Introduction
The oral cavity is the most prevalent area of
malignancy in the head and neck region, with the GLOBOCAN 2020 data
estimating the annual incidence and mortality as ~377,713 new cases
and 177,757 deaths, respectively, for lip and oral cavity cancer,
which is the fourth most common type of cancer among males in
Taiwan (1–3). Squamous cell carcinoma constitutes the
most commonly seen histological type in patients with oral cavity
cancer (4). Radical surgery with or
without adjuvant chemo-radiotherapy is part of the primary
management for patients with oral squamous cell carcinoma (OSCC),
and radical surgery has proven to be valuable in loco-regional
disease control (5). Although 80–90%
of early OSCC cases are cured, the prognosis for patients with
advanced-stage OSCC remains poor (6,7). For
patients who have undergone standard management, OSCC recurrence
varies between 18 and 76%, and recurrence has been identified as
the major cause of poor survival rates (8–11).
Previous studies have indicated that the median time to recurrence
is 7.5 months after therapy, with 86% of recurrences occurring
among 24 months (12–14).
In Taiwan, cisplatin is the mainstay of chemotherapy
for locally advanced oral cancer treatment. Its crucial cytotoxic
activity is due to the formation of DNA adducts, which result in
inter-strand and intra-strand cross-linking (15,16).
These DNA cross-links are identified and eliminated by the
nucleotide excision repair (NER) pathway protecting the integrity
of the genome (17,18). Tumor resistance to this platinum
complex seems to be multifactorial, with the NER pathway serving a
crucial role (19). NER is a
stepwise procedure of recognition, incision, excision, DNA
synthesis and ligation, which is executed by a multienzyme complex
(20,21). Previous studies have investigated the
association between gene expression and the effects of various
chemotherapeutic agents in cancer (22–24),
such as excision repair cross-complementing group 1 (ERCC1) being
identified as a marker for resistance to cisplatin in
non-small-cell lung cancer. Platinum resistance has been attributed
to enhanced repair of DNA damage via the NER pathway, which
consists of X-ray cross-complementing 1 (XRCC1), ERCC1 and ERCC2
(25–28).
Concurrent chemo-radiotherapy is regularly used for
locally advanced oral cancer treatment. DNA single-strand breaks
may take place directly from the damage to deoxyribose, or
indirectly as the ordinary intermediates of DNA base excision
repair (29). Since single-strand
breaks are provoked by endogenous reactive molecules, such as
reactive oxygen species, these injuries create a sustained threat
to genetic integrity (29). In
mammalian cells, the XRCC1 protein plays a leading part in the
repair of single-strand breaks via its capability to interact with
multiple enzymatic complexes of restoration (29). Hypersensitivity to oxidative stress,
ionizing radiation and alkylating agents has been observed in cells
lacking XRCC1 (29). A previous
study has indicated that high expression of both XRCC1 and ERCC1 is
significantly associated with radioresistant laryngeal carcinoma
(30).
XRCC1, ERCC1 and ERCC2 are well-reported DNA repair
proteins, implying their involvement in resistance to
chemo-radiotherapy; however, to the best of our knowledge, no
studies have explored the association of these NER
pathway-associated proteins with recurrence and prognosis in
patients with OSCC. Therefore, the current study hypothesized that
high protein expression levels of XRCC1, ERCC1 and ERCC2 may lead
to treatment resistance and recurrence or poor clinical outcomes in
patients with OSCC.
Materials and methods
Cell culture
Human oral keratinocytes (HOK; ScienCell Research
Laboratories, Inc.) were incubated with Oral Keratinocyte Medium
(ScienCell Research Laboratories, Inc.) in plates pre-coated with 2
µg/cm2 poly-L-lysine. DOK oral precancerous cells were
grown in DMEM supplemented with 10% FBS (both Thermo Fisher
Scientific, Inc.), penicillin (100 U/ml), streptomycin (100 µg/ml),
glutamine (2 mM) and hydrocortisone (5 µg/ml). Oral cancer SAS,
OECM1, HSC-3 and Cal-27 cells were grown in Eagle's Minimum
Essential Medium (Thermo Fisher Scientific, Inc.) with glutamine (2
mM) and FBS (10%), while human malignant glioma U87MG cells
(American Type Culture Collection; glioblastoma of unknown origin)
were grown in DMEM supplemented with 10% FBS. All cells were
incubated in 5% CO2 at 37°C.
