Introduction
Breast cancer is the most common cancer in the
world, and in the United States breast cancer alone accounted for
30% of all new cancer diagnoses in women in 2019 (1). The 5-year survival of patients with
breast cancer has been improved due to recent advances in surgical
therapy, radiotherapy, hormone therapy and immunotherapy. The death
rate for patients with breast cancer dropped by 40% between 1989
and 2016 (1,2). The main causes of mortality are
attributed to distant metastasis and disease recurrence (3). Early diagnosis of breast cancer is
crucial for effective treatment (4).
Determining some novel biomarkers and targets remains therefore
crucial to develop efficient therapies for breast cancer.
DEK proto-oncogene (DEK) is a highly conserved
endogenous DNA-binding chromatin nuclear factor that encodes a 375
amino acid protein (5). DEK was
initially described as part of the protein product of the DEK-CAN
fusion oncogene generated by a t (6;9) translocation in a subset of
patients with acute myelogenous leukemia (6,7). DEK is
one of only two known secreted nuclear chromatin factors. Its
ability to bind nucleic acids leads to the regulation of numerous
cellular processes, including the regulation of hematopoiesis,
global heterochromatin integrity, DNA replication, gene
transcription and DNA repair (5,8–12). DEK has therefore been associated with
tumor formation and development. DEK has been reported to be
overexpressed in numerous types of tumor, including lung cancer,
gastric adenocarcinoma, cervical cancer, pancreatic cancer,
hepatocellular carcinoma, ovarian cancer and oropharyngeal squamous
cell carcinoma (13–22). In addition, high expression of DEK
has been associated with low overall survival in patients with lung
cancer (23). Silencing of DEK and
inhibitor of nuclear factor kappa B can block the cell cycle in the
G0/G1 phase, with a corresponding decrease in
the G2/M phase, increased apoptosis and induced cell
senescence in CaSki cervical cancer cells (24). These findings indicate that DEK might
have an oncogenic role in tumorigenesis and neoplastic progression;
however, the protein expression and role of DEK in breast cancer
have not been extensively investigated.
In the present study, the association between DEK
expression and the clinicopathological characteristics of patients
with breast cancer was determined. In addition, the function of DEK
in the proliferative and invasive abilities of MCF7 breast cancer
cells was investigated to elucidate the significance of DEK in the
progression of breast cancer.
Materials and methods
Patients and clinicopathological
characteristics
A total of 110 patients with invasive ductal cancer
were randomly selected and included in the present study. These
patients underwent surgical resection at the First Affiliated
Hospital of China Medical University (Shenyang, Liaoning, China)
between January 2011 and December 2016. Tissues from 50 cases of
breast cancer were matched with adjacent normal breast tissues
(>2-cm away from the tumor). All patients were women and none
underwent chemotherapy or radiotherapy prior to surgical resection.
All tissue specimens were fixed with 10% neutral formalin at room
temperature after surgery for pathological examination and were
diagnosed as invasive ductal cancers by pathological examination.
The mean age of patients was 58 years (age range, 31–85 years). The
clinicopathological characteristics of patients, including age,
tumor differentiation, histological grade, lymph node metastasis,
tumor-node-metastasis (TNM) stage, and expression of estrogen
receptor (ER), progesterone receptor (PR), human epidermal growth
factor receptor 2 (Her-2) and Ki-67, were retrospectively
investigated. The TNM stage of patients with breast cancer was
classified as stages I–II (n=62) and stages III–IV (n=48) according
to the TNM staging system of the International Union Against Cancer
(25). The research protocol was
reviewed and approved by the local Institutional Review Board of
the China Medical University.
Immunohistochemistry
For immunohistochemical analysis, all tumor
specimens were fixed in 10% neutral formalin for 24 h at room
temperature and embedded in paraffin blocks. Sections (4-µm thick)
were cut and placed onto glass slides precoated with 2%
3-aminopropyl triethoxysilane for 1 h at room temperature (Fuzhou
Maixin Biotech Co., Ltd.). Immunostaining was performed using the
streptavidin-peroxidase complex method. The sections were
deparaffinized in xylene at room temperature, rehydrated in an
85–95% anhydrous alcohol gradient series and boiled in 0.01 M
citrate buffer (pH 6.0) for 2 min in an autoclave. Endogenous
peroxidase activity was blocked using 0.3% hydrogen peroxide at
37°C for 10 min and sections were subsequently incubated with 10%
normal goat serum (Fuzhou Maixin Biotech Co., Ltd.) at 37°C to
reduce nonspecific binding. The sections were incubated with the
rabbit polyclonal antibody against DEK (cat. no. 16448-1-AP; 1:150;
ProteinTech Group, Inc.) or the ready-to-use primary antibodies
against ER (cat. no. MAB-0062), PR (cat. no. MAB-0675), Her-2 (cat.
