Introduction
Breast cancer is one of the most common malignancies
affecting women worldwide, whereby ~1 in every 8 women are
diagnosed with breast cancer in their lifetime (1,2).
Currently, treatment options for breast cancer include surgery,
chemotherapy, radiotherapy and hormone treatment (3). Advancements in the early detection and
effective management of breast cancer have resulted in a worldwide
decrease in mortalities (3),
particularly in western countries, including North America and
Europe (1–4). However, breast cancer-associated
mortalities continue to increase in developing countries and in
patients suffering from different treatment-resistant subtypes of
breast cancer (5). In addition,
early diagnosis is one of the important factors influencing the
long-term survival rate in most types of cancer (3); however, the pathogenesis of breast
cancer is complex and heterogenous (6). Computed tomography (CT), magnetic
resonance imaging (MRI) and histopathological tools are commonly
used for the clinical diagnosis of breast cancer; however, early
diagnosis remains relatively uncommon (7). Thus, identifying novel biomarkers and
targets for the diagnosis and treatment of breast cancer remains
urgent.
Circular RNAs (circRNAs) are a group of non-coding
RNAs, which were first discovered in 1976 in RNA viruses via
electron microscopy (8). Their
unique closed circular structures (lacking 5′-3′ untranslated
regions and a poly A tail) meant they were initially regarded as
functionless by-products of errors in splicing (9). However, several studies have
demonstrated that circRNAs influence a number of biological
functions, such as microRNA (miRNA) sponging, regulation of
transcription, immune regulation and serving as templates for
protein translation (8,10,11),
which may explain their effects on different types of diseases,
including cardiovascular diseases, neurological dystrophy and
breast, liver, colorectal and lung cancers (12–16).
In breast cancer, extensive dysregulation of several
CircRNAs, such as hsa_circ_0104824, CircPVT1 and circRNA
0001073 (17–19), has been reported, and their potential
diagnostic and therapeutic values have been demonstrated. For
example, 1,705 differentially expressed circRNAs have been
identified in breast cancer tissues, and hsa_circ_0001982 has been
confirmed to be significantly upregulated in both breast cancer
tissues and cell lines (20). In
addition, hsa_circ_0001982 knockdown suppresses breast cancer cell
proliferation and invasiveness, and induces apoptosis by targeting
miR-143, which provides a novel insight into the molecular
mechanism underlying breast cancer tumorigenesis (20). Notably, Yin et al (21) identified 41 aberrantly expressed
circRNAs in plasma specimens between five breast cancer samples and
tissues from paired healthy volunteers; however, only three
circRNAs were validated to be significantly dysregulated, and
hsa_circ_0001785 exhibited a better diagnostic value compared with
the other two candidates (hsa_circ_0108942 and hsa_circ_0068033),
and carcinoembryonic antigen (21).
Furthermore, hsa_circ_0001785 plasma level was significantly
associated with tumor-node-metastasis (TNM) staging, grading of the
breast tumor and extent of distant metastasis (21). Thus, the potential role of other
circRNAs as diagnostic and prognostic biomarkers, as well as
therapeutic targets in breast cancer, warrants further
research.
In the present study, the novel circRNA
hsa_circ_0000129, located at 151145974-151149507 in chromosome 1,
was selected using a Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111504)
microarray dataset (GSE111504) (22). The present study investigated the
expression level of hsa_circ_0000129 in breast cancer patients and
breast cancer cell lines, and the results demonstrated that its
levels were significantly enhanced in breast cancer tissues and
cells compared with their corresponding controls. The effects of
hsa_circ_0000129 on the proliferation, migration and colony
formation abilities of breast cancer cells and the expression of
enhancer of zeste homolog 2 (EZH2) were also studied, aiming to
reveal the role of hsa_circ_0000129 in breast cancer
progression.
Materials and methods
Collection of specimens
Breast cancer tissues and adjacent paracancerous
tissues (collected ≥5 cm from the tumor border) were collected from
68 patients with breast cancer at the Jing'an District Centre
Hospital of Shanghai (Shangai, China) between February 2016 and May
2018. The fresh biopsy tissues were preserved in RNA fixer reagent
(Bioteke Corporation) and stored at −80°C prior to subsequent
experimentation. Clinical and demographic data were collected from
all patients, and no patients had received chemotherapy or
radiotherapy prior to surgical resection. Tumor size was determined
using CT scan images and both MRI and CT scans were used to
diagnose the extent of lymph node metastasis. Tumor staging and
grading were assessed according to the cancer staging manual of
American Joint Committee on Cancer (7th edition) (23). The present study was approved by the
Human Research Ethics Committee of Huashan Hospital (Shanghai,
China; approval no. 2016MS02), and written informed consent was
provided by all patients prior to the study start.
