Introduction
Non-small cell lung cancer (NSCLC) is an aggressive
malignancy worldwide (1,2), with an incidence rate of ~0.02%
(3). Surgery in combination with
chemotherapy and/or radiotherapy is the primary therapeutic regimen
for patients with NSCLC in the clinic (4); however, the 5-year survival rate for
these patients remains low at ~15% (5), which is due to the aggressive nature of
NSCLC (6). Thus, further
investigations into factors which may be involved in the process of
metastasis remain a priority to determine novel therapies for
patients with NSCLC.
MicroRNAs (miRNAs/miRs) are a group of endogenous
and non-coding RNAs, ~22 nucleotides in length, which control the
translation of target genes post-transcriptionally (7). During the initiation and development of
different types of cancer, including lung cancer and gastric
cancer, miRNAs have been reported to play crucial roles in
downregulating oncogenes or tumor suppressors, thus regulating the
cell proliferation, apoptosis and metastasis of cancer cells
(8,9). Notably, miR-217 has been identified to
act as a tumor suppressor in lung cancer cells (10). In addition, miR-217 expression is
downregulated by cigarette smoke extract (CSE) in human bronchial
epithelial (HBE) cells (11).
As a NAD+-dependent histone deacetylase,
sirtuin 1 (SIRT1) is universally expressed in the intracellular and
extracellular matrix (12). Notably,
SIRT1 expression is associated with a poor prognosis in patients
with NSCLC (13). In addition, SIRT1
expression is upregulated in the brain tissues of patients with
metastatic NSCLC, which subsequently promotes NSCLC cell migration
(14). miR-217 has been demonstrated
to decrease the development of osteosarcoma through SIRT1 (15). However, whether miR-217 inhibits the
progression of NSCLC by targeting SIRT1 remains unknown. Taken
together, the results of the present study demonstrated that
miR-217 inhibited cell proliferation and invasion, and induced cell
apoptosis of NSCLC cell lines by targeting SIRT1.
Materials and methods
Cell culture
HBE cells and the four NSCLC cell lines (H23, H292,
H1299 and A549) were purchased from the American Type Culture
Collection. All cells were maintained in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS) and 1%
penicillin/streptomycin (all purchased from Invitrogen; Thermo
Fisher Scientific, Inc.), at 37°C with 5% CO2.
Cell transfection
pcDNA3.1 or pcDNA3.1-SIRT1 reconstructed vectors
(Guangzhou RiboBio Co., Ltd.) were transfected into H1299 and A549
cells using Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol.
For the mimic transfections, cells were seeded into
6-well plates at a density of 1×105 cells/well and
cultured in complete culture medium (Thermo Fisher Scientific,
Inc.) overnight at 37°C with 5% CO2. Following
incubation, 50 nM miR-217 mimic or miR-negative control (NC) mimic
(Shanghai GenePharma Co., Ltd.) were transfected into H1299 and
A549 cells using Lipofectamine® 2000 reagent, according
to the manufacturer's protocol. Following incubation for 24 h at
37°C, cells were harvested for subsequent experimentation.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was assessed via the CCK-8 assay
(Dojindo Molecular Technologies, Inc.), according to the
manufacturer's protocol. Briefly, H1299 and A549 cells were seeded
into a 96-well plate at a density of 5×103 cells/well
and 10 µl CCK-8 solution was added to each well for 3, 0, 24 and 48
h post-transfection. Cell proliferation was measured at a
wavelength of 450 nm, using a microplate reader (Bio-Rad
Laboratories, Inc.).
Apoptosis analysis
H1299 and A549 cells (1×106 cells/ml)
were resuspended in 100 µl binding buffer provided in the
FITC-Annexin V apoptosis detection kit (cat. no. 556570; BD
Biosciences), and subsequently treated with 5 µl FITC-Annexin V or
propidium iodide for 15 min at room temperature in the dark. The
reaction was terminated following addition of 400 µl binding
buffer, and apoptotic cells were subsequently analyzed using a
FACSCalibur cytometer (BD Biosciences) and CellQuest software
(version 5.1; Becton, Dickinson and Company).