Western blotting
Total cell lysates were extracted using RIPA buffer
(Thermo Fisher Scientific, Inc.) and protein concentrations were
detected using the BCA protein assay (Bio-Rad Laboratories, Inc.).
Protein lysates (20 µg/lane) were separated via 10% SDS-PAGE and
transferred to a PVDF membrane, which was then blocked for 1 h at
room temperature with 5% non-fat milk in TBS-0.1% Tween-20 (TBST).
Subsequently, the membrane was incubated overnight at 4°C with
primary antibodies including XRCC1 monoclonal antibody (1:1,000;
cat. no. GTX83411; GeneTex, Inc.), ERCC1 monoclonal antibody
(1:1,000; cat. no. GTX22356; GeneTex, Inc.), ERCC2 polyclonal
antibody (1:1,000; cat. no. GTX105357; GeneTex, Inc.) and β-actin
antibody (1:10,000; cat. no. GTX629630; GeneTex, Inc.). After
washing three times with TBST for 10 min, the membrane was
incubated with HRP-conjugated secondary antibody (1:5,000; cat. no.
GTX213110-01; GeneTex, Inc.) for 2 h at room temperature. The
immunoblots were visualized using Chemiluminescence Reagent Plus
(PerkinElmer, Inc.) and quantified using Image Lab™ software
version 5.1 (Bio-Rad Laboratories, Inc.).
Oncomine™ platform
To study the mRNA expression levels of XRCC1, ERCC1
and ERCC2 in oral cancer and normal oral tissues, Oncomine™ [Estilo
Head-Neck: XRCC1160033_s_at (31),
Peng Head-Neck: XRCC13864445 (32),
Cromer Head-Neck: ERCC11902_at (33), Ginos Head-Neck: ERCC1203720_s_at
(34), Peng Head-Neck: ERCC13865378
(32), Peng Head-Neck: ERCC23865301
(32), Ginos Head-Neck:
ERCC2213468_at (34), Cromer
Head-Neck: ERCC241095_at (33)], an
integrated cancer microarray database and web-based data-mining
platform (35), was used.
Patients
Between September 2002 and December 2011, a total of
98 patients with OSCC (92 men and 6 women) were enrolled from the
Department of Oral and Maxillofacial Surgery of Kaohsiung Medical
University Hospital (Kaohsiung, Taiwan), with a median follow-up
time of 40 months (range, 2.4–137.4 months). G*Power (version
3.1.9.4; http://ps-power-and-sample-size-calculation.software.informer.com/3.1/),
a freely available windows application software, was used for
sample size and power estimation, with α=0.05 and estimated effect
size w=0.46. A total of 98 patients were recruited in the
present study to achieve sufficient power of ≥90%. The current
study was approved by the Institutional Review Board of Kaohsiung
Medical University Hospital (approval no. 20140158) and patient
informed consent was waived by the Institutional Review Board due
to the retrospective nature of the study. OSCC pathology was
determined by two pathologists independently, and the final
diagnoses were made using clinical and histological data. Patients
without previous history of any treatment for oral cancer were
included. Patients who were <18 or >80 years old were
excluded. Baseline characteristic data included patient age, sex,
tumor location, grade, tumor size, lymph node metastases and tumor
stage; additionally, substance use, such as alcohol consumption,
betel nut chewing or cigarette smoking, and adjunct treatment
details were recorded (Table I). The
mean age of the study group was 51.4 years and the median age was
51 years (age range, 31–76 years). Clinical staging of the patients
was determined using the TNM staging system according to the 1992
criteria of the American Joint Committee on Cancer/Union for
International Cancer Control (36).
The primary tumor locations were buccal mucosa (77.6%) and tongue
(22.4%). All of the patients received surgery as primary treatment,
and some patients received adjuvant treatment, such as radiotherapy
and chemotherapy. A total of 62 patients received adjuvant
chemotherapy, with the chemotherapy regimen consisting of cisplatin
or carboplatin with or without the addition of 5-fluorouracil or
paclitaxel. A total of 45 patients received adjuvant
intensity-modulated radiotherapy, and the scheduled doses were
given once per day, 5 days per week. Postoperative patients
received the planned course of adjuvant radiotherapy of 60–66 Gy in
2-Gy fractions to the post-operative high-risk region.
 | Table I.Clinicopathological characteristics
of 98 patients with oral squamous cell carcinoma. |
Table I.