Cell culture
The breast cancer cell lines (MCF-7, MDA-MB-231 and
MDA-MB-468) and normal epithelial breast cell line (MCF10A) were
purchased from the American Type Culture Collection. All cells were
maintained in DMEM supplemented with 10% fetal bovine serum (FBS)
and 1% penicillin-streptomycin (all purchased from Gibco; Thermo
Fisher Scientific, Inc.), at 37°C with 5% CO2.
Cells successfully transfected with the
overexpression vector or interference sequence of hsa_circ_0000129
were cultured in complete DMEM at 37°C.
Transient transfection
MCF-7 and MDA-MB-468 cells were seeded into 6-well
plates at a density of 4.0×105 cells/well. After 24 h,
MCF-7 cells were transfected with the overexpression vector for
hsa_circ_0000129 (Geneseed Biotech, Inc.) using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol, while
a control vector (Geneseed Biotech, Inc.) was used as the negative
control (NC). MDA-MB-468 cells were transfected with an
interference sequence for hsa_circ_0000129 or NC sequence (Geneseed
Biotech, Inc., http://www.geneseed.com.cn), using
Lipofectamine® 2000 reagent. All cells were transfected
for 24 h at 37°C and another 48 h later, transfection efficiency
was determined via reverse transcription-quantitative (RT-q)PCR
analysis.
RT-qPCR
Total RNA was extracted from breast cancer tissues
and adjacent paracancerous tissues using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Total RNA was reverse transcribed into
double-strand cDNA using the ReverTra Ace qPCR RT kit (cat. no.
FSQ-101; Toyobo Life Science), according to the manufacturer's
protocol. qPCR was subsequently performed using the SYBR Premix Ex
Taq™ II kit (cat. no. RR820A; Takara Biotechnology Co., Ltd.), 0.5
µl cDNA, 0.5 µl each primer and 5 µl SYBR Green. The following
primer sequences were designed by Sangon Biotech Co., Ltd., and
used for qPCR: GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and
reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′; hsa_circ_0000129 forward,
5′-AAGAGGGAAATCCCAGCAGA-3′ and reverse, 5′-GCATGAGGAGTCAATGCAGA-3′;
and EZH2 forward, 5′-AATCAGAGTACATGCGACTGAGA-3′ and reverse,
5′-GCTGTATCCTTCGCTGTTTCC-3′. The following thermocycling conditions
were used for qPCR: 94°C for 5 min, 42 cycles of 94°C for 5 sec and
60°C for 1 min. Relative expression levels were calculated using
the 2−ΔΔCq method (24)
and normalized to the internal reference gene GAPDH.
Cell proliferation assay
MCF-7 and MDA-MB-468 cells transfected with NC or
hsa_circ_0000129 sequences were seeded into 96-well plates at a
density of 3.5×103 cells/well and incubated at 37°C for
1–7 days. Subsequently, cells were incubated with 5 mg/ml MTT
reagent (Sigma-Aldrich; Merck KGaA) for 3 h at 37°C. Following the
MTT incubation, the supernatant was removed and the purple formazan
crystals were dissolved using dimethyl sulfoxide (Sigma-Aldrich;
Merck KGaA). Cell proliferation was subsequently analyzed at a
wavelength of 492 nm, between 0–7 days. Cell proliferation was
assessed using the following formula: Proliferation rate = A
(sample)/A (control), where A is the absorbance measured at 492 nm
and control is the sample which was analyzed on day 0.
Colony formation assay
MCF-7 and MDA-MB-468 cells transfected with NC or
hsa_circ_0000129 sequences were seeded into 6-well plates at a
density of 6.0×102 cells/well and incubated for 14 days
at 37°C. The cell media were replaced every 48 h. The plates were
subsequently washed three times with PBS and the cell colonies were
fixed with 100% methanol at room temperature for 15 min, prior to
staining with 1% crystal violet for 30 min at room temperature.
Stained colonies (with >50 cells) were observed under a light
microscope (magnification, ×200).
Migration assay
The migratory ability MCF-7 and MDA-MB-468 cells
transfected with NC or hsa_circ_0000129 sequences was assessed
using 24-well Transwell inserts (8-µm pore size; EMD Millipore),
according to the manufacturer's protocol. A total of
1.0×104 cells were plated in the upper chambers of
Transwell plates in 100 µl serum-free DMEM. DMEM (600 µl)
supplemented with 10% FBS was plated in the lower chambers.
Following incubation at 37°C for 24 h, the non-migratory cells in
the upper chambers were removed using cotton swabs, while the
migratory cells in the lower chambers were fixed with 100% methanol
at room temperature for 15 min and stained with 1% crystal violet
for 30 min at room temperature. Stained cells were counted in five
randomly selected fields using a light microscope (magnification,
×200).