Cell invasion assay
Cell invasion was determined using Transwell
chambers (8-µm pore; BD Biosciences). Briefly, 5×104
H1299 and A549 cells were plated in the Matrigel-coated (at 37°C
for 5 h) upper chambers, in serum-free RPMI-1640 medium. RPMI-1640
medium (500 µl) supplemented with 10% FBS was plated in the lower
chambers. Following incubation for 24 h at 37°C, the non-invasive
cells were removed from the upper chambers using cotton swabs,
while the invasive cells in the lower chambers were fixed with 100%
methanol for 1 min at room temperature and stained with 0.1%
crystal violet for 15 min at room temperature. Stained cells were
counted in five randomly selected fields using a light microscope
(magnification, ×100).
Dual-luciferase reporter assay
The binding site between miR-217 and the
3′-untranslated region (UTR) of SIRT1 was predicted using the
online TargetScan database (www.targetscan.org). The 3′-UTRs of wild-type (WT) and
mutant (MT) SIRT1 were reconstructed into the XbaI site of
the pGL3 vector (Promega Corporation). Subsequently, cells were
seeded into a 24-well plate at a density of 1×104
cells/well and co-transfected with 0.4 mg pGL3-WT-SIRT1 or
pGL3-MT-SIRT1 and/or 20 nM miR-217 or miR-NC, using
Lipofectamine® 2000 reagent, according to the
manufacturer's protocol. Following incubation for 48 h at 37°C,
cells were harvested and luciferase activities were detected using
the Dual-Luciferase® Reporter Assay kit (cat. no. E1910;
Promega Corporation). Firefly luciferase activity was normalized to
Renilla luciferase activity.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from HBE, H23, H292, H1299
and A549 cells using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). Total RNA was reverse transcribed into
cDNA using the cDNA Reverse Transcription kit (cat. no. 4368813,
Applied Biosystems; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. miR-217 expression was detected using
the mirVana™ qRT-PCR miRNA Detection kit (cat. no. AM1558, Ambion;
Thermo Fisher Scientific, Inc.), and normalized to the internal
reference gene U6. PCNA, E-cadherin, vimentin and SIRT1 expression
levels were detected using the SYBR Premix Ex Taq™ kit (cat. no.
DRR041A, Takara Bio, Inc.), and normalized to the internal
reference gene β-actin. qPCR was performed using the Applied
Biosystems 7900 Fast Real-Time PCR system (Thermo Fisher
Scientific, Inc.) The following thermocycling conditions were used
for qPCR: Initial denaturation at 95°C for 5 min; followed by 40
cycles of 95°C for 15 sec, 60°C for 15 sec and 72°C for 32 sec. The
following primer sequences were used for qPCR: miR-217 forward,
5′-AGGTTAGTCAAGGACTAGCTCA-3, and reverse,
5′-TGAGCTAGTCCTTGACTAACCT-3′; SIRT1 forward,
5′-TAGCCTTGTCAGATAAGGAAGGA-3′ and reverse,
5′-ACAGCTTCACAGTCAACTTTGT-3′; proliferating cell nuclear antigen
(PCNA) forward, 5′-TCAAGAAAATAAAACTAAGCTCT-3′ and reverse,
5′-CTTCTAGGTTAACTAACCACA−3′; E-cadherin forward,
5′-TGTAACTTGCAATGGGCAGC-3′ and reverse, 5′-CAAGCTCTCCTGCCATCTCC-3′;
vimentin forward, 5′-ACGTCTTGACCTTGAACGCA-3′ and reverse,
5′-TCTTGGCAGCCACACTTTCA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; and β-actin forward,
5′-CATGTACGTTGCTATCCAGGC−3′ and reverse,
5′-CTCCTTAATGTCACGCACGAT−3′. Relative expression levels were
quantified using the 2−ΔΔCq method (16).