Clinicopathological characteristics
of 98 patients with oral squamous cell carcinoma.
|
Characteristics | N (%) |
|---|
| Age, years |
|
|
<50 | 47 (48.0) |
|
≥50 | 51 (52.0) |
| Sex |
|
|
Male | 92 (93.9) |
|
Female | 6 (6.1) |
| Cigarette
smoking |
|
| No | 11 (11.1) |
|
Yes | 87 (88.9) |
| Betel quid
chewing |
|
| No | 30 (30.6) |
|
Yes | 68 (69.4) |
| Alcohol
consumption |
|
| No | 35 (35.7) |
|
Yes | 63 (34.3) |
| Tumor location |
|
| Buccal
mucosa | 76 (77.6) |
|
Tongue | 22 (22.4) |
| Grade |
|
| Grade
0 | 86 (87.8) |
| Grade
I+II+III | 12 (12.2) |
| Tumor size |
|
| T1 | 22 (22.4) |
| T2 | 30 (30.6) |
| T3 | 6 (6.1) |
| T4 | 40 (40.8) |
| Lymph node
metastases |
|
| N0 | 50 (51.0) |
| N1 | 30 (30.6) |
| N2 | 18 (18.4) |
| Tumor stage |
|
| I | 18 (18.4) |
| II | 17 (17.3) |
|
III | 18 (18.4) |
| IV | 45 (45.9) |
| RT |
|
| No | 53 (54.1) |
|
Yes | 45 (45.9) |
| CT |
|
| No | 36 (36.7) |
|
Yes | 62 (63.3) |
| Recurrence |
|
| No | 29 (29.6) |
|
Yes |
|
|
Pre-CT/RT
recurrence | 22 (22.4) |
|
Post-CT/RT
recurrence | 47 (48.0) |
Survival endpoints and recurrence
Follow-up data was retrieved and updated from case
records obtained from the medical records department until October
2014. Any patient not followed up within the last six months was
contacted by phone to determine their current health status.
Primary endpoints were disease-free survival (DFS) and overall
survival (OS), with DFS being measured from the date of surgery to
the date of locoregional recurrence, distant metastases or death
from any cause, while OS was measured from the date of surgery to
the date of death from any cause. Postoperative recurrence was
defined as a lesion that exhibited postoperative regrowth at the
same site after confirmation of healing of the surgical wounds.
Immunohistochemical (IHC)
analysis
To determine the expression levels of XRCC1, ERCC1
and ERCC2 in OSCC tissues by IHC staining, the tissues were fixed
in 10% neutral buffered formalin for 48 h at room temperature to
prepare paraffin-embedded tumor tissue blocks for IHC sections
(4-mm-thick). For the normal control, the oral mucosa tissue from a
delinked patient with fibroma was used after Institutional Review
Board approval (approval no. KMUH-IRB-20140158). Sections were
deparaffinized and rehydrated following standard methods. Briefly,
the sections were deparaffinized with xylene for 5 min for three
times and rehydrated in graded ethanol (80–100%) for 5 min. A
microwave antigen retrieval procedure was performed for 20 min in
citrate buffer (pH 6.0), and 3% hydrogen peroxide was used to block
non-specific peroxidase reactions at room temperature for 10 min.
After washing twice with TBS, non-specific blocking was performed
with Protein Block (Novolink Polymer Detection System; Leica
Microsystems, Inc.) for 5 min at room temperature and washed twice
with TBS. Following washing with TBS, sections were incubated with
the aforementioned XRCC1 monoclonal antibody (1:100), ERCC1
monoclonal antibody (1:200) and ERCC2 polyclonal antibody (1:100)
at 4°C overnight. The sections were subsequently incubated with the
Post Primary rabbit anti-mouse IgG antibody (Novolink Polymer
Detection System; Leica Microsystems, Inc.) for 30 min at room
temperature as secondary antibody application. The staining
intensity of tumor tissues was determined as score 0 (negative),
score 1 (weak), score 2 (moderate) and score 3 (strong) for
antigens present in the cytoplasm and nucleus of cells using a
light microscope (magnification, ×200), determined separately by
two independent pathologists.
Statistical analysis
Statistical analyses were performed using SPSS 19.0
(IBM Corp.). Descriptive statistics were used and compared using
independent t-test, χ2 test or Fisher's exact test.