no. MAB-0198) and Ki-67 (cat. no. MAB-0672) (ready to use; all
Fuzhou Maixin Biotech Co., Ltd.) at 4°C overnight. Section stained
with PBS only was considered as a negative control. After washing
with PBS, the sections were incubated for 30 min at 37°C with
secondary biotinylated goat anti-rabbit serum IgG antibody (cat.
no. SPKIT-C2) and horseradish peroxidase-conjugated
streptavidin-biotin (cat. no. SPKIT-A2) (ready to use; all Fuzhou
Maixin Biotech Co., Ltd.). The staining was visualized using
3,3-diaminobenzidine (Fuzhou Maixin Biotech Co., Ltd.). Sections
were then stained with hematoxylin for 10 min at 37°C and observed
under ×400 magnification using a light microscope (Olympus
Corporation).
The evaluation of immunostaining was performed
semi-quantitatively. Nuclear staining of the tumor cells was
considered to be DEK positive. In total, 10 high-power
representative fields were selected per slide, and the staining
intensity and positive rate of tumor cells were scored. The
intensity of the staining was scored as follows: 0, negative; 1,
weak; 2, intermediate; and 3, strong. The positive rate for each
case was obtained by calculating the percentage of positively
stained tumor cells on each slide and was scored as follows: 0,
negative; 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, >75%. The
scores for each tumor sample were multiplied to obtain a final
score of 0 to 12. A final score ≥6 was defined as high DEK
expression whereas a score <6 was defined as low DEK expression
(23,26).
The Cancer Genome Atlas (TCGA) data
collection and analysis
The expression data of DEK in breast cancers
molecular subtypes were obtained using the online database UALCAN
(http://ualcan.path.uab.edu) (TCGA
dataset of breast invasive carcinoma, n=719) (27). Correlation analysis between DEK
expression and Ki-67 and PCNA expression in breast cancers was
retrieved from cBioPortal database (http://www.cbioportal.org/) [mRNA expression
(microarray), n=1,904] (28,29). Survival curve for patients with
breast cancer with high or low DEK expression (n=4,929) was
obtained from Kaplan-Meier plotter (http://kmplot.com/analysis/) (30).
Cell lines and transfection
The human breast cancer cell line MCF7 (luminal A
subtype) was purchased from The Cell Bank of Type Culture
Collection of The Chinese Academy of Sciences. Cells were cultured
in DMEM (Invitrogen, Thermo Fisher Scientific, Inc.), supplemented
with 10% FBS (cat. no. FB15015; Clark Bioscience) and placed at
37°C in a humidified incubator containing 5% CO2.
For transfection, MCF7 cells were seeded into 6-well
plates for 24 h at 37°C and cultured to 70–80% confluence before
transfection. The plasmids containing the DEK gene (pCMV6-DEK) or
DEK short hairpin RNA (shRNA) sequences (pCMV6-shDEK) were
synthesized by GENECHEM (Shanghai GeneChem Co., Ltd.). The
corresponding empty vector pCMV6 or plasmid containing scrambled
shRNA sequences served as negative controls. The plasmids (2.5 µg)
were transfected into cells using Lipofectamine™ 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to manufacturers'
instructions at 37°C. Subsequent experiments were performed 24 h
after transfection.
Western blotting
Cells were lysed using RIPA lysis buffer at 4°C
(Pierce; Thermo Fisher Scientific, Inc.) and proteins were
quantified using the Bradford method (31). Proteins (60 µg) were separated by 10%
SDS-PAGE and transferred onto PVDF membranes. Membranes were
blocked in 5% non-fat milk for 1 h at room temperature. The
membranes were incubated with primary antibodies against DEK (cat.
no. 16448-1-AP; 1:1,000; ProteinTech Group, Inc.), glycogen
synthase kinase-3β (Gsk-3β; cat. no. 5676; 1:1,000; Cell Signaling
Technology, Inc.), cyclin D1 (cat. no. SC-8396; 1:100; Santa Cruz
Biotechnology, Inc.), β-catenin (cat. no. 17565-1-AP), active
β-catenin (cat. no. 51067-2-AP), c-Myc (cat. no. 67447-1-Ig) and
GAPDH (cat. no. 60004-1-Ig) (all 1:1,000; ProteinTech Group, Inc.)
overnight at 4°C. Membranes were washed with tris-buffered saline
containing 0.1% Tween-20 and were incubated with IgG antibody (cat.
no. SA00001-1/2; 1:2,000; ProteinTech Group, Inc.) at 37°C for 2 h.
Bands were detected using enhanced chemiluminescence substrate
(Pierce; Thermo Fisher Scientific, Inc.) and detected using a
bioimaging system (DNR Bio-Imaging System, Ltd.). All the western
blotting bands were probed from the same membrane. After each
probing, the membranes were stripped using a stripping buffer
(Beyotime Institute of Biotechnology) according to the
manufacturers' protocol and re-probed for other proteins. Relative
expression levels were normalized to endogenous control GAPDH,
which were analyzed with ImageJ software (version 1.47; National
Institutes of Health).