Western blotting
Total protein was extracted from HBE, H1299 and A549
cells using RIPA lysis buffer (Beyotime Institute of
Biotechnology). Protein concentration was determined using the BCA
method (Beyotime Institute of Biotechnology). Protein samples (15
µg/lane) were separated via 10% SDS-PAGE, transferred onto PVDF
membranes and subsequently blocked with 5% non-fat milk for 1 h at
room temperature. The membranes were incubated with primary
antibodies against: SIRT1 (1:1,000; cat. no. 9475), PCNA (1:1,000;
cat. no. 13110), E-cadherin (1:1,000; cat. no. 3195), vimentin
(1:1,000; cat. no. 5741), p-AMPKα (1:1,000; cat. no. 2537), p-mTOR
(1:1,000; cat. no. 5536), AMPKα (1:1,000; cat. no. 5832), mTOR
(1:1,000; cat. no. 2983) and β-actin (1:2,000; cat. no. 4970)
overnight at 4°C (all purchased from Cell Signaling Technology,
Inc.). Following the primary incubation, membranes were incubated
with anti-rabbit IgG, HRP-linked secondary antibodies (1:3,000;
cat. no. 7074; Cell Signaling Technology, Inc.) for 2 h at room
temperature. Protein bands were visualized using enhanced
chemiluminescence (Amersham; Cytiva).
Statistical analysis
Statistical analysis was performed using SPSS 15.0
software (SPSS, Inc.). All experiments were performed in triplicate
and data are presented as the mean ± standard deviation. Student's
t-test was used to compare differences between two groups, while
one-way ANOVA and Tukey's post hoc test were used to compare
differences between multiple groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-217 expression in the NSCLC cell
lines
The results demonstrated that miR-217 expression was
significantly downregulated in the NSCLC cells (H23, H292, H1299
and A549) compared with HBE cells (P<0.05 and P<0.01;
Fig. 1). Among the four NSCLC cell
lines, H1299 and A549 cells had the lowest miR-217 expression
levels; thus, these cell lines were selected for subsequent
experimentation.
SIRT1 is the target gene of
miR-217
TargetScan analysis demonstrated that miR-217 was
complementary to the 3′-UTR of SIRT1 (Fig. 2A). The results of the dual-luciferase
reporter assay further validated that co-transfection with miR-217
mimic decreased the relative luciferase activity of cells
transfected with WT-SIRT1 (P<0.05); however, no significant
changes were observed following transfection with MT-SIRT1
(Fig. 2B).
SIRT1 expression in the NSCLC cell
lines
The results demonstrated that both SIRT1 mRNA
(Fig. 3A) and protein (Fig. 3B and C) expression levels were
significantly upregulated in H1299 and A549 cells compared with HBE
cells (P<0.01).
Transfection with miR-217 mimic in
NSCLC cells
No significant differences in miR-217 expression
were observed between the control (un-transfected group) and miR-NC
mimic groups; however, miR-217 expression significantly increased
following transfection with miR-217 mimic (P<0.01; Fig. 4A). In addition, no significant
differences in SIRT1 mRNA and protein expression were observed
between the control and miR-NC mimic groups; however, SIRT1 mRNA
and protein expression significantly decreased following
transfection with miR-217 mimic (P<0.01; Fig. 4B-D).
