK
signaling pathway (17).
After silencing ONECUT2 expression, the migration
rate and invasion rate of cancer cells decreases significantly.
ONECUT2 silencing upregulates the E-cadherin level and
downregulates the N-cadherin level, and the change in EMT-related
markers is consistent with the characteristics of EMT. ONECUT2
silencing also significantly inhibits the expression of VEGF. These
results suggest that ONECUT2 may promote tumor migration and
invasion by activating EMT and promoting angiogenesis (15).
ONECUT2 induces stem cell properties after
chemotherapy in breast cancer
Cancer stem cells (CSCs) have been proposed as the
driving force of tumorigenesis and as the seeds of metastases
(43), and they tend to be resistant
to chemoradiotherapy and molecular targeted therapy. ONECUT2 may be
involved in inducing the stemness of tumor cells.
Chemotherapy induces breast cancer cells to secrete
a variety of miRNAs, including miR-9-5p, miR-195-5p and
miR-203a-3p. These miRNAs are wrapped in extracellular vesicles
(EVs), which enable cancer cells to communicate with each other and
with non-cancerous cells in tumor pathogenesis and in response to
therapies, resulting in resistance to treatment. The aforementioned
three miRNAs target ONECUT2 at the same time and downregulate its
expression, leading to induction of CSC traits and upregulation of
the expression of stem cell characteristic-related genes. The
results suggest a possible mechanism of cancer cells communicating
with each other and adapting themselves to survive in response to
cytotoxic therapy. Inhibition of these miRNAs or recovery of
ONECUT2 expression can eliminate the CSC stimulation of EVs by
cancer cells after chemotherapy. This suggests a potential
strategy, namely, blocking the EV-miRNA ONECUT2 axis to maximize
the anticancer effect of chemotherapy and reduce chemoresistance in
cancer management (19).
ONECUT2 promotes RAS-driven lung cancer and
promotes stem cell properties
Lung cancer encompasses a heterogenous group of
malignancies. ONECUT2 expression across various subtypes of lung
cancer was analyzed using the publicly available dataset GSE30219
(16). According to this study,
ONECUT2 is aberrantly activated in lung cancer, including small
cell lung cancer (SCLC), large cell NE lung cancer, lung carcinoid
tumor and lung ADC (16). The
upregulation of ONECUT2 is significantly associated with shorter
overall survival times in 60 patients with RAS-driven ADC and 115
patients with RAS-activated ADC (16). Overexpression of ONECUT2 can promote
the malignant growth, invasion and adhesion of A549 lung cancer
cells in vitro (16). A549 is
a human lung ADC cell line with a homozygous KRAS mutation. In
vivo, ONECUT2 overexpression can enhance xenograft
tumorigenesis and bone metastasis (16). Further integrative transcriptomics
and epigenomics analyses indicate that ONECUT2 promotes the
transdifferentiation of lung cancer cells by preferentially
targeting and regulating the activity of bivalent chromatin domains
through modulation of polycomb repressive complex 2 occupancy
(16). The specific mechanism behind
the oncogenic role of ONECUT2 in the RAS-driven progression and
metastasis of lung cancer should be further investigated, which may
provide potential therapies for RAS-driven lung cancer.
lncRNA plays an important role in the occurrence and
development of tumors due to its transcription, epigenetic
modification or chromosomal abnormalities (44). The lncRNA THUMPD3 antisense RNA1
(THUMPD3-AS1) has oncogene activity and is aberrantly upregulated
in non-SCLC (NSCLC) tissues, correlating with TNM stage and
recurrence. THUMPD3-AS1 promotes NSCLC proliferation and
self-renewal capacity by acting as an endogenous sponge of miR-543,
and ONECUT2 is a direct downstream target of miR-543, which
suggests that ONECUT2 can promote stem cell properties in NSCLC
(45).