Colony formation assay
Cells were seeded in 6 cm cell culture dishes (1,000
cells per dish) 24 h after transfection and were cultured for 10
days. The medium was changed every 3 days. Cells were then washed
with PBS and stained with hematoxylin for 10 min at room
temperature. The number of colonies with >50 cells were counted
using a bioimaging system (version 5.2.1; DNR Bio-Imaging Systems,
Ltd.).
Cell proliferation assay
Cell proliferation was detected using Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.). Briefly, 24 h
after transfection, cells were seeded into 96-well plates (3,000
cells per well) in 100 ul medium containing 10% FBS. CCK-8 reagent
was added to each well (1:10, v/v) and incubated for 2 h at 37°C.
Cell proliferation was assessed at days 1, 2, 3, 4 and 5. The
absorbance was read at 450 nm using a microplate reader.
Cell migration and invasion
assays
Cell migratory and invasive abilities were assessed
using 24-well transwell chambers containing inserts of 8 µm pore
size (Costar; Corning, Inc.). For the invasion assay only, the
upper side of the inserts were coated with Matrigel for at least 2
h at 37°C. (1:8; BD Biosciences). MCF7 cells were seeded
(1.5×105 cells/well) in the upper chambers in 100 µl
medium supplemented with 2% FBS. To attract cells, the lower
chambers were filled with 600 µl medium containing 20% FBS. After
20 h, cells that had migrated to the lower chambers were fixed with
4% paraformaldehyde and stained with hematoxylin for 10 min at
37°C. The non-invading cells on the upper surface were cleared
using a cotton swab. A total of 10 randomly selected high-power
fields were observed under light microscopy (magnification, ×200),
and the numbers of migrated or invaded cells were counted (Motic
Image Plus 2.0; Motic (Xiamen) Medical Diagnostic Systems Co. Ltd.)
All experiments described were performed independently and in
triplicate.
Statistical analysis
Statistical analysis was performed using SPSS
version 17.0 (IBM Corp.). Associations between DEK expression and
the clinicopathological characteristics of patients were analyzed
using χ2 test and Student's t-test. Comparisons among
two experimental groups were performed using two-tailed Student's
t-test. The P-value threshold was adjusted using Bonferroni
correction when comparing more than two groups using Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
DEK protein expression is higher in
breast cancer tissues compared with normal breast tissues
Immunohistochemistry was performed for 110 cases of
breast cancer and 50 cases of adjacent normal breast tissues. The
results demonstrated that DEK protein was mainly expressed in the
nuclei of cancer cells. Furthermore, high DEK expression was
observed in 62.7% (69/110) of breast cancer tissues, which was
significantly higher than that in normal breast tissues (12.0%;
P=0.002; Table I and Fig. 1A-C).
 | Table I.Association between DEK expression
and the clinicopathological characteristics of patients with breast
cancer. |
Table I.
Association between DEK expression
and the clinicopathological characteristics of patients with breast
cancer.
|
|
| DEK |
|
|---|
|
|
|
|
|
|---|
| Variable | Cases | High
expression | Low expression | P-value |
|---|
| Tissue |
|
|
| 0.002 |
| Normal
breast tissue | 50 | 6 | 44 |
|
| Breast
cancer | 110 | 69 | 41 |
|
| Histological
grade |
|
|
| 0.010 |
| I | 16 | 5 | 11 |
|
|
II–III | 94 | 64 | 30 |
|
| TNM stage |
|
|
| 0.030 |
|
I–II | 62 | 33 | 29 |
|
|
III–IV | 48 | 36 | 12 |
|
| Lymph node
metastasis |
|
|
| 0.003 |
|
Negative | 68 | 35 | 33 |
|
|
Positive | 42 | 34 | 8 |
|
| Ki-67 index |
|
|
| 0.028 |
|
High | 31 | 25 | 6 |
|
|
Low | 79 | 44 | 35 |
|
| ER expression |
|
|
| 0.070 |
|
Positive | 70 | 39 | 31 |
|
|
Negative | 40 | 30 | 10 |
|
| PR expression |
|
|
| 0.510 |
|
Positive | 64 | 38 | 26 |
|
|
Negative | 46 | 31 | 15 |
|
| HER-2
expression |
|
|
| 0.420 |
|
Positive | 48 | 27 | 20 |
|
|
Negative | 63 | 42 | 21 |
|
| Age, years |
|
|
| 0.290 |
|
<51 | 78 | 46 | 33 |
|
|
≥51 | 32 | 23 | 8 |
|
High DEK expression is associated with
certain clinicopathological characteristics and poor prognosis of
patients
The expression of DEK was associated with
histological grade (P=0.01), lymph node metastasis (P=0.003), TNM
stage (P=0.030) and Ki-67 expression (P=0.028); however, DEK
expression was not associated with ER expression (P=0.070), PR
expression (P=0.510), Her-2 expression (P=0.420) or patient age
(P=0.290; Table I and Fig. 1). Furthermore, according to the
cBioPortal database, it was also found that DEK expression was
positively correlated with the expression of Ki-67 (P<0.001;
n=1,904) or proliferating cell nuclear antigen (PCNA; P<0.001;
n=1,904) (Fig. 2A and B), which are
both indexes of proliferation (32).