Survival analyses were evaluated using the Kaplan-Meier method with
the log-rank test, while the Cox proportional-hazards model was
used for multivariate analysis. Furthermore, hazard ratios (HRs)
and 95% CIs were calculated by multivariable Cox regression models
and used to investigate the association between clinicopathological
characteristics and survival. DFS was defined as the time after
surgery during which the patient survived with no sign of
recurrence. OS was defined as the time elapsed between surgery and
death. All P-values were two-sided, with P<0.05 considered to
indicate a statistically significant difference.
Results
Expression profiles of XRCC1, ERCC1
and ERCC2 in oral cancer cell lines
Since DNA repair proteins are essential for oral
cancer cells in response to chemo-radiotherapy, the expression
levels of XRCC1, ERCC1 and ERCC2 in HOK normal oral epithelial
cells, DOK oral pre-cancer cells and oral cancer SAS, OECM1, HSC-3
and Cal-27 cell lines were first examined using western blotting
(Fig. 1), with U87MG glioblastoma
cells being included as a positive control for XRCC1, ERCC1 and
ERCC2 proteins (37). The results
revealed that the expression levels of XRCC1 and ERCC1 were weakly
detected in all oral cancer cells, while ERCC2 expression was
detected in all cell lines (Fig. 1).
Notably, HOK cells had lower ERCC2 protein expression compared with
DOK cells and the oral cancer cell lines.
 | Figure 1.XRCC1, ERCC1 and ERCC2 protein
expression in primary oral epithelial cells, oral pre-cancer cells
and various oral cancer cells. The cell lysates extracted from HOK
primary oral epithelial cells, DOK oral pre-cancer cells and oral
cancer cell lines, including SAS, OECM1, HSC-3 and Cal-27,
underwent western blotting for detecting the expression levels of
XRCC1, ERCC1 and ERCC2. Glioblastoma U87MG cell line served as a
positive control. The numbers correspond to the grey density values
of a single replicate. XRCC1, X-ray repair cross-complementing
group 1; ERCC1/2, excision repair cross-complementing group
1/2. |
Association of XRCC1, ERCC1 and ERCC2
expression in OSCC tissues with clinicopathological
characteristics
In the present study, 98 patients with OSCC (92 men
and 6 women) were included. The clinicopathological characteristics
of these patients, including age, sex, tumor location, grade, tumor
size, lymph node metastases, tumor stage, radiotherapy,
chemotherapy, smoking habit and recurrence, are shown in Table I. XRCC1, ERCC1 and ERCC2 expression
in oral cancer tissues, as well as in normal oral mucosa tissue of
a patient with fibroma, was analyzed using an online database and
IHC analysis. The Oncomine database revealed that the mRNA
expression levels of XRCC1, ERCC1 and ERCC2 were significantly
increased in oral cancer tissues compared with in normal epithelial
tissues (Fig. S1), except in the
Peng Head-Neck XRCC1-3864445 dataset, where there was no
significant difference, but there was still a trend toward XRCC1
elevation in oral cancer tissues (P=0.061; Fig. S1A). Notably, IHC staining without
addition of the primary antibody was used as a negative control for
XRCC1, ERCC1 and ERCC2 staining (Fig.
S2). According to the staining intensity in tumor tissues, the
expression levels of the three proteins were categorized into four
scores (0–3). XRCC1 and ERCC1 proteins were predominantly stained
in the nuclei, while ERCC2 protein was predominantly stained in the
cytoplasm of oral cancer tissues (Fig.
2). To analyze the association between the protein expression
levels of the three proteins in OSCC tissues and
clinicopathological characteristics, the expression levels of the
three proteins were further divided into high expression (score 3)
and low expression (score 0–2) groups. High expression groups of
XRCC1 (P=0.020), ERCC1 (P=0.006) or ERCC2 (P<0.001) were
significantly associated with OSCC recurrence (Tables II–IV). Furthermore, univariate and
multivariate analyses were used to explore OSCC recurrence
predictors in patients with OSCC. In univariate analysis, lymph
node metastases (N1+N2), high XRCC1 expression, high ERCC1
expression and high ERCC2 expression were significantly associated
with OSCC recurrence (P=0.006, P=0.020, P=0.006 and P<0.001,
respectively; Table V). In
multivariate analysis, lymph node metastases (N1+N2) (2.38-fold;
95% CI, 1.02–5.58; P=0.045) and high ERCC2 expression (4.84-fold;
95% CI, 2.56–9.16; P<0.001) were significantly associated with
increased risk of OSCC recurrence (Table
V).