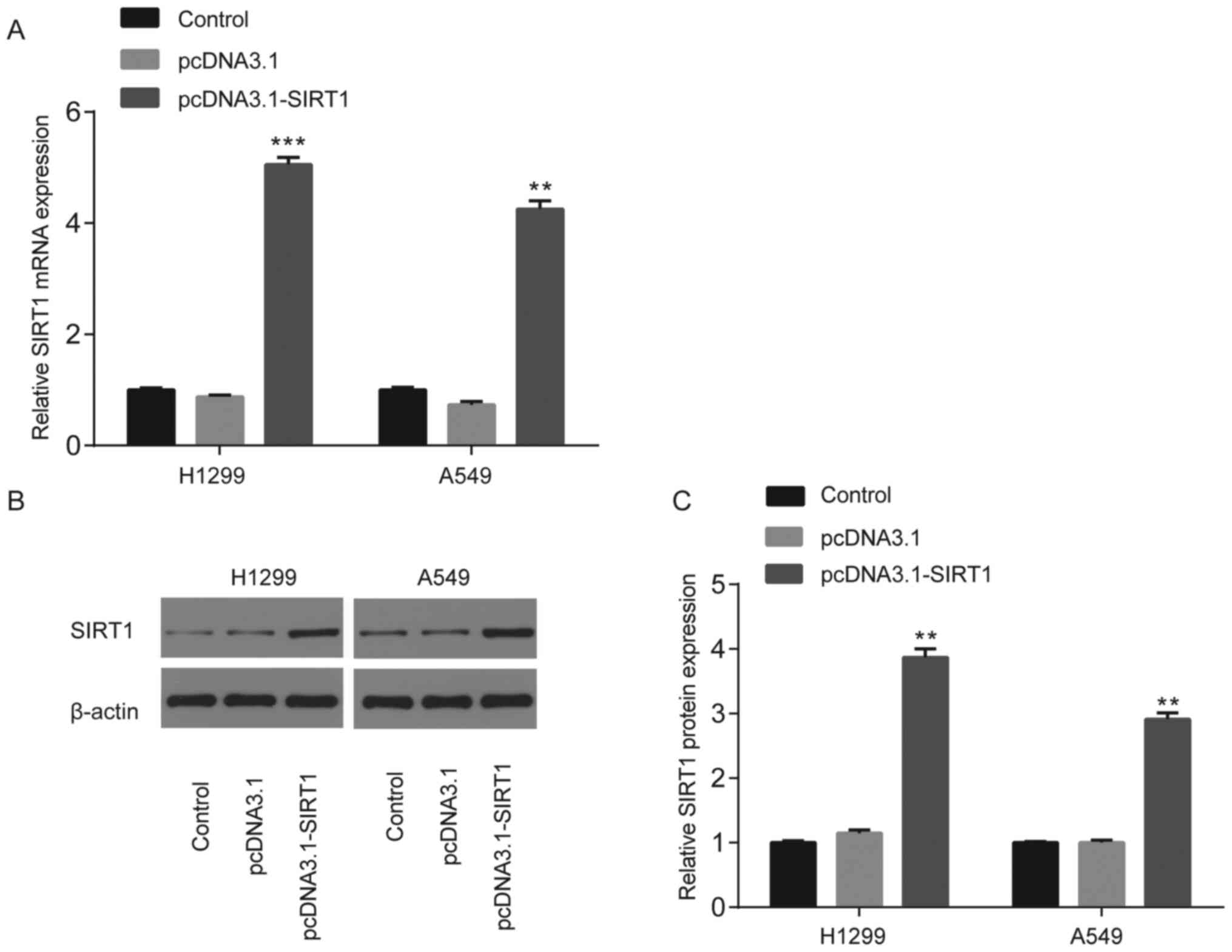
Transfection with pcDNA3.1-SIRT1 in
NSCLC cells
No significant differences in SIRT1 mRNA or protein
expression were observed between the control (un-transfected group)
and pcDNA3.1 groups; however, SIRT1 mRNA and protein expression
significantly increased following transfection with pcDNA3.1-SIRT1
(P<0.01 and P<0.001; Fig.
5A-C).
SIRT1 reverses the miR-217
mimic-induced inhibition of NSCLC cell proliferation and
invasion
To further determine whether the effects of miR-217
on NSCLC cell proliferation and invasion were mediated by SIRT1,
miR-217 mimic and pcDNA3.1-SIRT1 were co-transfected into H1299 and
A549 cells. The results demonstrated that miR-217 mimic
significantly decreased (P<0.01) cell proliferation (Fig. 6A and B), and PCNA mRNA (Fig. 6C) and protein expression levels of
H1299 and A549 cells (Fig. 6D and
E), the effects of which were partially rescued following
overexpression of SIRT1 (P<0.05).
miR-217 mimic significantly decreased (P<0.01)
cell invasion (Fig. 7A) and
downregulated vimentin mRNA levels (Fig.