Western blotting
Cell lysates were extracted from MCF-7 and
MDA-MB-468 cells transfected with NC or hsa_circ_0000129 sequences
using cell lysis buffer (Sigma-Aldrich; Merck KGaA), and
centrifuged at 13,800 × g for 5 min at 4°C. Total protein was
quantified using the BCA protein assay kit (cat. no. 70-PQ0011;
Hangzhou MultiSciences Biotech Co., Ltd., http://www.liankebio.com), according to the
manufacturer's protocol, and 20 µg protein/lane was separated via
SDS-PAGE on a 12% gel. The separated proteins were subsequently
transferred onto polyvinylidene fluoride membranes and blocked with
5% non-fat milk at room temperature for 2 h. The membranes were
incubated with primary monoclonal antibodies against EZH2 (cat. no.
5246; 1:1,000) and β-actin (cat. no. 3700, 1:1,000) overnight at
4°C. Following the primary incubation, membranes were incubated
with HRP-linked secondary antibody (cat. no. 7076, 1:1,000), at
room temperature for 1 h and subsequently visualized using the
Ultra-sensitive ECL chemiluminescence kit (cat. no. P0018AS;
Beyotime Institute of Biotechnology). All antibodies were purchased
from Cell Signaling Technology, Inc.
Statistical analysis
Statistical analysis was performed using SPSS 16.0
(IBM Corp.) and GraphPad Prism 5.0 (GraphPad Software, Inc.). All
experiments were performed in triplicate and data are presented as
the mean ± standard deviation. Paired and unpaired Student's
t-tests were used to compare differences between two groups, while
ANOVA followed by Tukey's post hoc test were used to compare
differences between multiple groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
hsa_circ_0000129 expression is
upregulated in breast cancer
RT-qPCR analysis was performed to detect
hsa_circ_0000129 expression in breast cancer tissues and adjacent
normal tissues. The results demonstrated that hsa_circ_0000129
expression was significantly higher in tumor tissues compared with
normal tissues (P<0.001; Fig.
1A). Similarly, hsa_circ_0000129 expression was compared
between the breast cancer cell lines (MCF-7, MDA-MB-231 and
MDA-MB-468) and normal MCF10A cells. The results demonstrated that
hsa_circ_0000129 expression was significantly higher in all three
cancer cell lines (MCF-7, MDA-MB-231 and MDA-MB-468) compared with
MCF10A cells (P<0.05, P<0.01 and P<0.001, respectively;
Fig. 1B).
Association between hsa_circ_0000129
expression and the clinicopathological characteristics of patients
with breast cancer
As presented in Table