ONECUT2 is a promising predictor of bladder
cancer
Hematuria is the most common finding in bladder
cancer (BCa), and it is estimated that approximately 3–15% of the
patients with hematuria have urinary tract cancer (with the most
common being BCa) (46). Cystoscopy
is considered the gold standard for the detection of BCa in
patients with asymptomatic hematuria (47), however it is expensive and invasive
in nature. Several studies have aimed to establish urine testing
methods to screen patients with asymptomatic hematuria for the
detection of bladder cancer. DNA hypermethylation is a common
epigenetic regulation in malignancies. Beukers et al
(20) stratified 167 patients by age
and analyzed the methylation of five genes, ONECUT2 included. The
results showed that ONECUT2 methylation is present in patients with
urothelial carcinoma and that the methylation rate is higher in
elderly individuals than in the young. van Kessel et al
(21) included 200 patients from
three European countries and extracted DNA from urine for
methylation analysis. Combining the methylation markers (OTX1,
ONECUT2 and TWIST1) and mutation markers (FGFR3, TERT and HRAS)
with age contributed to a validated accurate prediction model,
which was suggested to reduce the requirement for diagnostic
cystoscopy by 77%. Wu et al (22) collected and evaluated 192 urine
samples from patients with hematuria in China. A combination
methylation assay of four genes (HOXA9, PCDH17, POU4F2 and ONECUT2)
was developed and indicated that ~60% of cystoscopies are
unnecessary. ONECUT2 is a promising predictor of bladder cancer.
However, thus far, studies on the mechanism of ONECUT2 in the
development of bladder cancer are lacking, and further
investigation is needed.
Future prospects
Increasing evidence demonstrates that the expression
of ONECUT2 affects the development and progression of malignant
tumors, and that expression level may display great potential in
the diagnosis, prognosis and treatment of tumors. Research into the
regulatory mechanics of the ONECUT2 gene has enormous potential.
ONECUT2 has been identified as a driver of NEPC. However, further
validation in genetically engineered mouse models or in
pre-clinical PCa organoid models is warranted. Establishing which
subtype of prostate cancer responds to ONECUT2 inhibitors in
clinical treatment will be critical for the majority of CRPC
patients. CSCs play a potential key role in the progression,
metastasis and chemoresistance of tumors. ONECUT2 may be involved
in inducing the stemness of tumor cells. Regulation of ONECUT2 via
miRNA targeting and other mechanisms would reprogram the levels of
gene expression towards a pluripotent state. Future studies
deciphering the spatiotemporal pattern of ONECUT2 expression and
its regulation during tumor progression may provide novel insights
into improved cancer management. ONECUT2 represents a good
candidate as a reliable biomarker for diagnosis in bladder cancer.
However, at present, it can only be used in combination with other
biomarkers for bladder cancer diagnosis, and the clinical
application of ONECUT2 still requires further investigation and
optimization.
Conclusions
ONECUT2 is a transcription factor associated with
development, and recent studies have shown that the aberrant
expression of ONECUT2 in cancer is relevant with cancer
progression. ONECUT2 has gained attention from research into
prostate cancer and it interacts with AR to promote prostate cancer
cell proliferation. Meanwhile, in CRPC, ONECUT2 inhibits the AR
axis and drives NE trans-differentiation. This dual role of ONECUT2
could help explain how certain prostate cancer types evade hormone
therapy and become more aggressive, and indicates that ONECUT2 is a
new target for the treatment of CRPC. ONECUT2 also plays a role in
the progression of other tumors by regulating cell proliferation,
metastasis and EMT. CSCs are associated with tumor metastasis, and
ONECUT2 may be involved in inducing the characteristics of CSCs
during chemoradiotherapy resistance. Blocking the effect of ONECUT2
may provide a new method for the treatment of cancer and the
elimination of chemotherapy resistance.
Acknowledgements
Not applicable.
Funding
This study was supported by the Fundamental Research
Funds for the Central Universities (grant no. 22120180603) and the
Important Weak Subject Construction Project of Pudong Health and
Family Planning Commission of Shanghai (grant no.
PWZbr2017-06).