7B), while significantly upregulating E-cadherin mRNA levels
(Fig. 7C) of H1299 and A549 cells.
These effects were partially rescued following overexpression of
SIRT1 (P<0.05). The protein profiles of vimentin and E-cadherin
were similar to that of their mRNA levels (Fig. 7D-F).
SIRT1 reverses the miR-217
mimic-induced NSCLC cell apoptosis
To further determine whether the effects of miR-217
on NSCLC cell apoptosis were mediated by SIRT1, miR-217 mimic and
pcDNA3.1-SIRT1 were co-transfected into H1299 and A549 cells. The
results demonstrated that miR-217 mimic significantly increased
(P<0.01) cell apoptosis in H1299 (Fig. 8A and B) and A549 (Fig. 8C and D) cells, the effects of which
were partially rescued following overexpression of SIRT1
(P<0.01).
SIRT1 reverses the miR-217
mimic-induced decrease of phosphorylated (p)-AMPKα and increase of
p-mTOR in NSCLC cells
To further determine whether the effects of miR-217
and SIRT1 on NSCLC progression involve the p-AMPKα and p-mTOR
signaling pathways, miR-217 mimic and pcDNA3.1-SIRT1 were
co-transfected into H1299 and A549 cells. The results demonstrated
that miR-217 mimic significantly downregulated (P<0.01) p-AMPKα
protein expression (Fig. 9A and B)
and upregulated p-mTOR protein expression (Fig. 9A and C) in H1299 and A549 cells.
These effects were partially rescued following overexpression of
SIRT1 (P<0.05).
Discussion
miR-217 has been reported to act as a tumor
suppressor in several types of diseases. For example, miR-217
represses HeLa cell migration and invasion through MAPK1 in
cervical cancer (17); it inhibits
M2-like macrophage polarization by decreasing IL-6 in ovarian
cancer (18); and it inhibits the
proliferation and promotes apoptosis in hepatocytes by targeting
NAT2 in liver injury (19). In
addition, miR-217 expression is downregulated in lung cancer, while
overexpression of miR-217 has been demonstrated to inhibit the
proliferation, migration and invasion, and induce apoptosis of the
lung cancer cell lines, SPC-A-1 and A549 by targeting KRAS
(10). miR-217 expression is also
downregulated by CSE in HBE cells, while miR-217 regulation of
EZH2/H3K27me3 by MALAT1 is involved in CSE-induced malignant
transformation of HBE cells (11).
Furthermore, circ_0023404 promotes cell proliferation, migration
and invasion of the NSCLC cell lines, A549 and H1299 by targeting
the miR-217/ZEB1 axis (20). The
inhibitory effects of miR-217 overexpression on proliferation and
invasion have been previously reported, along with the promotional
effects of miR-217 overexpression on the apoptosis of the NSCLC
cell lines, H1299 and A549 (10,20). The
present study aimed to investigate other potential mRNA targets of
miR-217 in NSCLC.
Increasing evidence has indicated that SIRT1 serves
as an oncogene. For example, SIRT1 inactivates p53, thus inhibiting
the functions of several tumor suppressors (21), and SIRT1 has also been demonstrated
to regulate multiple tumor suppressors epigenetically (22,23).