I, significant associations were observed between
hsa_circ_0000129 expression and lymph node metastasis (P<0.05)
and TNM stage (P<0.01). However, no significant associations
were observed between hsa_circ_0000129 expression and age,
menopause status and tumor size.
 | Table I.Association between hsa_circ_0000129
expression and the clinicopathological characteristics of patients
with breast cancer (n=68). |
Table I.
Association between hsa_circ_0000129
expression and the clinicopathological characteristics of patients
with breast cancer (n=68).
| Characteristic | Number of cases,
n | hsa_circ_0000129
expression, mean ± standard deviation | P-value |
|---|
| Age, years |
|
| 0.499 |
|
≤50 | 36 | 2.15±1.47 |
|
|
>50 | 32 | 1.87±1.93 |
|
| Menopause |
|
| 0.348 |
| No | 40 | 1.97±1.45 |
|
|
Yes | 28 | 2.37±2.07 |
|
| Tumor size, cm |
|
| 0.847 |
| ≤2 | 30 | 1.98±1.41 |
|
|
>2 | 38 | 2.06±1.91 |
|
| Lymph node
metastasis |
|
| 0.036a |
| No | 33 | 1.58±1.36 |
|
|
Yes | 35 | 2.44±1.89 |
|
| TNM stage |
|
| 0.002b |
|
I–II | 37 | 1.47±1.33 |
|
|
III–IV | 31 | 2.68±1.87 |
|
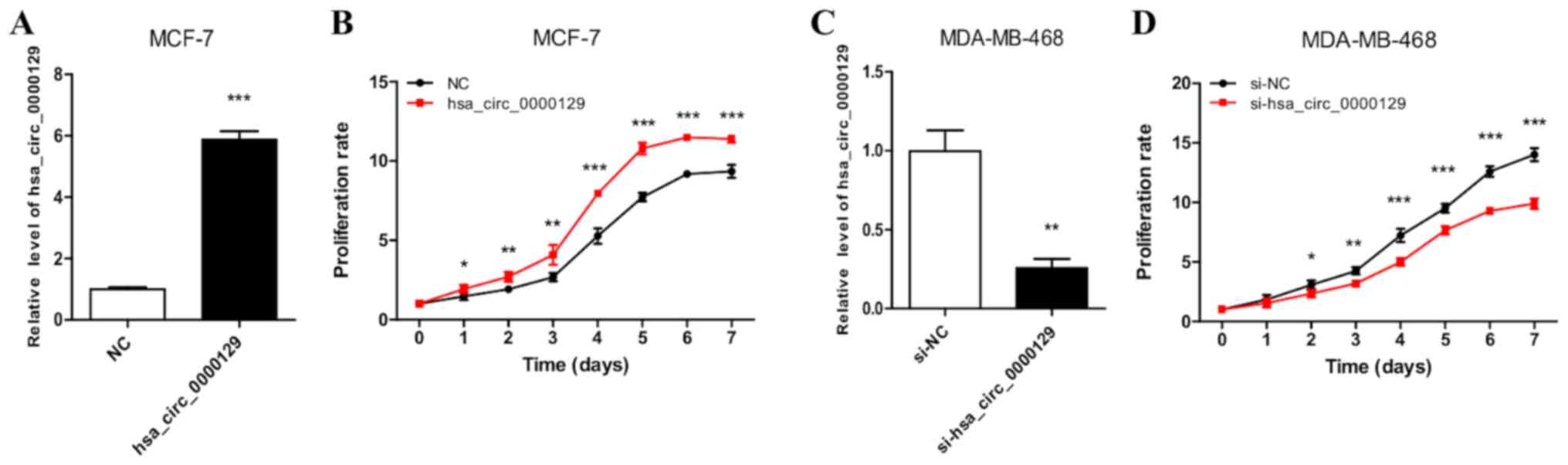
hsa_circ_0000129 regulates the
proliferation of breast cancer cells
RT-qPCR analysis demonstrated the successful
overexpression of hsa_circ_0000129 in MCF-7 cells (Fig. 2A). MCF-7 cells transfected with
hsa_circ_0000129 sequence exhibited significantly increased cell
proliferation compared with cells in the NC group (P<0.001 at
days 4, 5, 6 and 7; Fig. 2B).
RT-qPCR analysis also demonstrated the successful knockdown of
hsa_circ_0000129 in MDA-MB-468 cells (Fig. 2C). The results of the MTT assay
demonstrated that hsa_circ_0000129 knockdown significantly
decreased the proliferation rate of MDA-MB-468 cells compared with
cells in the si-NC group (P<0.001 at days 4, 5, 6 and 7
post-incubation; Fig. 2D).
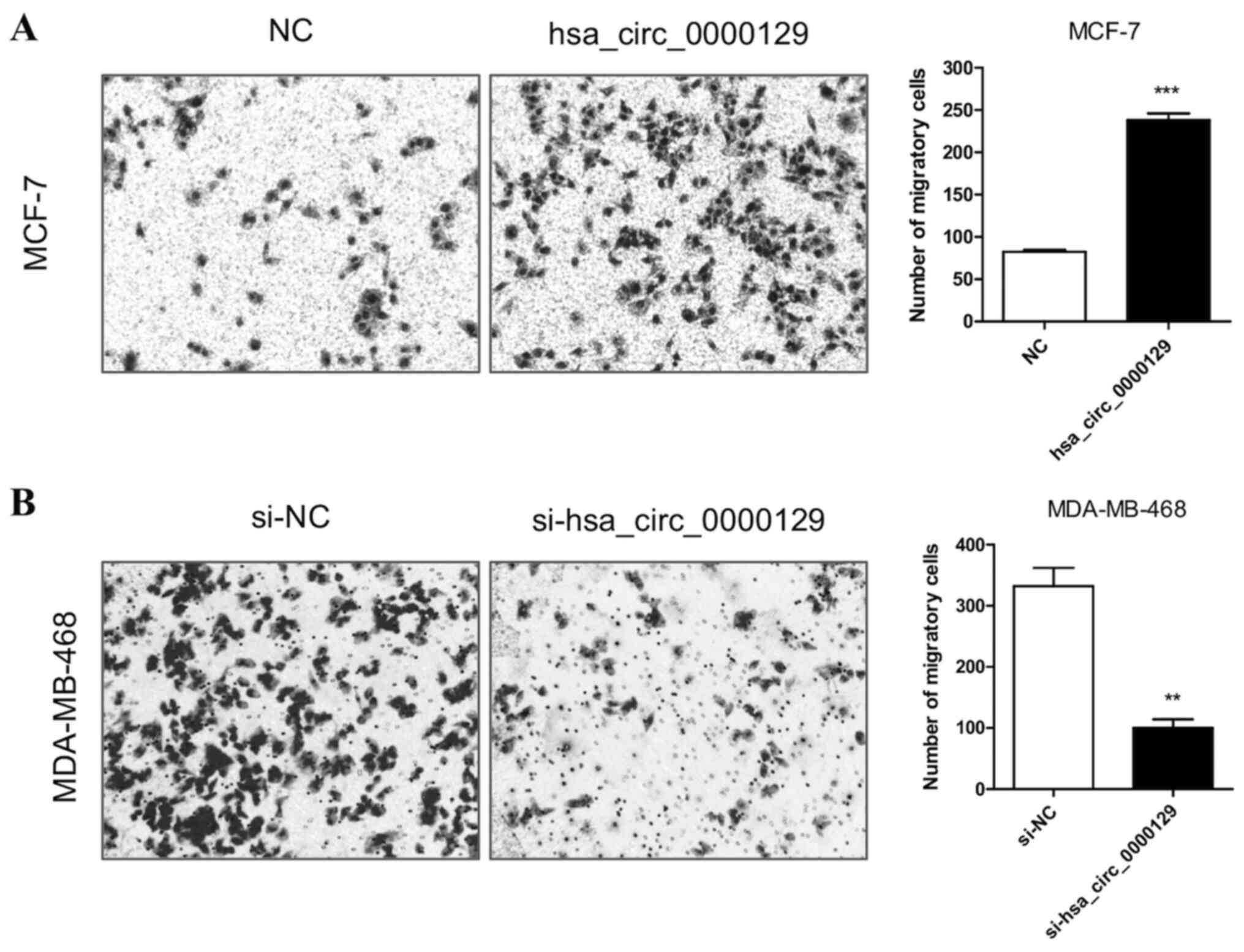
hsa_circ_0000129 regulates the
migration of breast cancer cells
The results demonstrated that the migratory ability
was significantly enhanced in MCF-7 cells transfected with
hsa_circ_0000129 vector compared with the NC group (P<0.001;
Fig. 3A). Conversely, the migratory
ability was significantly attenuated in MDA-MB-468 cells
transfected with si-hsa_circ_0000129 compared with the si-NC group
(P<0.01; Fig. 3B).
hsa_circ_0000129 regulates colony
formation in breast cancer cells
The effect of hsa_circ_0000129 on colony formation
ability was assessed. The results demonstrated that the colony
formation ability of MCF-7 cells was significantly enhanced