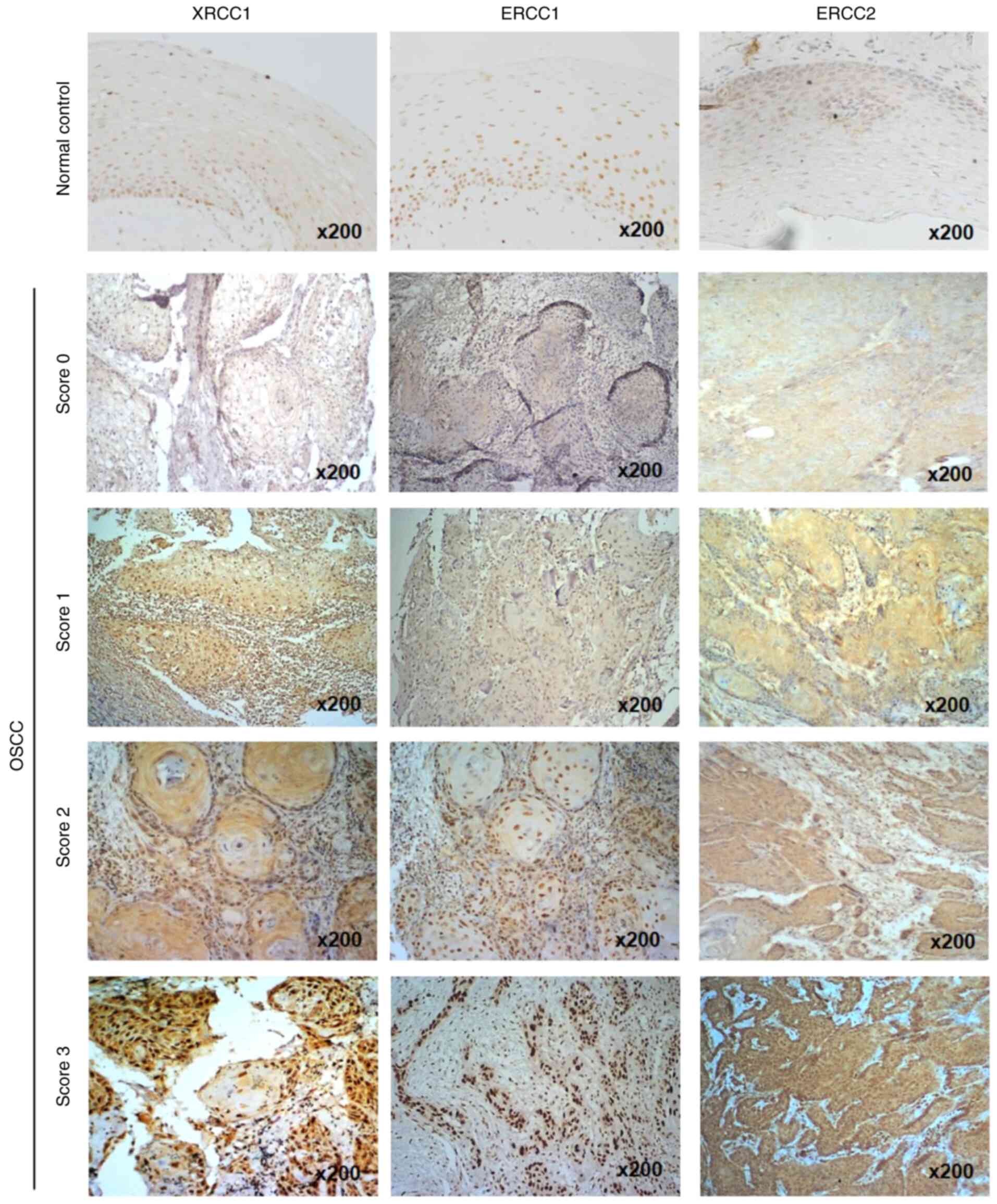 | Figure 2.Immunohistochemical staining of
XRCC1, ERCC1 and ERCC2 expression in OSCC tissues. ERCC1, ERCC2 or
XRCC1 expression in oral normal tissues and cancer tissues,
determined by immunohistochemistry, was divided into 4 categories
according to the staining intensity: Score 0, no staining; score 1,
weak staining; score 2, moderate staining; and score 3, strong
staining. Original magnification, ×200. XRCC1, X-ray repair
cross-complementing group 1; ERCC1/2, excision repair
cross-complementing group 1/2; OSCC, oral squamous cell
carcinoma. |
 | Table II.Association between clinical
characteristics of patients with oral squamous cell carcinoma and
XRCC1 expression. |
Table II.