Furthermore, SIRT1 was identified to be overexpressed in mouse and
human prostate cancer (24), while
inhibition of SIRT1 has been reported to repress the progression of
several types of tumors, including colon, prostate and melanoma
cancer (25). SIRT1 is associated
with the poor prognosis of patients with NSCLC (13), and is overexpressed in metastatic
brain tissues of patients with NSCLC, where it promotes migration
of A549 cells (14). The results of
the present study confirmed that SIRT1 is a target of miR-217, and
serves as an oncogene in NSCLC. Overexpression of SIRT1 reversed
the inhibitory effects of miR-217 mimic on cell proliferation and
migration, and the promotional effects of miR-217 mimic on cell
apoptosis in the NCSLC cell lines, H1299 and A549. Notably, during
the process of the present study, a newly published study
demonstrated the involvement of miR-217 and SIRT1 in lung cancer,
suggesting that miR-217 inhibits the proliferation, migration, and
invasion of the human pulmonary adenocarcinoma brain metastasis
cell line PC-14/B, by targeting SIRT1 and activating the P53/KAI1
signaling pathway (26). Other
studies reported that downregulation of miR-217 attenuates
atherosclerosis by inhibiting macrophage apoptosis and inflammation
via SIRT1 (27), miR-217 decreases
proliferation, migration, and invasion of osteosarcoma by targeting
SIRT1 (15), and downregulation of
miR-217 mitigates inflammation and myocardial injury in children
with viral myocarditis by regulating the AMPK/NF-κB signaling
pathway via SIRT1 (28). Consistent
with previous findings, the results of the present study verified
the interaction between miR-217 and SIRT1 in the proliferation,
invasion and apoptosis of NSCLC cells. However, the downstream
molecules responsible for the aforementioned effects of SIRT1 are
yet to be elucidated.
As a downstream effector of SIRT1, AMPK functions as
a metabolic stress sensor in HepG2 cells (29). In addition to AMPK-mediated lysosome
biogenesis in the tumor growth of lung cancer (30), AMPK is indispensable for the
Ginsenoside metabolite compound K induced apoptosis of NSCLC cells
(31), as well as downregulation of
TUFM induced invasion of NSCLC cells (32). mTOR is an oncogene that mediates cell
survival (33) and is indispensable
for apoptosis (34), and the
MetaLnc9 induced metastasis of NSCLC (35). Activation of AMPK negatively
regulates mTOR expression (36).
Notably, this association has been previously observed in NSCLC
(31,37), suggesting the negative regulation of
mTOR via the LKB1-AMPK signaling pathway. Taken together, the
results of the present study suggest that miR-217 may inhibit AMPK
phosphorylation and promote mTOR phosphorylation by targeting
SIRT1. However, further studies are required to assess other key
factors involved in mTOR signalization. Also, the present study
failed to confirm the function of miR-217 in the in vivo
experiments, which will be investigated in the future.
In conclusion, the results of the present study
demonstrated that miR-217 inhibited NSCLC cell proliferation and
invasion, and induced NCLC cell apoptosis by targeting SIRT1 and
inactivating the SIRT1-mediated p-AMPK/p-mTOR signaling
pathway.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Author's contributions
GL and SZ conceived the present study, performed the
experiments, and analyzed the data. SZ supervised and drafted the
initial manuscript. Both authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Brahmer JR, Govindan R, Anders RA, Antonia
SJ, Sagorsky S, Davies MJ, Dubinett SM, Ferris A, Gandhi L, Garon
EB, et al: The Society for Immunotherapy of Cancer consensus
statement on immunotherapy for the treatment of nonsmall cell lung
cancer (NSCLC). J Immunother Cancer. 6:752018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang L, Shi SB, Zhu Y, Qian TT and Wang
HL: Long non-coding RNA ASAP1-IT1 promotes cell proliferation,
invasion and metastasis through the PTEN/AKT signaling axis in
non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 22:142–149.
2018.PubMed/NCBI
|
|
3
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
NSCLC Meta-analyses Collaborative Group
and Arriagada R, ; Auperin A, Burdett S, Higgins JP, Johnson DH, Le
Chevalier T, Le Pechoux C, Parmar MK, Pignon JP, et al: Adjuvant
chemotherapy, with or without postoperative radiotherapy, in
operable non-small-cell lung cancer: Two meta-analyses of
individual patient data. Lancet. 375:1267–1277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeng H, Zheng R, Guo Y, Zhang S, Zou X,
Wang N, Zhang L, Tang J, Chen J, Wei K, et al: Cancer survival in
China, 2003–2005: A population-based study. Int J Cancer.
136:1921–1930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu S, Zhao X, Wu S, Du R, Zhu Q, Fang H,
Zhang X, Zhang C, Zheng W, Yang J and Feng H: Overexpression of
B7-H3 correlates with aggressive clinicopathological
characteristics in non-small cell lung cancer. Oncotarget.