following overexpression of hsa-circ-0000129 (P<0.001; Fig. 4A). Conversely, hsa_circ_0000129
knockdown significantly inhibited the colony formation ability of
MDA-MB-468 cells compared with the si-NC group (P<0.01; Fig. 4B).
Effect of hsa_circ_0000129 on EZH2
expression
EZH2 expression was detected to determine its
underlying molecular mechanism in relation to hsa_circ_0000129.
RT-qPCR and western blot analyses demonstrated that EZH2 expression
was significantly higher in MCF-7 and MDA-MB-468 cells compared
with MCF-10A cells (P<0.01 and P<0.001, respectively;
Fig. 5A and B). RT-qPCR and western
blot analyses also demonstrated that EZH2 expression was
significantly higher in MCF-7 cells overexpressing hsa_circ_0000129
compared with cells in the NC group (P<0.01; Fig. 5C and D). Conversely, RT-qPCR and
western blot analyses demonstrated that EZH2 expression was
significantly downregulated in MDA-MB-468 cells following
hsa_circ_0000129 knockdown compared with cells in the si-NC group
according (P<0.001; Fig. 5E and
F).
Discussion
The present study aimed to investigate the role and
determine the underlying molecular mechanism of hsa_circ_0000129 in
breast cancer. The results demonstrated that hsa_circ_0000129
expression was significantly higher in breast cancer tissues
compared with adjacent paracancerous tissues, and hsa_circ_0000129
expression was significantly associated with lymph node metastasis
and high TNM staging, which suggests that high hsa_circ_0000129
expression may predict a higher malignancy index in patients with
breast cancer.
Similarly, the results of the present study
demonstrated that hsa_circ_0000129 expression was significantly
higher in all three breast cancer cell lines (MCF-7, MDA-MB-231 and
MDA-MB-468) compared with the normal epithelial breast cancer cell
line, MCF10A. Notably, hsa_circ_0000129 expression was higher in
MDA-MB-468 cells compared with MDA-MB-231 cells, and exhibited the
lowest expression in MCF-7 cells. The differences in expression
between the three breast cancer cell lines may be attributed to the
differences in cellular characteristics. First, although all three
cell lines originate from the metastatic pleural effusions of
patients with breast cancer, the malignancy degrees may vary
(25). Secondly, the results of the
present study demonstrated that high hsa_circ_0000129 expression in
the clinical samples was associated with a higher degree of
malignancy, which may also be applicable to these cell lines.