In addition, according to Kaplan-Meier analysis
using the online database KM-plotter, patients with high expression
of DEK presented a significant shorter overall survival compared
with patients with low DEK expression (P<0.01; median survival,
163.46 months for the high expression cohort vs. 216.66 months for
the low expression cohort; Fig. 2C).
As seen in the UALCAN database, the expression of DEK was
significantly higher in triple negative breast cancer (TNBC)
compared with luminal breast cancer (P<0.001) and Her-2 positive
breast cancer (P<0.001). However, the expression level of DEK
was not significant different between luminal and Her2 positive
subtypes of breast cancer (P=0.123; Fig.
2D).
DEK regulates the expression of
β-catenin and target genes of Wnt signaling pathway
Enhanced DEK expression by DEK gene transfection in
MCF7 cells (MCF7-DEK) increased the expression of active-β-catenin
and inhibited the expression of Gsk-3β (P<0.05). The expression
of cyclin D1 and c-Myc, which are target genes of the Wnt signaling
pathway, was also significantly increased in MCF7-DEK cells
(P<0.05). However, the level of total β-catenin was not markedly
changed following DEK overexpression (P>0.05; Fig. 3A and B). Conversely, following DEK
knockdown by shRNA interference (MCF7-ShDEK), the expression of
active-β-catenin, cyclin D1 and c-Myc was significantly
downregulated, whereas the expression of Gsk-3β was significantly
increased in MCF7 cells (P<0.05). The level of total β-catenin
was not changed following DEK knockdown (P>0.05; Fig. 3C and D).
DEK overexpression promotes the
proliferation, colony formation and migratory and invasive
abilities of breast cancer cells
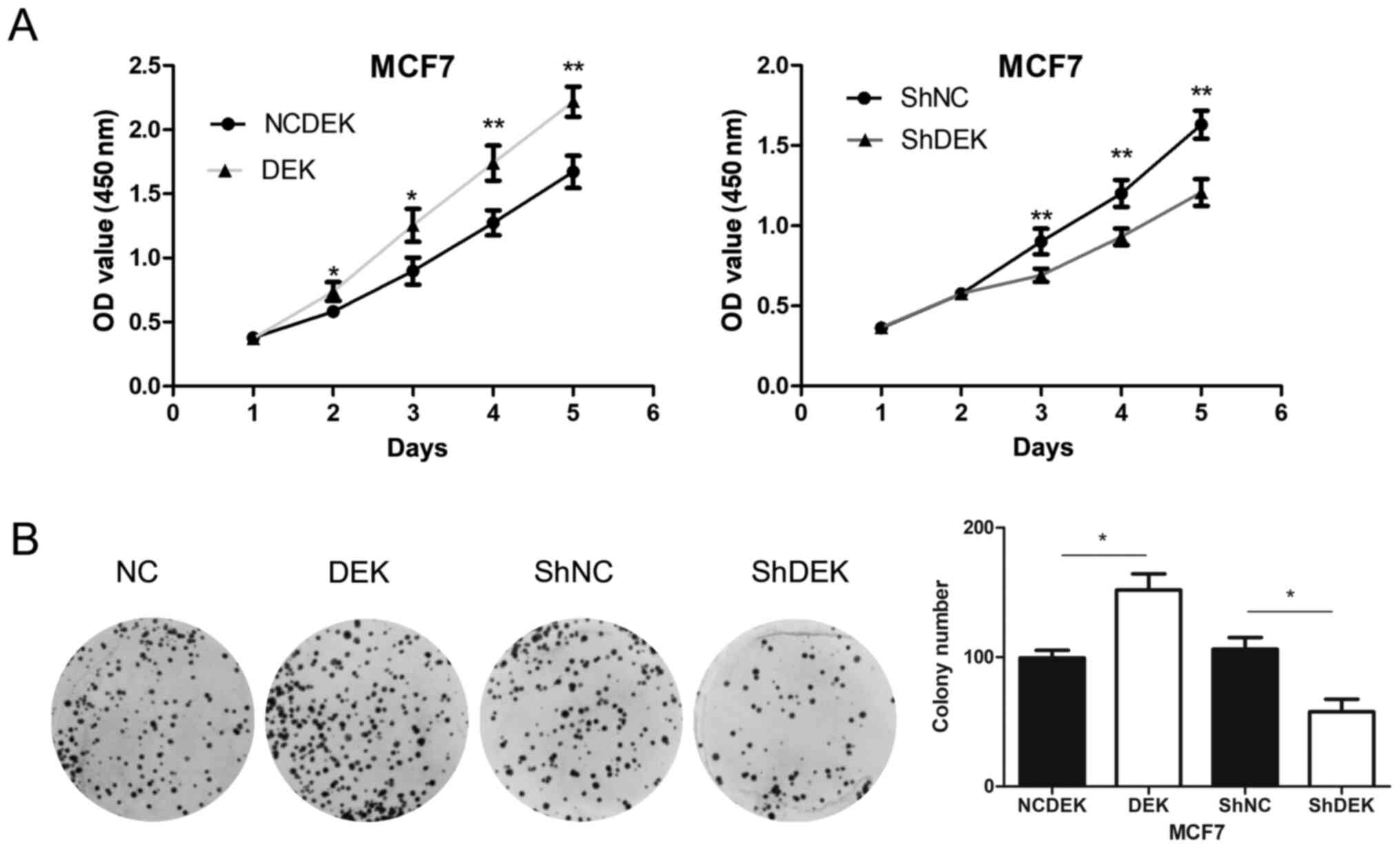
Overexpression of DEK enhanced the proliferation
rate (P<0.05 for days 1 and 2; P<0.01 for days 3 and 4) and
the colony formation (P<0.05) of MCF7 cells compared with
control cells. Conversely, following DEK knockdown, the
proliferation rate (P<0.01 for days 3, 4 and 5) and colony
formation (P<0.05) of MCF7 cells were significantly inhibited
compared with control cells (Fig. 4A and
B).
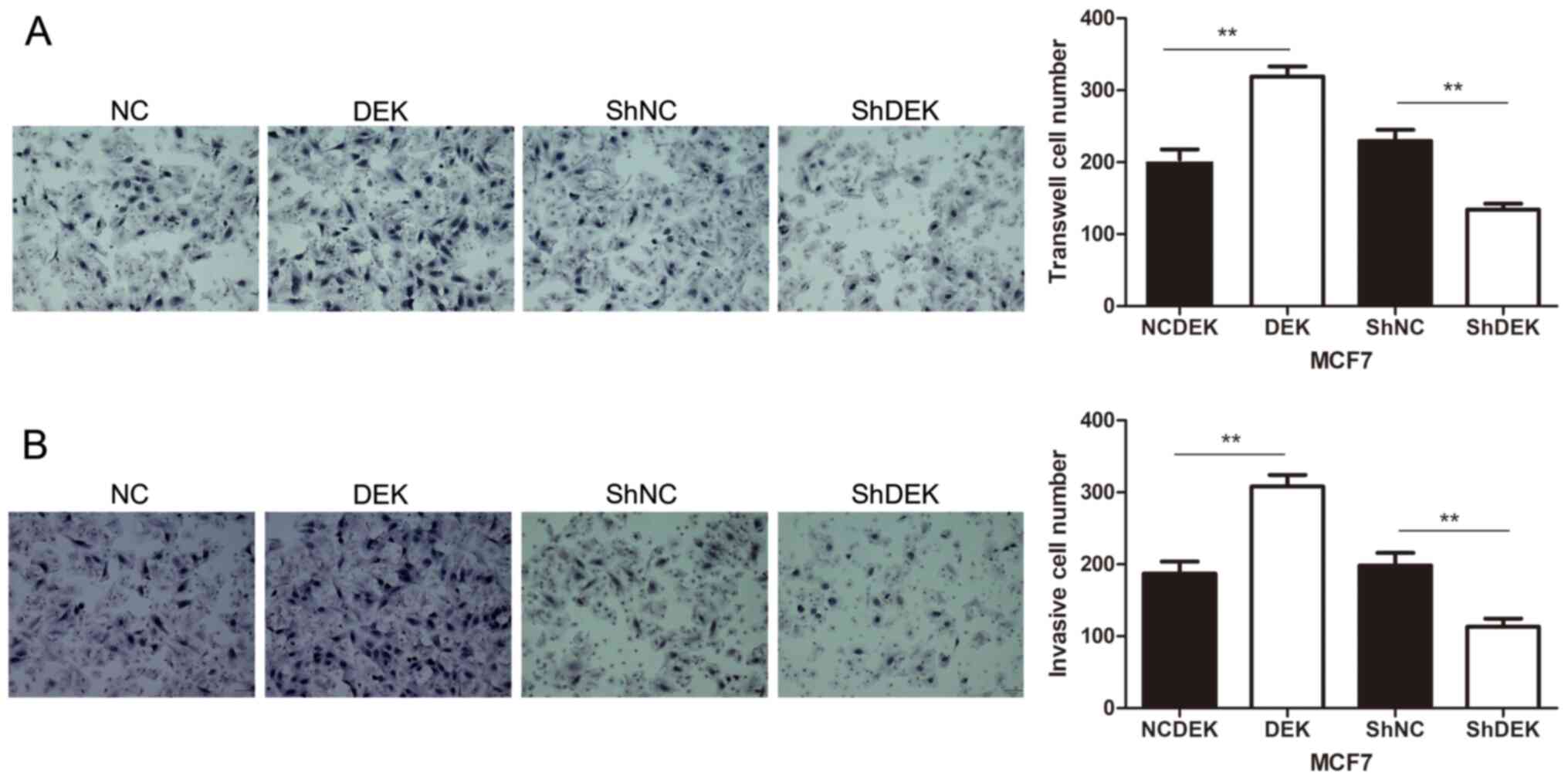
Furthermore, DEK overexpression promoted the
migratory and invasive abilities of MCF7 cells compared with
control cells (P<0.01). Conversely, DEK downregulation inhibited
the migratory and invasive abilities of MCF7 cells (P<0.01;
Fig. 5A and B).