Availability of data and materials
Not applicable.
Authors' contributions
JY and DL searched the literature and wrote the
manuscript. HJ revised the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jacquemin P, Lannoy VJ, Rousseau GG and
Lemaigre FP: OC-2, a novel mammalian member of the ONECUT class of
homeodomain transcription factors whose function in liver partially
overlaps with that of hepatocyte nuclear factor-6. J Biol Chem.
274:2665–2671. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dusing MR, Maier EA, Aronow BJ and
Wiginton DA: Onecut-2 knockout mice fail to thrive during early
postnatal period and have altered patterns of gene expression in
small intestine. Physiol Genomics. 42:115–125. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maier EA, Dusing MR and Wiginton DA:
Temporal regulation of enhancer function in intestinal epithelium:
A role for Onecut factors. J Biol Chem. 281:32263–32271. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Margagliotti S, Clotman F, Pierreux CE,
Beaudry JB, Jacquemin P, Rousseau GG and Lemaigre FP: The Onecut
transcription factors HNF-6/OC-1 and OC-2 regulate early liver
expansion by controlling hepatoblast migration. Dev Biol.
311:579–589. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Clotman F, Jacquemin P, Plumb-Rudewiez N,
Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG
and Lemaigre FP: Control of liver cell fate decision by a gradient
of TGF beta signaling modulated by Onecut transcription factors.
Genes Dev. 19:1849–1854. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Laudadio I, Manfroid I, Achouri Y, Schmidt
D, Wilson MD, Cordi S, Thorrez L, Knoops L, Jacquemin P, Schuit F,
et al: A feedback loop between the liver-enriched transcription
factor network and miR-122 controls hepatocyte differentiation.
Gastroenterology. 142:119–129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Goetz JJ, Martin GM, Chowdhury R and
Trimarchi JM: Onecut1 and Onecut2 play critical roles in the
development of the mouse retina. PLoS One. 9:e1101942014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sapkota D, Chintala H, Wu F, Fliesler SJ,
Hu Z and Mu X: Onecut1 and Onecut2 redundantly regulate early
retinal cell fates during development. Proc Natl Acad Sci USA.
111:E4086–E4095. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van der Raadt J, van Gestel SHC, Nadif
Kasri N and Albers CA: ONECUT transcription factors induce neuronal
characteristics and remodel chromatin accessibility. Nucleic Acids
Res. 47:5587–5602. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamamoto F and Yamamoto M: Scanning copy
number and gene expression on the 18q21-qter chromosomal region by
the systematic multiplex PCR and reverse transcription-PCR methods.
Electrophoresis. 28:1882–1895. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Leyten GH, Hessels D, Smit FP, Jannink SA,
de Jong H, Melchers WJ, Cornel EB, de Reijke TM, Vergunst H, Kil P,
et al: Identification of a candidate gene panel for the early
diagnosis of prostate cancer. Clin Cancer Res. 21:3061–3070. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu
Z, Li X and Wu M: MiR-429 inhibits cells growth and invasion and
regulates EMT-related marker genes by targeting Onecut2 in
colorectal carcinoma. Mol Cell Biochem. 390:19–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ranković B, Zidar N, Zlajpah M and
Bostjancic E: Epithelial-mesenchymal transition-related MicroRNAs
and their target genes in colorectal cancerogenesis. J Clin Med.
8:16032019. View Article : Google Scholar
|
|
14
|
Zhang J, Cheng J, Zeng Z, Wang Y, Li X,
Xie Q, Jia J, Yan Y, Guo Z, Gao J, et al: Comprehensive profiling
of novel microRNA-9 targets and a tumor suppressor role of
microRNA-9 via targeting IGF2BP1 in hepatocellular carcinoma.