7:81750–81756. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu T, Wu X, Chen T, Luo Z and Hu X:
Downregulation of DNMT3A by miR-708-5p inhibits lung cancer stem
cell-like phenotypes through repressing Wnt/β-catenin signaling.
Clin Cancer Res. 24:1748–1760. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Paliouras AR, Monteverde T and Garofalo M:
Oncogene-induced regulation of microRNA expression: Implications
for cancer initiation, progression and therapy. Cancer Lett.
421:152–160. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo J, Feng Z, Huang Z, Wang H and Lu W:
MicroRNA-217 functions as a tumour suppressor gene and correlates
with cell resistance to cisplatin in lung cancer. Mol Cells.
37:664–671. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu L, Luo F, Liu Y, Liu X, Shi L, Lu X and
Liu Q: Posttranscriptional silencing of the lncRNA MALAT1 by
miR-217 inhibits the epithelial-mesenchymal transition via enhancer
of zeste homolog 2 in the malignant transformation of HBE cells
induced by cigarette smoke extract. Toxicol Appl Pharmacol.
289:276–285. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, Bi Y, Chen X, Li C, Li Y, Zhang Z,
Wang J, Lu Y, Yu Q, Su H, et al: Histone deacetylase SIRT1
negatively regulates the differentiation ofinterleukin-9-producing
CD4+ T cells. Immunity. 44:1337–1349. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grbesa I, Pajares MJ, Martínez-Terroba E,
Agorreta J, Mikecin AM, Larráyoz M, Idoate MA, Gall-Troselj K, Pio
R and Montuenga LM: Pression of sirtuin 1 and 2 is associated with
poor prognosis in non-small cell lung cancer patients. PLoS One.
10:e01246702015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Han L, Liang XH, Chen LX, Bao SM and Yan
ZQ: SIRT1 is highly expressed in brain metastasis tissues of
non-small cell lung cancer (NSCLC) and in positive regulation of
NSCLC cell migration. Int J Clin Exp Pathol. 6:2357–2365.
2013.PubMed/NCBI
|
|
15
|
He S, Wang Z, Tang H, Dong J, Qu Y and Lv
J: miR-217 inhibits proliferation, migration, and invasion by
targeting SIRT1 in osteosarcoma. Cancer Biother Radiopharm.
34:264–270. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu L, Yang S and Wang J: miR-217 inhibits
the migration and invasion of HeLa cells through modulating MAPK1.
Int J Mol Med. 44:1824–1832. 2019.PubMed/NCBI
|
|
18
|
Jiang B, Zhu SJ, Xiao SS and Xue M:
miR-217 inhibits M2-Like macrophage polarization by suppressing
secretion of interleukin-6 in ovarian cancer. Inflammation.
42:1517–1529. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang CL, Zheng XL, Ye K and Xue M: Effects
of microRNA-217 on proliferation, apoptosis, and autophagy of
hepatocytes in rat models of CCL4-induced liver injury by targeting
NAT2. J Cell Physiol. 234:3410–3424. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu C, Zhang Z and Qi D: Circular RNA
hsa_circ_0023404 promotes proliferation, migration and invasion in
non-small cell lung cancer by regulating miR-217/ZEB1 axis. Onco
Targets Ther. 12:6181–6189. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vaziri H, Dessain SK, Ng Eaton E, Imai SI,
Frye RA, Pandita TK, Guarente L and Weinberg RA: hSIR2(SIRT1)
functions as an NAD-dependent p53 deacetylase. Cell. 107:149–159.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wong S and Weber JD: Deacetylation of the
retinoblastoma tumour suppressor protein by SIRT1. Biochem J.
407:451–460. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang C, Chen L, Hou X, Li Z, Kabra N, Ma
Y, Nemoto S, Finkel T, Gu W, Cress WD and Chen J: Interactions
between E2F1 and SirT1 regulate apoptotic response to DNA damage.