Association between clinical
characteristics of patients with oral squamous cell carcinoma and
XRCC1 expression.
|
| XRCC1
expression |
|
|---|
|
|
|
|
|---|
|
Characteristics | Low, n (%) | High, n (%) | P-value |
|---|
| Age, years |
|
<50 | 13 (46.4) | 34 (48.6) | 0.850 |
|
≥50 | 15 (53.6) | 36 (51.4) |
|
| Sex |
|
Male | 28 (100.0) | 64 (91.4) | 0.180a |
|
Female | 0 (0.0) | 6 (8.6) |
|
| Cigarette
smoking |
| No | 1 (3.6) | 10 (14.3) | 0.170a |
|
Yes | 27 (96.4) | 60 (85.7) |
|
| Betel quid
chewing |
| No | 7 (25.0) | 23 (32.9) | 0.450 |
|
Yes | 21 (75.0) | 47 (37.1) |
|
| Alcohol
consumption |
| No | 6 (21.4) | 29 (41.4) | 0.070 |
|
Yes | 22 (78.6) | 41 (58.6) |
|
| Tumor location |
| Buccal
mucosa | 24 (85.7) | 52 (74.3) | 0.290a |
|
Tongue | 4 (14.3) | 18 (25.7) |
|
| Grade |
| Grade
I | 26 (92.9) | 60 (85.7) | 0.500a |
| Grade
II+III | 2 (7.1) | 10 (14.3) |
|
| Tumor size |
|
T1+T2 | 16 (57.1) | 36 (51.4) | 0.610 |
|
T3+T4 | 12 (42.9) | 34 (48.6) |
|
| Lymph node
metastases |
| N0 | 15 (53.6) | 35 (50.0) | 0.750 |
|
N1+N2 | 13 (46.4) | 35 (50.0) |
|
| Tumor stage |
|
I+II | 11 (39.3) | 24 (34.3) | 0.640 |
|
III+IV | 17 (60.7) | 46 (65.7) |
|
| Radiotherapy |
| No | 18 (64.3) | 35 (50.0) | 0.200 |
|
Yes | 10 (35.7) | 35 (50.0) |
|
| Chemotherapy |
| No | 12 (42.9) | 24 (34.3) | 0.430 |
|
Yes | 16 (57.1) | 46 (65.7) |
|
| Recurrence |
| No | 13 (46.4) | 16 (22.9) | 0.020 |
|
Yes | 15 (53.6) | 54 (77.1) |
|
 | Table IV.Association between clinical
characteristics of patients with oral squamous cell carcinoma and
ERCC2 expression. |
Table IV.
Association between clinical
characteristics of patients with oral squamous cell carcinoma and
ERCC2 expression.
|
| ERCC2
expression |
|
|---|
|
|
|
|
|---|
|
Characteristics | Low, n (%) | High, n (%) | P-value |
|---|
| Age, years |
|
<50 | 21 (53.8) | 26 (44.1) | 0.340 |
|
≥50 | 18 (46.2) | 33 (55.9) |
|
| Sex |
|
Male | 36 (92.3) | 56 (94.9) | 0.680a |
|
Female | 3 (7.7) | 3 (5.1) |
|
| Smoking |
| No | 5 (12.8) | 6 (10.2) | 0.680 |
|
Yes | 34 (87.2) | 53 (89.8) |
|
| Betel quid
chewing |
| No | 8 (20.5) | 22 (37.3) | 0.080 |
|
Yes | 31 (79.5) | 37 (62.7) |
|
| Alcohol
consumption |
| No | 12 (30.8) | 23 (39.0) | 0.410 |
|
Yes | 27 (39.2) | 36 (61.0) |
|
| Tumor location |
| Buccal
mucosa | 28 (71.8) | 48 (81.4) | 0.270 |
|
Tongue | 11 (28.2) | 11 (18.6) |
|
| Grade |
| Grade
I | 34 (87.2) | 52 (88.1) | >0.999 |
| Grade
II+III | 5 (12.8) | 7 (11.9) |
|
| Tumor size |
|
T1+T2 | 24 (61.5) | 28 (47.5) | 0.170 |
|
T3+T4 | 15 (38.5) | 31 (52.5) |
|
| Lymph node
metastases |
| N0 | 24 (61.5) | 26 (44.1) | 0.090 |
|
N1+N2 | 15 (38.5) | 33 (55.9) |
|
| Tumor stage |
|
I+II | 18 (46.2) | 17 (28.8) | 0.080 |
|
III+IV | 21 (53.8) | 42 (71.2) |
|
| Radiotherapy |
| No | 20 (51.3) | 33 (55.9) | 0.650 |
|
Yes | 19 (48.7) | 26 (44.1) |
|
| Chemotherapy |
| No | 18 (46.2) | 18 (30.5) | 0.120 |
|
Yes | 21 (53.8) | 41 (69.5) |
|
| Recurrence |
| No | 23 (59.0) | 6 (10.2) | <0.001 |
|
Yes | 16 (41.0) | 53 (89.8) |
|
 | Table V.Univariate and multivariate analyses
of recurrence predictors in 98 patients with oral squamous cell
carcinoma. |
Table V.