Discussion
In order to accurately diagnose and treat cancers,
researchers are working to determine novel effective markers to
improve the clinical evaluation of outcomes and to develop targeted
therapy. DEK has been reported to be overexpressed in numerous
types of human cancer, including lung cancer, pancreatic cancer,
gastric adenocarcinoma, ovarian cancer, cervical cancer,
hepatocellular cancer and TNBC (13,15–19,21,33). DEK
plays an active role in tumor initiation and maintenance. The
expression of DEK has been associated with the clinicopathological
characteristics of patients with hepatocellular cancers and
pancreatic adenocarcinomas and described as an indicator of poor
prognosis (11,16), suggesting that DEK may be considered
as a potential prognostic biomarker in various types of cancer.
Although DEK overexpression has been reported in
breast cancer (34–36), the expression pattern of DEK and its
association with the clinicopathological characteristics of
patients remain unclear. The present study demonstrated that DEK
protein was present in cancer cell nucleus and that its expression
was higher in breast cancer tissues compared with normal breast
tissues. Furthermore, high DEK expression was associated with a
high grade, advanced TNM stage and high index of proliferation,
which is characterized by a high expression of Ki67 or PCNA, in
patients with breast cancer. In addition, high expression of DEK
could predict a poor prognosis in patients with breast cancer,
suggesting that DEK may be considered as a potentially valuable
prognostic marker in breast cancer. As seen in UALCAN database, DEK
expression level was significantly higher in TNBC compared with
luminal and Her-2 positive breast cancers; however, similar results
were not observed in the population from the present study; which
may be due to the limited sample size and the different types of
detection methods. The present study only examined the protein
expression of DEK using immunohistochemistry in paraffin embedded
samples. The mRNA expression of DEK was not detected in the present
study and should be investigated in the future using fresh breast
cancer tissues.
DEK can regulate the proliferation, migration,
invasion and apoptosis of cancer cells and be subjected to a
variety of tumor-associated modifications (37,38). It
has been reported that DEK knockdown can inhibit the proliferation
of ovarian, lung and cervical cancers (15,23,39). The
present study demonstrated that DEK overexpression promoted the
proliferation, colony formation and invasive and migratory
abilities of MCF7 cells, which was consistent with the results
in vivo.
DEK is involved in cancer progression through the
regulation of numerous signaling pathways. For example, DEK
expression is regulated by the transcription factors Nuclear
Factor-Y and Yin Yang-1 (40) and
can be induced by high-risk human papillomavirus E7 to overcome
cellular senescence (41). In
addition, DEK is a regulator of the G1 to S transition
and a potential target gene of the p16-pRB-E2F pathway (42). DEK regulates apoptosis in
glioblastomas partly through modulating p53 by inhibiting its
transcription activity and protein stability (38). Furthermore, blocking the
PI3K/AKT/mammalian target of rapamycin pathway using specific
inhibitors can significantly attenuate DEK-enhanced migration and
angiogenesis in TNBCs (33). In
cervical cancer, DEK promotes Hela cell metastasis via upregulation
of the Wnt pathway and matrix metalloproteinase-9 expression
(39). The results from the present
study demonstrated that DEK could upregulate the expression of
active β-catenin and Wnt target genes, such as cyclin D1 and c-Myc.
DEK may therefore promote the proliferation and invasive ability of
breast cancer by activating the Wnt signaling pathway. Taken
together, these findings suggested that DEK may act as an oncogene
and promote breast cancer development; however, the underlying
oncogenic mechanism of DEK in breast cancer requires further
investigation.
In conclusion, the results from the present
demonstrated that high expression of DEK was common in breast
cancer tissues. In addition, DEK overexpression promoted the
proliferation and invasive ability of breast cancer cells in
vitro, and was associated with high grade, advanced TNM stage
and poor prognosis in patients with breast cancer.