Nat Cell Biol. 8:1025–1031. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huffman DM, Grizzle WE, Bamman MM, Kim JS,
Eltoum IA, Elgavish A and Nagy TR: SIRT1 is significantly elevated
in mouse and human prostate cancer. Cancer Res. 67:6612–6618. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Alves-Fernandes DK and Jasiulionis MG: The
role of SIRT1 on DNA damage response and epigenetic alterations in
cancer. Int J Mol Sci. 20:31532019. View Article : Google Scholar
|
|
26
|
Jiang W, Hou L, Wei J, Du Y, Zhao Y, Deng
X and Lin X: Hsa-miR-217 inhibits the proliferation, migration, and
invasion in non-small cell lung cancer cells via targeting SIRT1
and P53/KAI1 signaling. Balkan Med J. 37:208–214. 2020.PubMed/NCBI
|
|
27
|
Zhang L, Chen J, He Q, Chao Z, Li X and
Chen M: MicroRNA-217 is involved in the progression of
atherosclerosis through regulating inflammatory responses by
targeting sirtuin 1. Mol Med Rep. 20:3182–3190. 2019.PubMed/NCBI
|
|
28
|
Xia K, Zhang Y and Sun D: miR-217 and
miR-543 downregulation mitigates inflammatory response and
myocardial injury in children with viral myocarditis by regulating
the SIRT1/AMPK/NF-κB signaling pathway. Int J Mol Med. 45:634–646.
2020.PubMed/NCBI
|
|
29
|
Salomone F, Barbagallo I, Godos J, Lembo
V, Currenti W, Cinà D, Avola R, D'Orazio N, Morisco F, Galvano F
and Li Volti G: Silibinin restores NAD(+) levels and induces the
SIRT1/AMPK pathway in non-alcoholic fatty liver. Nutrients.
9:102017. View Article : Google Scholar
|
|
30
|
Patra KC, Weerasekara VK and Bardeesy N:
AMPK-mediated lysosome biogenesis in lung cancer growth. Cell
Metab. 29:238–240. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li C, Dong Y, Wang L, Xu G, Yang Q, Tang
X, Qiao Y and Cong Z: Ginsenoside metabolite compound K induces
apoptosis and autophagy in non-small cell lung cancer cells via
AMPK-mTOR and JNK pathways. Biochem Cell Biol. 97:406–414. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
He K, Guo X, Liu Y, Li J, Hu Y, Wang D and
Song J: TUFM downregulation induces epithelial-mesenchymal
transition and invasion in lung cancer cells via a mechanism
involving AMPK-GSK3β signaling. Cell Mol Life Sci. 73:2105–2121.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan G, Ru Y, Wu K, Yan F, Wang Q, Wang J,
Pan T, Zhang M, Han H, Li X and Zou L: GOLM1 promotes prostate
cancer progression through activating PI3K-AKT-mTOR signaling.
Prostate. 78:166–177. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu G, Pei F, Yang F, Li L, Amin AD, Liu
S, Buchan JR and Cho WC: Role of autophagy and apoptosis in
non-small-cell lung cancer. Int J Mol Sci. 18:3672017. View Article : Google Scholar
|
|
35
|
Yu T, Zhao Y, Hu Z, Li J, Chu D, Zhang J,
Li Z, Chen B, Zhang X, Pan H, et al: MetaLnc9 facilitates lung
cancer metastasis via a PGK1-activated AKT/mTOR pathway. Cancer
Res. 77:5782–5794. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao W, Peng F, Shu M, Liu H, Hou X, Wang
X, Ye J, Zhao B, Wang K, Zhong C, et al: Isogambogenic acid
inhibits the growth of glioma through activation of the AMPK-mTOR
pathway. Cell Physiol Biochem. 44:1381–1395. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dong LX, Sun LL, Zhang X, Pan L, Lian LJ,
Chen Z and Zhong DS: Negative regulation of mTOR activity by
LKB1-AMPK signaling in non-small cell lung cancer cells. Acta
Pharmacol Sin. 34:314–318. 2013. View Article : Google Scholar : PubMed/NCBI
|