Univariate and multivariate analyses
of recurrence predictors in 98 patients with oral squamous cell
carcinoma.
|
| Recurrence | Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
|
Characteristics | Yes | No | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| Age, years |
|
≥50 | 32 (46.4) | 19 (65.5) | 0.080 | 1.0 | 0.370 |
|
<50 | 37 (53.6) | 10 (34.5) |
| 1.28
(0.75–2.20) |
|
| Sex |
|
Female | 3 (4.3) | 3 (10.3) | 0.360 | 1.0 | 0.290 |
|
Male | 66 (95.7) | 26 (89.7) |
| 0.48
(0.13–1.83) |
|
| Smoking |
| No | 8 (11.6) | 3 (10.3) | >0.999 | 1.0 | 0.400 |
|
Yes | 61 (88.4) | 26 (89.1) |
| 0.64
(0.22–1.84) |
|
| Betel quid
chewing |
| No | 23 (33.3) | 7 (24.1) | 0.370 | 1.0 | 0.330 |
|
Yes | 46 (66.7) | 22 (75.9) |
| 1.39
(0.72–2.71) |
|
| Alcohol
consumption |
| No | 27 (39.1) | 8 (27.6) | 0.370 | 1.0 | 0.960 |
|
Yes | 42 (60.9) | 21 (72.4) |
| 0.98
(0.54–1.81) |
|
| Tumor location |
| Buccal
mucosa | 53 (76.8) | 23 (79.3) | 0.790 | 1.0 | 0.190 |
|
Tongue | 16 (23.2) | 6 (20.7) |
| 0.64
(0.32–1.25) |
|
| Grade |
| Grade
I | 60 (87.0) | 26 (89.7) | >0.999 | 1.0 | 0.680 |
| Grade
II+III | 9 (13.0) | 3 (10.3) |
| 0.86
(0.40–1.81) |
|
| Tumor size |
|
T1+T2 | 34 (49.3) | 18 (62.1) | 0.250 | 1.0 | 0.150 |
|
T3+T4 | 35 (50.7) | 11 (37.9) |
| 1.78
(0.81–3.88) |
|
| Lymph node
metastases |
| N0 | 29 (42) | 21 (72.4) | 0.006 | 1.0 | 0.045 |
|
N1+N2 | 40 (58) | 8 (27.6) |
| 2.38
(1.02–5.58) |
|
| Tumor stage |
|
I+II | 21 (30.4) | 14 (48.3) | 0.090 | 1.0 | 0.460 |
|
III+IV | 48 (69.6) | 15 (51.7) |
| 0.65
(0.20–1.71) |
|
| XRCC1
expression |
|
Low | 15 (21.7) | 13 (44.8) | 0.020 | 1.0 | 0.450 |
|
High | 54 (78.3) | 16 (55.2) |
| 1.31
(0.65–2.66) |
|
| ERCC1
expression |
|
Low | 22 (31.9) | 18 (62.1) | 0.006 | 1.0 | 0.460 |
|
High | 47 (68.1) | 11 (37.9) |
| 1.25
(0.69–2.25) |
|
| ERCC2
expression |
|
Low | 16 (23.2) | 23 (79.3) | <0.001 | 1.0 | <0.001 |
|
High | 53 (76.8) | 6 (20.7) |
| 4.84
(2.56–9.16) |
|
Univariate and multivariate analyses were also used
to explore OSCC survival predictors in patients with OSCC. In
univariate analysis, tumor size, lymph node metastases (N1+N2),
tumor stage, high XRCC1 expression, high ERCC1 expression and high
ERCC2 expression were significantly associated with a worse
survival in patients with OSCC (P=0.001, P=0.005, P=0.001, P=0.002,
P=0.014 and P<0.001, respectively; Table VI). In multivariate analysis, tumor
location (4.11-fold; 95% CI, 1.392–12.14; P=0.01) and high ERCC2
expression (15.55-fold; 95% CI, 4.34–55.67; P<0.001) were
significantly associated with increased risk of death (Table VI).
 | Table VI.Univariate and multivariate analysis
of survival predictors in 98 patients with oral squamous cell
carcinoma. |
Table VI.