Oncotarget. 6:42040–42052. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu T, Wu B, Yu Y, Zhu W, Zhang S, Zhang Y,
Guo J and Deng N: Blockade of ONECUT2 expression in ovarian cancer
inhibited tumor cell proliferation, migration, invasion and
angiogenesis. Cancer Sci. 109:2221–2234. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma Q, Wu K, Li H, Li H, Zhu Y, Hu G, Hu L
and Kong X: ONECUT2 overexpression promotes RAS-driven lung
adenocarcinoma progression. Sci Rep. 9:200212019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abid MR, Guo S, Minami T, Spokes KC, Ueki
K, Skurk C, Walsh K and Aird WC: Vascular endothelial growth factor
activates PI3K/Akt/forkhead signaling in endothelial cells.
Arterioscler Thromb Vasc Biol. 24:294–300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rotinen M, You S, Yang J, Coetzee SG,
Reis-Sobreiro M, Huang WC, Huang F, Pan X, Yáñez A, Hazelett DJ, et
al: ONECUT2 is a targetable master regulator of lethal prostate
cancer that suppresses the androgen axis. Nat Med. 24:1887–1898.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shen M, Dong C, Ruan X, Yan W, Cao M,
Pizzo D, Wu X, Yang L, Liu L, Ren X and Wang SE:
Chemotherapy-Induced Extracellular Vesicle miRNAs promote breast
cancer stemness by targeting ONECUT2. Cancer Res. 79:3608–3621.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Beukers W, Hercegovac A, Vermeij M,
Kandimalla R, Blok AC, van der Aa MM, Zwarthoff EC and Zuiverloon
TC: Hypermethylation of the polycomb group target gene PCDH7 in
bladder tumors from patients of all ages. J Urol. 190:311–316.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
van Kessel KE, Beukers W, Lurkin I,
Ziel-van der Made A, van der Keur KA, Boormans JL, Dyrskjot L,
Marquez M, Ørntoft TF, Real FX, et al: Validation of a DNA
methylation-mutation urine assay to select patients with hematuria
for cystoscopy. J Urol. 197:590–595. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu Y, Jiang G, Zhang N, Liu S, Lin X,
Perschon C, Zheng SL, Ding Q, Wang X, Na R, et al: HOXA9, PCDH17,
POU4F2, and ONECUT2 as a urinary biomarker combination for the
detection of bladder cancer in chinese patients with hematuria. Eur
Urol Focus. 6:284–291. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo H, Ci X, Ahmed M, Hua JT, Soares F,
Lin D, Puca L, Vosoughi A, Xue H, Li E, et al: ONECUT2 is a driver
of neuroendocrine prostate cancer. Nat Commun. 10:2782019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chung S, Nakagawa H, Uemura M, Piao L,
Ashikawa K, Hosono N, Takata R, Akamatsu S, Kawaguchi T, Morizono
T, et al: Association of a novel long non-coding RNA in 8q24 with
prostate cancer susceptibility. Cancer Sci. 102:245–252. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pienta KJ and Bradley D: Mechanisms
underlying the development of androgen-independent prostate cancer.
Clin Cancer Res. 12:1665–1671. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Harris WP, Mostaghel EA, Nelson PS and
Montgomery B: Androgen deprivation therapy: Progress in
understanding mechanisms of resistance and optimizing androgen
depletion. Nat Clin Pract Urol. 6:76–85. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aggarwal R, Huang J, Alumkal JJ, Zhang L,
Feng FY, Thomas GV, Weinstein AS, Friedl V, Zhang C, Witte ON, et
al: Clinical and genomic characterization of treatment-emergent
small-cell neuroendocrine prostate cancer: A Multi-institutional
prospective study. J Clin Oncol. 36:2492–2503. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lapuk AV, Wu C, Wyatt AW, McPherson A,
McConeghy BJ, Brahmbhatt S, Mo F, Zoubeidi A, Anderson S, Bell RH,
et al: From sequence to molecular pathology, and a mechanism
driving the neuroendocrine phenotype in prostate cancer. J Pathol.