Acknowledgements
Not applicable.
Funding
This study was supported by the Youth Scientific
Research Foundation of Shenyang Medical College (grant no.
20182048) and the Natural Science Foundation of Liaoning Province
(grant no. 2020-MS-179).
Availability of data and materials
The datasets used during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
HTX designed the study. MQY and HTX participated in
drafting the manuscript. MQY, LLB, ZW, LL, YWZ, ZHL, WJH and CCL
performed the experiments. MQY and HTX confirm the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The research protocol was reviewed and approved by
the local institutional review board at the China Medical
University. Written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ER
|
estrogen receptor
|
|
Gsk
|
glycogen synthase kinase
|
|
Her-2
|
human epidermal growth factor receptor
2
|
|
PR
|
progesterone receptor
|
|
TCGA
|
The Cancer Genome Atlas
|
|
TNM
|
tumor-node-metastasis
|
|
TNBC
|
triple negative breast cancer
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gonzalez-Angulo AM, Morales-Vasquez F and
Hortobagyi GN: Overview of resistance to systemic therapy in
patients with breast cancer. Adv Exp Med Biol. 608:1–22. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Redig AJ and McAllister SS: Breast cancer
as a systemic disease: A view of metastasis. J Intern Med.
274:113–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kappes F, Scholten I, Richter N, Gruss C
and Waldmann T: Functional domains of the ubiquitous chromatin
protein DEK. Mol Cell Biol. 24:6000–6010. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fu GK, Grosveld G and Markovitz DM: DEK,
an autoantigen involved in a chromosomal translocation in acute
myelogenous leukemia, binds to the HIV-2 enhancer. Proc Natl Acad
Sci USA. 94:1811–1815. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boer J, Mahmoud H, Raimondi S, Grosveld G
and Krance R: Loss of the DEK-CAN fusion transcript in a child with
t(6;9) acute myeloid leukemia following chemotherapy and allogeneic
bone marrow transplantation. Leukemia. 11:299–300. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McGarvey T, Rosonina E, McCracken S, Li Q,
Arnaout R, Mientjes E, Nickerson JA, Awrey D, Greenblatt J,
Grosveld G and Blencowe BJ: The acute myeloid leukemia-associated
protein, DEK, forms a splicing-dependent interaction with
exon-product complexes. J Cell Biol. 150:309–320. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kappes F, Burger K, Baack M, Fackelmayer
FO and Gruss C: Subcellular localization of the human
proto-oncogene protein DEK. J Biol Chem. 276:26317–26323. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Waldmann T, Eckerich C, Baack M and Gruss
C: The ubiquitous chromatin protein DEK alters the structure of DNA
by introducing positive supercoils. J Biol Chem. 277:24988–24994.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee SY, Jung W, Lee J, Kim A, Kim HK and
Kim BH: High expression of DEK is associated with poor prognosis in
hepatocellular carcinoma. Histol Histopathol. 34:1279–1288.
2019.PubMed/NCBI
|
|
12
|
Capitano ML, Mor-Vaknin N, Saha AK, Cooper
S, Legendre M, Guo H, Contreras-Galindo R, Kappes F, Sartor MA, Lee
CT, et al: Secreted nuclear protein DEK regulates hematopoiesis
through CXCR2 signaling. J Clin Invest. 129:2555–2570. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smith EA, Kumar B, Komurov K, Smith SM,
Brown NV, Zhao S, Kumar P, Teknos TN and Wells SI: DEK associates
with tumor stage and outcome in HPV16 positive oropharyngeal
squamous cell carcinoma. Oncotarget. 8:23414–23426. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Riveiro-Falkenbach E, Ruano Y,
Garcia-Martin RM, Lora D, Cifdaloz M, Acquadro F, Ballestín C,
Ortiz-Romero PL, Soengas MS and Rodríguez-Peralto JL: DEK oncogene
is overexpressed during melanoma progression. Pigment Cell Melanoma
Res. 30:194–202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hacker KE, Bolland DE, Tan L, Saha AK,
Niknafs YS, Markovitz DM and McLean K: The DEK oncoprotein
functions in ovarian cancer growth and survival. Neoplasia.
20:1209–1218. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao T, Qiu B, Zhou S, Ding G, Cao L and
Wu Z: Expression of DEK in pancreatic cancer and its correlation
with clinicopathological features and prognosis. J Cancer.
10:911–917. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Serrano-Lopez J, Nattamai K, Pease NA,
Shephard MS, Wellendorf AM, Sertorio M, Smith EA, Geiger H, Wells
SI, Cancelas JA and Privette Vinnedge LM: Loss of DEK induces
radioresistance of murine restricted hematopoietic progenitors. Exp
Hematol. 59:40–50.e3. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou QC, Deng XF, Yang J, Jiang H, Qiao
MX, Liu HH, Qian Z, Hou LL and Hu HG: Oncogene DEK is highly
expressed in lung cancerous tissues and positively regulates cell
proliferation as well as invasion. Oncol Lett. 15:8573–8581.
