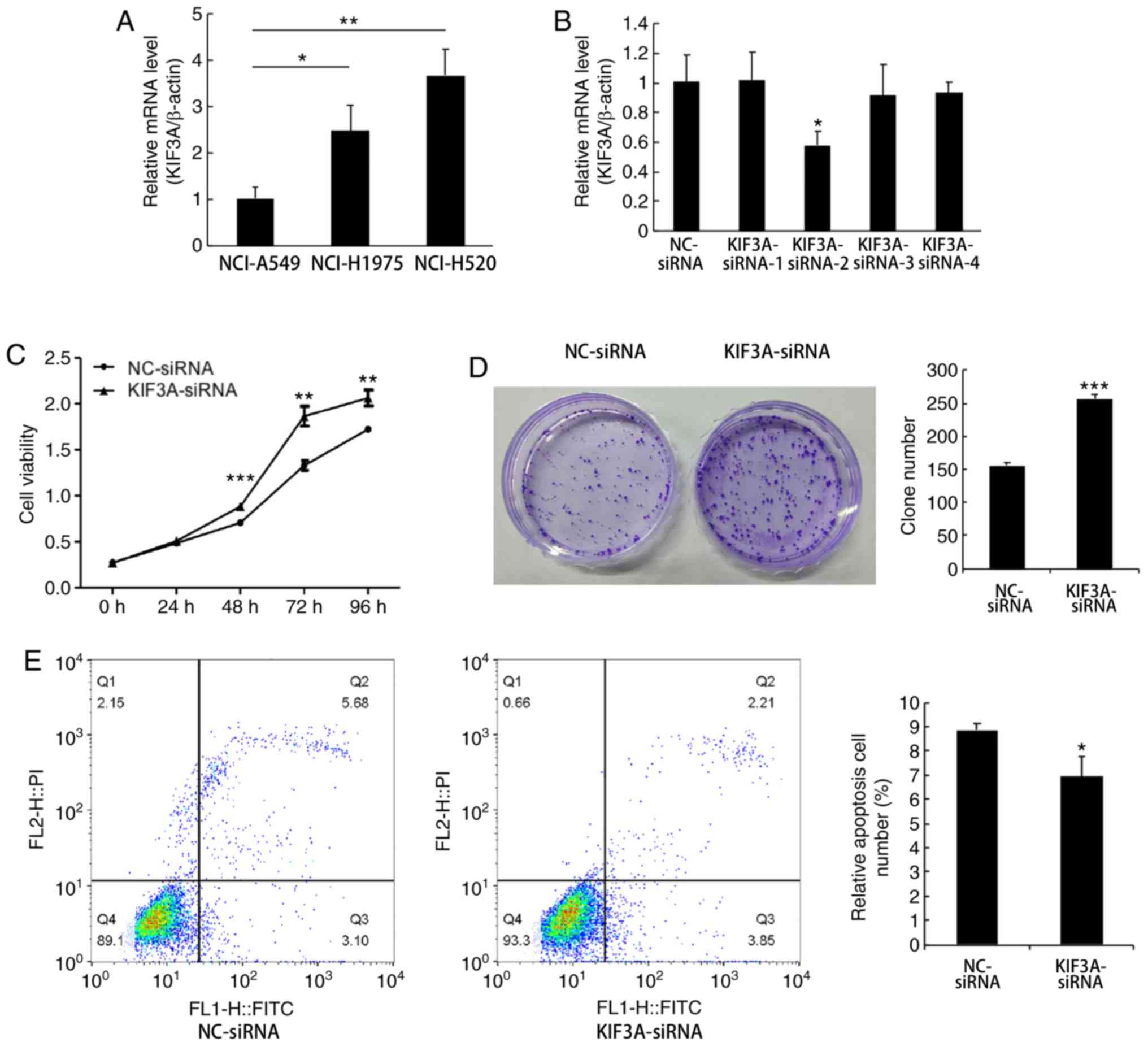
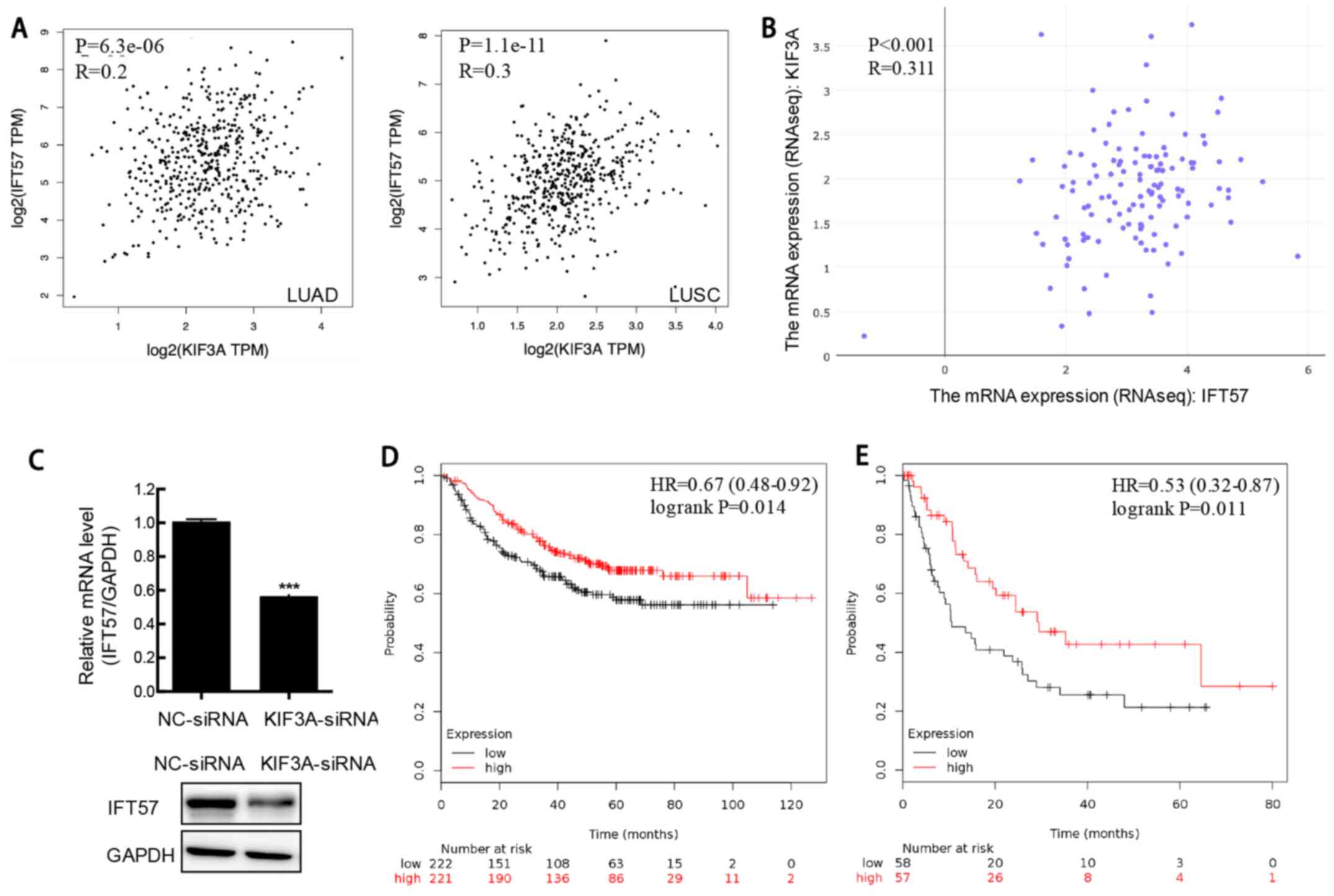
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gridelli C, Rossi A, Carbone DP, Guarize
J, Karachaliou N, Mok T, Petrella F, Spaggiari L and Rosell R:
Non-small-cell lung cancer. Nat Rev Dis Primer. 1:150092015.
View Article : Google Scholar
|
|
3
|
Boloker G, Wang C and Zhang J: Updated
statistics of lung and bronchus cancer in United States (2018). J
Thorac Dis. 10:1158–1161. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kolpakova-Hart E, Jinnin M, Hou B, Fukai N
and Olsen BR: Kinesin-2 controls development and patterning of the
vertebrate skeleton by Hedgehog- and Gli3-dependent mechanisms. Dev
Biol. 309:273–284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rosenbaum JL and Witman GB: Intraflagellar
transport. Nat Rev Mol Cell Biol. 3:813–825. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hirokawa N: Kinesin and dynein superfamily
proteins and the mechanism of organelle transport. Science.
279:519–526. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Barakat MT, Humke EW and Scott MP: Kif3a
is necessary for initiation and maintenance of medulloblastoma.
Carcinogenesis. 34:1382–1392. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Z, Rebowe RE, Wang Z, Li Y, Wang Z,
DePaolo JS, Guo J, Qian C and Liu W: KIF3a promotes proliferation
and invasion via Wnt signaling in advanced prostate cancer. Mol
Cancer Res. 12:491–503. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim M, Suh YA, Oh JH, Lee BR, Kim J and
Jang SJ: KIF3A binds to β-arrestin for suppressing Wnt/β-catenin
signalling independently of primary cilia in lung cancer. Sci Rep.