227:286–297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sharma NL, Massie CE, Ramos-Montoya A,
Zecchini V, Scott HE, Lamb AD, MacArthur S, Stark R, Warren AY,
Mills IG and Neal DE: The androgen receptor induces a distinct
transcriptional program in castration-resistant prostate cancer in
man. Cancer Cell. 23:35–47. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim J, Jin H, Zhao JC, Yang YA, Li Y, Yang
X, Dong X and Yu J: FOXA1 inhibits prostate cancer neuroendocrine
differentiation. Oncogene. 36:4072–4080. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ooi L and Wood IC: Chromatin crosstalk in
development and disease: Lessons from REST. Nat Rev Genet.
8:544–554. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Akamatsu S, Wyatt AW, Lin D, Lysakowski S,
Zhang F, Kim S, Tse C, Wang K, Mo F, Haegert A, et al: The
placental gene PEG10 promotes progression of neuroendocrine
prostate cancer. Cell Rep. 12:922–936. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cully M: Anticancer drugs: Cutting down on
prostate cancer metastases. Nat Rev Drug Discov. 18:172018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
O'Brien SJ, Carter JV, Burton JF, Oxford
BG, Schmidt MN, Hallion JC and Galandiuk S: The role of the miR-200
family in epithelial-mesenchymal transition in colorectal cancer: A
systematic review. Int J Cancer. 142:2501–2511. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hui Z, Zhanwei W, Xi Y, Jin L, Jing Z and
Shuwen H: Construction of ceRNA coexpression network and screening
of molecular targets in colorectal cancer. Dis Markers.
2020:28605822020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang JD and Roberts LR: Epidemiology and
management of hepatocellular carcinoma. Infect Dis Clin North Am.
24899–919. (viii)2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guo LM, Pu Y, Han Z, Liu T, Li YX, Liu M,
Li X and Tang H: MicroRNA-9 inhibits ovarian cancer cell growth
through regulation of NF-kappaB1. FEBS J. 276:5537–5546. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wan HY, Guo LM, Liu T, Liu M, Li X and
Tang H: Regulation of the transcription factor NF-kappaB1 by
microRNA-9 in human gastric adenocarcinoma. Mol Cancer. 9:162010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Song Y, Li J, Zhu Y, Dai Y, Zeng T, Liu L,
Li J, Wang H, Qin Y, Zeng M, et al: MicroRNA-9 promotes tumor
metastasis via repressing E-cadherin in esophageal squamous cell
carcinoma. Oncotarget. 5:11669–11680. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu W, Gao G, Hu X, Wang Y, Schwarz JK,
Chen JJ, Grigsby PW and Wang X: Activation of miR-9 by human
papillomavirus in cervical cancer. Oncotarget. 5:11620–11630. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Medema JP: Cancer stem cells: The
challenges ahead. Nat Cell Biol. 15:338–344. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xie Y, Dang W, Zhang S, Yue W, Yang L,
Zhai X, Yan Q and Lu J: The role of exosomal noncoding RNAs in
cancer. Mol Cancer. 18:372019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hu J, Chen Y, Li X, Miao H, Li R, Chen D
and Wen Z: THUMPD3-AS1 is correlated with non-small cell lung
cancer and regulates self-renewal through miR-543 And ONECUT2. Onco
Targets Ther. 12:9849–9860. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Grossfeld GD, Litwin MS, Wolf JS, Jr,
Hricak H, Shuler CL, Agerter DC and Carroll PR: Evaluation of
asymptomatic microscopic hematuria in adults: The American
Urological Association best practice policy-part II: Patient
evaluation, cytology, voided markers, imaging, cystoscopy,
nephrology evaluation, and follow-up. Urology. 57:604–610. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Babjuk M, Burger M, Comperat EM, Gontero
P, Mostafid AH, Palou J, van Rhijn BWG, Roupret M, Shariat SF,
Sylvester R, et al: European association of urology guidelines on
Non-muscle-invasive bladder cancer (TaT1 and Carcinoma in
situ)-2019 update. Eur Urol. 76:639–657. 2019. View Article : Google Scholar : PubMed/NCBI
|

