2018.PubMed/NCBI
|
|
19
|
Ou Y, Xia R, Kong F, Zhang X, Yu S, Jiang
L, Zheng L and Lin L: Overexpression of DEK is an indicator of poor
prognosis in patients with gastric adenocarcinoma. Oncol Lett.
11:1823–1828. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Matrka MC, Watanabe M, Muraleedharan R,
Lambert PF, Lane AN, Romick-Rosendale LE and Wells SI:
Overexpression of the human DEK oncogene reprograms cellular
metabolism and promotes glycolysis. PLoS One. 12:e01779522017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qiao MX, Li C, Zhang AQ, Hou LL, Yang J
and Hu HG: Regulation of DEK expression by AP-2α and methylation
level of DEK promoter in hepatocellular carcinoma. Oncol Rep.
36:2382–2390. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu Y, Liang Z, Li C, Yang Z and Chen L:
LCMR1 interacts with DEK to suppress apoptosis in lung cancer
cells. Mol Med Rep. 16:4159–4164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Sun L, Yang M, Luo W, Gao Y, Liu
Z, Qiu X and Wang E: DEK depletion negatively regulates
Rho/ROCK/MLC pathway in non-small cell lung cancer. J Histochem
Cytochem. 61:510–521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu K, Feng T, Liu J, Zhong M and Zhang S:
Silencing of the DEK gene induces apoptosis and senescence in CaSki
cervical carcinoma cells via the up-regulation of NF-κB p65. Biosci
Rep. 32:323–332. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cserni G, Chmielik E, Cserni B and Tot T:
The new TNM-based staging of breast cancer. Virchows Arch.
472:697–703. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Lei L, Zheng YW, Zhang L, Li ZH,
Shen HY, Jiang GY, Zhang XP, Wang EH and Xu HT: Odd-skipped related
1 inhibits lung cancer proliferation and invasion by reducing Wnt
signaling through the suppression of SOX9 and β-catenin. Cancer
Sci. 109:1799–1810. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Juríková M, Danihel Ľ, Polák Š and Varga
I: Ki67, PCNA, and MCM proteins: Markers of proliferation in the
diagnosis of breast cancer. Acta Histochem. 118:544–552. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang Y, Gao M, Lin Z, Chen L, Jin Y, Zhu
G, Wang Y and Jin T: DEK promoted EMT and angiogenesis through
regulating PI3K/AKT/mTOR pathway in triple-negative breast cancer.
Oncotarget. 8:98708–98722. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu S, Wang X, Sun F, Kong J, Li Z and Lin
Z: DEK overexpression is correlated with the clinical features of
breast cancer. Pathol Int. 62:176–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ying G and Wu Y: DEK: A novel early
screening and prognostic marker for breast cancer. Mol Med Rep.
12:7491–7495. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Privette Vinnedge LM, McClaine R, Wagh PK,
Wikenheiser-Brokamp KA, Waltz SE and Wells SI: The human DEK
oncogene stimulates β-catenin signaling, invasion and mammosphere
formation in breast cancer. Oncogene. 30:2741–2752. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Y, Liu J, Wang S, Luo X, Li Y, Lv Z,
Zhu J, Lin J, Ding L and Ye Q: The DEK oncogene activates VEGF
expression and promotes tumor angiogenesis and growth in
HIF-1α-dependent and -independent manners. Oncotarget.
7:23740–23756. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Feng T, Liu Y, Li C, Li Z and Cai H: DEK
proto-oncogene is highly expressed in astrocytic tumors and
regulates glioblastoma cell proliferation and apoptosis. Tumour
Biol. 39:10104283177162482017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu X, Zou L, Yao Q, Zhang Y, Gan L and
Tang L: Silencing DEK downregulates cervical cancer tumorigenesis
and metastasis via the DEK/p-Ser9-GSK-3β/p-Tyr216-GSK-3β/β-catenin
axis. Oncol Rep. 38:1035–1042. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sitwala KV, Adams K and Markovitz DM: YY1
and NF-Y binding sites regulate the transcriptional activity of the
dek and dek-can promoter. Oncogene. 21:8862–8870. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wise-Draper TM, Allen HV, Thobe MN, Jones
EE, Habash KB, Münger K and Wells SI: The human DEK proto-oncogene
is a senescence inhibitor and an upregulated target of high-risk
human papillomavirus E7. J Virol. 79:14309–14317. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Carro MS, Spiga FM, Quarto M, Di Ninni V,
Volorio S, Alcalay M and Müller H: DEK Expression is controlled by
E2F and deregulated in diverse tumor types. Cell Cycle.
5:1202–1207. 2006. View Article : Google Scholar : PubMed/NCBI
|