6:327702016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Travis WD, Brambilla E, Burke AP, Marx A
and Nicholson AG: WHO Classification of Tumours of the Lung,
Pleura, Thymus and Heart. 4th edition. International Agency for
Research on Cancer; Lyon: 2015
|
|
11
|
Amin MB, Edge S, Greene F, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC Cancer Staging Manual. 8th edition.
Springer International Publishing; New York, NY: 2017, View Article : Google Scholar
|
|
12
|
Liu X, Li C, Yang Y, Liu X, Li R, Zhang M,
Yin Y and Qu Y: Synaptotagmin 7 in twist-related protein 1-mediated
epithelial-mesenchymal transition of non-small cell lung cancer.
EBioMedicine. 46:42–53. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gyorffy B, Surowiak P, Budczies J and
Lanczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Broglio KR and Berry DA: Detecting an
overall survival benefit that is derived from progression-free
survival. J Natl Cancer Inst. 101:1642–1649. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mizuno H, Kitada K, Nakai K and Sarai A:
PrognoScan: A new database for meta-analysis of the prognostic
value of genes. BMC Med Genomics. 2:182009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jensen LJ, Kuhn M, Stark M, Chaffron S,
Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et
al: STRING 8-a global view on proteins and their functional
interactions in 630 organisms. Nucleic Acids Res. 37:D412–D416.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ghandi M, Huang FW, Jane-Valbuena J,
Kryukov GV, Lo CC, McDonald ER III, Barretina J, Gelfand ET,
Bielski CM, Li H, et al: Next-generation characterization of the
cancer cell line encyclopedia. Nature. 569:503–508. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Raponi M, Zhang Y, Yu J, Chen G, Lee G,
Taylor JM, Macdonald J, Thomas D, Moskaluk C, Wang Y and Beer DG:
Gene expression signatures for predicting prognosis of squamous
cell and adenocarcinomas of the lung. Cancer Res. 66:7466–7472.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wilkerson MD, Yin X, Hoadley KA, Liu Y,
Hayward MC, Cabanski CR, Muldrew K, Miller CR, Randell SH, Socinski
MA, et al: Lung squamous cell carcinoma mRNA expression subtypes
are reproducible, clinically important, and correspond to normal
cell types. Clin Cancer Res. 16:4864–4875. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wouters MW, Siesling S, Jansen-Landheer
ML, Elferink MA, Belderbos J, Coebergh JW and Schramel FM:
Variation in treatment and outcome in patients with non-small cell
lung cancer by region, hospital type and volume in the Netherlands.
Eur J Surg Oncol. 36 (Suppl 1):S83–S92. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barua S, Fang P, Sharma A, Fujimoto J,
Wistuba I, Rao AUK and Lin SH: Spatial interaction of tumor cells
and regulatory T cells correlates with survival in non-small cell
lung cancer. Lung Cancer. 117:73–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Y, Deng L, Zhao X, Li B, Ren D, Yu L,
Pan H, Gong Q, Song L, Zhou X and Dai T: Tripartite
motif-containing 37 (TRIM37) promotes the aggressiveness of
non-small-cell lung cancer cells by activating the NF-KB pathway. J
Pathol. 246:366–378. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guo X, Macleod GT, Wellington A, Hu F,
Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL and
Zinsmaier KE: The GTPase dMiro is required for axonal transport of
mitochondria to Drosophila synapses. Neuron. 47:379–393.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Geng G, Du Y, Dai J, Tian D, Xia Y and Fu
Z: KIF3A knockdown sensitizes bronchial epithelia to apoptosis and
aggravates airway inflammation in asthma. Biomed Pharmacother.
97:1349–1355. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee J, Oh DH, Park KC, Choi JE, Kwon JB,
Lee J, Park K and Sul HJ: Increased primary cilia in idiopathic
pulmonary fibrosis. Mol Cells. 41:224–233. 2018.PubMed/NCBI
|
|
31
|
Taschner M and Lorentzen E: The
intraflagellar transport machinery. Cold Spring Harb Perspect Biol.
8:a0280922016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Prevo B, Scholey JM and Peterman EJG:
Intraflagellar transport: Mechanisms of motor action, cooperation,
and cargo delivery. FEBS J. 284:2905–2931. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nakayama K and Katoh Y: Ciliary protein
trafficking mediated by IFT and BBSome complexes with the aid of
kinesin-2 and dynein-2 motors. J Biochem. 163:155–164. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Funabashi T, Katoh Y, Okazaki M, Sugawa M
and Nakayama K: Interaction of heterotrimeric kinesin-II with
IFT-B-connecting tetramer is crucial for ciliogenesis. J Cell Biol.
217:2867–2876. 2018. View Article : Google Scholar : PubMed/NCBI
|