Furthermore, certain studies investigating these cell lines have
reported that the phenotypes of the cells are notably different,
whereby the migratory and invasive abilities, and stemness of
MDA-MB-231 cells are higher compared with MCF-7 cells (25–27). In
addition, the migratory, invasive and colony formation abilities of
MDA-MB-468 cells have been demonstrated to be significantly higher
compared with MDA-MB-231 cells (28), which are consistent with the results
of the present study and suggest that MDA-MB-468 cells are more
malignant than MCF-7 cells.
The role of hsa_circ_0000129 in different phenotypes
of breast cancer cells was assessed in the present study. The
results demonstrated that cell proliferation, migration and colony
formation were significantly enhanced in MCF-7 cells overexpressed
with hsa_circ_0000129. Conversely, hsa_circ_0000129 knockdown
significantly inhibited cell proliferation, migration and colony
formation of MDA-MB-468 cells compared with the control cells.
Generally, higher proliferative, migratory and colony formation
abilities indicate a more malignant phenotype in cancer cells
(29,30). Thus, the results of the present
suggest that hsa_circ_0000129 expression level in breast cancer
plays an important role in cellular malignancy, which is closely
associated with the malignancy degree of breast cancer. Taken
together, the results of the present study suggest that
hsa_circ_0000129 may represent a promising prognostic biomarker for
breast cancer.
In accordance with the results of the present study,
several studies have investigated the role of circRNAs in breast
cancer and other types of cancer, including hepatocellular
carcinoma, clear cell renal cell carcinoma and colorectal cancer
(8), and demonstrated the potential
role of circRNAs as both diagnostic and prognostic markers. For
example, Nair et al (31)
cataloged the different circRNAs associated with three different
types of breast cancer, triple negative, estrogen receptor positive
and ErB2 overexpressing Her2-positive breast cancer cells. In
addition, Wang et al (32)
demonstrated that circ-UBE2D2 is upregulated in breast cancer cell
lines and tissues, and is also closely associated with aggressive
clinical characteristics and a poor prognosis (32). Furthermore, it has been reported that
silencing circ-UBE2D2 notably inhibits the proliferative, migratory
and invasive abilities of BC cells, and the in vivo delivery
of oligonucleotides that inhibits circ-UBE2D2 significantly delays
tumor growth (32). To the best of
our knowledge, the present study is the first to investigate the
role of hsa_circ_0000129 in breast cancer.
The potential underlying molecular mechanism of
hsa_circ_0000129 in breast cancer was assessed in the present
study. EZH2, a polycomb group protein, is known to be associated
with carcinogenesis (33,34). Polycomb group proteins are
evolutionarily conserved regulators and act by silencing different
growth regulatory genes (33,34).
There are two main families of polycomb repressive complexes, 1 and
2 (PRC2), and EZH2 is a catalytic subunit of PRC2 (34). Previous studies have demonstrated
that upregulated EZH2 expression is associated with different types
of carcinoma, including ovarian, cervical, glioma, breast, prostate
and renal cell cancer (33–35).
Previous studies have reported that upregulated EZH2
expression in breast cancer is associated with advanced form of the
disease, higher grade of tumor staging, increased proliferation of
tumor, increased chance of metastasis and poor long term overall
survival (36–38). Xue et al (39) reported that arsenite, a known
carcinogenic agent, upregulates hsa_circ_100284 expression in human
ketratocyte (HaCaT) cells. The underlying molecular mechanism was
also investigated, and it was demonstrated that hsa_circ_100284
serves as a sponge for miR-217 and indirectly upregulates EZH2.
Notably, EZH2 upregulates the expression levels of cyclin D1 and
K4, and promotes malignant changes in HaCaT cells. Furthermore,
while investigating the association between EZH2 expression and
risk of developing breast cancer in a case control study, Beca
et al (40) discovered that
EZH2 expression is an independent risk factor for breast
cancer.
Similarly, the underlying molecular mechanism of
hsa_circ_0000129 in breast cancer was investigated in the present
study, and the results demonstrated that EZH2 expression was
upregulated in all three breast cancer cell lines. In addition,
EZH2 expression was significantly enhanced in MCF-7 cells
transfected with hsa_circ_0000129 vector, and significantly
downregulated in MDA-MB-468 cells following hsa_circ_0000129
knockdown, compared with their respective control groups.
Collectively, the results of the present study suggest that
hsa_circ_0000129 exerts its role in breast cancer by regulating
expression of the oncogene, EZH2.
A limitation of the present study is that EZH2
expression was only assessed in breast cancer cell lines, while its
expression in human breast cancer tissues remains unknown. Thus,
this will be the focus of prospective studies.
In conclusion, the results of the present study
suggest that hsa_circ_0000129 may represent a prognostic marker for
patients with breast cancer. The role of hsa_circ_0000129 in breast
cancer cell lines reveals a novel mechanism for tumorigenesis, as
well as a potent target for interference of malignant progression.
In addition, the carcinogenic molecular mechanism of
hsa_circ_0000129 may be associated with upregulated EZH2 expression
in breast cancer.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Joint Medical
Research Program of Shanghai Jing'an District Science and
Technology Commission and Health Commission [grant no.
(JI)2016MS02] and the National Key R&D Program of China (grant
no. 2018YFC2002400).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon request.
Authors' contributions
ZZ, ZS and SZ conceived and designed the present
study, and analyzed and interpreted the data. XW and LH provided
administrative support and interpreted the data. QL and HW
performed the experiments, and collected and assembled the data.
All authors contributed to drafting the initial manuscript and
revising the manuscript for important intellectual content. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Human Research
Ethics Committee of Huashan Hospital (Shanghai, China; approval no.
2016MS02), and written informed consent was provided by all
patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EZH2
|
enhancer of zeste homolog 2
|
|
TNM
|
tumor-node-metastasis
|
|
RT
|
room temperature
|
References
|
1
|
Wang X and Fang L: Advances in circular
RNAs and their roles in breast cancer. J Exp Clin Cancer Res.
37:2062018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Key TJ, Verkasalo PK and Banks E:
Epidemiology of breast cancer. Lancet Oncol. 2:133–140. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harbeck N and Gnant M: Breast cancer.
Lancet. 389:1134–1150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Malvezzi M, Carioli G, Bertuccio P, Rosso
T, Boffetta P, Levi F, La Vecchia C and Negri E: European cancer
mortality predictions for the year 2016 with focus on leukemias.
Ann Oncol. 27:725–731. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brufsky AM: Long-term management of
patients with hormone receptor-positive metastatic breast cancer:
Concepts for sequential and combination endocrine-based therapies.
Cancer Treat Rev. 59:22–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Majeed W, Aslam B, Javed I, Khaliq T,
Muhammad F, Ali A and Raza A: Breast cancer: Major risk factors and
recent developments in treatment. Asian Pac J Cancer Prev.
15:3353–3358. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jafari SH, Saadatpour Z, Salmaninejad A,
Momeni F, Mokhtari M, Nahand JS, Rahmati M, Mirzaei H and Kianmehr
M: Breast cancer diagnosis: Imaging techniques and biochemical
markers. J Cell Physiol. 233:5200–5213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R
and Li H: The emerging functions and roles of circular RNAs in
cancer. Cancer Lett. 414:301–309. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Han YN, Xia SQ, Zhang YY, Zheng JH and Li
W: Circular RNAs: A novel type of biomarker and genetic tools in
cancer. Oncotarget. 8:64551–64563. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Z, Chen Z, Hu G and Jiang Y: Roles of
circular RNA in breast cancer: Present and future. Am J Transl Res.
11:3945–3954. 2019.PubMed/NCBI
|
|
11
|
Zhou SY, Chen W, Yang SJ, Xu ZH, Hu JH,
Zhang HD, Zhong SL and Tang JH: The emerging role of circular RNAs
in breast cancer. Biosci Rep. 39:BSR201906212019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ng WL, MohdMohidin TB and Shukla K:
Functional role of circular RNAs in cancer development and
progression. RNA Biol. 15:995–1005. 2018.PubMed/NCBI
|
|
13
|
Lukiw WJ: Circular RNA (circRNA) in
Alzheimer's disease (AD). Front Genet. 4:3072013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou J, Zhang WW, Peng F, Sun JY, He ZY
and Wu SG: Downregulation of hsa_circ_0011946 suppresses the
migration and invasion of the breast cancer cell line MCF-7 by
targeting RFC3. Cancer Manag Res. 10:535–544. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lü L, Sun J, Shi P, Kong W, Xu K, He B,
Zhang S and Wang J: Identification of circular RNAs as a promising
new class of diagnostic biomarkers for human breast cancer.
Oncotarget. 8:44096–44107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li X, Ma F, Wu L, Zhang X, Tian J, Li J,
Cao J, Ma Y, Zhang L and Wang L: Identification of Hsa_circ_0104824
as a potential biomarkers for breast cancer. Technol Cancer Res
Treat. 19:15330338209607452020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bian Q: Circular RNA PVT1 promotes the
invasion and epithelial-mesenchymal transition of breast cancer
cells through serving as a competing endogenous RNA for miR-204-5p.
Onco Targets Ther. 12:11817–11826. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yi Z, Li Y, Wu Y, Zeng B, Li H, Ren G and
Wang X: Circular RNA 0001073 attenuates malignant biological
behaviours in breast cancer cell and is delivered by nanoparticles
to inhibit mice tumour growth. Onco Targets Ther. 13:6157–6169.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R,
Yang SY, Yang DC and Wang XL: Circular RNA hsa_circ_0001982
promotes breast cancer cell carcinogenesis through decreasing
miR-143. DNA Cell Biol. 36:901–908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yin WB, Yan MG, Fang X, Guo JJ, Xiong W
and Zhang RP: Circulating circular RNA hsa_circ_0001785 acts as a
diagnostic biomarker for breast cancer detection. Clin Chim Acta.
487:363–368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J,
Wang Z, Wang Q, Li A, Marks JR, et al: CircIRAK3 sponges miR-3607
to facilitate breast cancer metastasis. Cancer Lett. 430:179–192.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hayes DF, Allred C, Anderson BO, et al:
Breast. American Joint Committee on Cancer (AJCC) Cancer Staging
Manual. 7th edition. Edge SB, Byrd DR and Compton CC: Springer; New
York, NY: pp. 347–376. 2009
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chekhun VF, Lukianova NY, Chekhun SV,
Bezdieniezhnykh NO, Zadvorniy TV, Borikun TV, Polishchuk LZ and
Klyusov OМ: Association of CD44+CD24−/low
with markers of aggressiveness and plasticity of cell lines and
tumors of patients with breast cancer. Exp Oncol. 39:203–211. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang T, Zhu Q, Li X, Zhu G, Deng S, Wang
Y, Ni L, Chen X, Zhang Y, Xia T, et al: Protease Nexin I is a
feedback regulator of EGF/PKC/MAPK/EGR1 signaling in breast cancer
cells metastasis and stemness. Cell Death Dis. 10:6492019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Croker AK, Goodale D, Chu J, Postenka C,
Hedley BD, Hess DA and Allan AL: High aldehyde dehydrogenase and
expression of cancer stem cell markers selects for breast cancer
cells with enhanced malignant and metastatic ability. J Cell Mol
Med. 13:2236–2252. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan
L, Andrews R, Zhong W, Zhang X, Song E, et al: Circular RNA
hsa_circ_001783 regulates breast cancer progression via sponging
miR-200c-3p. Cell Death Dis. 10:552019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tasharrofi B and Ghafouri-Fard S: Long
non-coding RNAs as regulators of the mitogen-activated protein
kinase (MAPK) pathway in cancer. Klin Onkol. 31:95–102. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Roversi FM, Olalla Saad ST and
Machado-Neto JA: Serine peptidase inhibitor Kunitz type 2 (SPINT2)
in cancer development and progression. Biomed Pharmacother.
101:278–286. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nair AA, Niu N, Tang X, Thompson KJ, Wang
L, Kocher JP, Subramanian S and Kalari KR: Circular RNAs and their
associations with breast cancer subtypes. Oncotarget.
7:80967–80979. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Y, Li J, Du C, Zhang L, Zhang Y,
Zhang J and Wang L: Upregulated circular RNA circ-UBE2D2 predicts
poor prognosis and promotes breast cancer progression by sponging
miR-1236 and miR-1287. Transl Oncol. 12:1305–1313. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Martinez AM and Cavalli G: The role of
polycomb group proteins in cell cycle regulation during
development. Cell Cycle. 5:1189–1197. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Margueron R and Reinberg D: The polycomb
complex PRC2 and its mark in life. Nature. 469:343–349. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang X, Hu B, Shen H, Zhou H, Xue X, Chen
Y, Chen S, Han Y, Yuan B, Zhao H, et al: Clinical and prognostic
relevance of EZH2 in breast cancer: A meta-analysis. Biomed
Pharmacother. 75:218–225. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Boostani F, Dolatkhah R, Fakhrjou A,
Farassati F and Sanaat Z: Association of clinicopathologic
characteristics and outcomes with EZH2 expression in patients with
breast cancer in East Azerbaijan, Iran. Onco Targets Ther.
11:449–457. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shen L, Cui J, Liang S, Pang Y and Liu P:
Update of research on the role of EZH2 in cancer progression. Onco
Targets Ther. 6:321–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pourakbar S, Pluard TJ, Accurso AD and
Farassati F: Ezh2. A novel target in detection and therapy of
breast cancer. Onco Targets Ther. 10:2685–2687. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xue J, Liu Y, Luo F, Lu X, Xu H, Liu X, Lu
L, Yang Q, Chen C, Fan W and Liu Q: Circ100284, via miR-217
regulation of EZH2, is involved in the arsenite-accelerated cell
cycle of human keratinocytes in carcinogenesis. Biochim Biophys
Acta Mol Basis Dis. 1863:753–763. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Beca F, Kensler K, Glass B, Schnitt SJ,
Tamimi RM and Beck AH: EZH2 protein expression in normal breast
epithelium and risk of breast cancer: Results from the Nurses'
health studies. Breast Cancer Res. 19:212017. View Article : Google Scholar : PubMed/NCBI
|