Univariate and multivariate analysis
of survival predictors in 98 patients with oral squamous cell
carcinoma.
|
| Death | Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
|
Characteristics | Yes | No | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| Age, years |
|
≥50 | 25 (54.3) | 26 (50.0) | 0.540 | 0.96
(0.48–19.2) | 0.910 |
|
<50 | 21 (45.7) | 26 (50.0) |
| 1 |
|
| Sex |
|
Female | 3 (6.5) | 3 (5.8) | 0.410 | 1 | 0.290 |
|
Male | 43 (93.5) | 49 (94.2) |
| 0.43
(0.09–2.04) |
|
| Smoking |
| No | 5 (10.9) | 6 (11.5) | 0.880 | 1 | 0.350 |
|
Yes | 41 (89.1) | 46 (88.5) |
| 0.53
(0.14–2.00) |
|
| Betel quid
chewing |
| No | 19 (41.3) | 15 (28.8) | 0.270 | 1 | 0.790 |
|
Yes | 27 (58.7) | 37 (71.2) |
| 1.11
(0.53–2.32) |
|
| Alcohol
consumption |
| No | 17 (37.0) | 18 (34.6) | 0.910 | 1 | 0.460 |
|
Yes | 29 (63.0) | 34 (65.4) |
| 1.33
(0.62–2.86) |
|
| Tumor location |
| Buccal
mucosa | 39 (84.8) | 37 (71.2) | 0.060 | 1 | 0.010 |
|
Tongue | 7 (15.2) | 15 (28.8) |
|
4.11(1.39–12.14) |
|
| Grade |
| Grade
I | 38 (82.6) | 48 (92.3) | 0.710 | 1 | 0.430 |
| Grade
II+III | 8 (17.4) | 4 (7.7) |
| 0.71
(0.30–1.67) |
|
| Tumor size |
|
T1+T2 | 17 (37.0) | 35 (37.3) | 0.001 | 1 | 0.430 |
|
T3+T4 | 29 (63.0) | 17 (32.7) |
| 1.55
(0.52–4.58) |
|
| Lymph node
metastases |
| N0 | 29 (63.0) | 19 (36.5) | 0.005 | 1 | 0.560 |
|
N1+N2 | 17 (37.0) | 33 (63.5) |
| 1.31
(0.54–31.8) |
|
| Tumor stage |
|
I+II | 9 (19.6) | 26 (50.0) | 0.001 | 1 | 0.500 |
|
III+IV | 37 (80.4) | 26 (50.0) |
| 1.70
(0.36–8.04) |
|
| XRCC1
expression |
|
Low | 5 (10.9) | 23 (44.2) | 0.002 | 1 | 0.100 |
|
High | 41 (89.1) | 29 (41.4) |
| 2.65
(0.82–8.52) |
|
| ERCC1
expression |
|
Low | 10 (21.7) | 30 (57.7) | 0.014 | 1 | 0.900 |
|
High | 36 (78.3) | 22 (42.3) |
| 1.06
(0.45–2.50) |
|
| ERCC2
expression |
|
Low | 3 (6.5) | 36 (69.2) | <0.001 | 1 | <0.001 |
|
High | 43 (93.5) | 16 (60.2) |
| 15.55
(4.34–55.67) |
|
The association between the expression levels of the
three proteins and DFS or OS rates in patients with OSCC was
analyzed using the Kaplan-Meier method. Significantly decreased DFS
rates were observed in patients with high expression levels of
XRCC1, ERCC1 and ERCC2 (P=0.020, P=0.040 and P<0.001,
respectively), as well as significantly decreased OS rates in
patients with high expression levels of XRCC1, ERCC1 and ERCC2
(P=0.002, P=0.014 and P<0.001, respectively), as determined
using the log-rank test (Fig. 3).
The findings of the survival analysis indicating that high ERCC2
expression indicated a poor prognosis were consistent with the
results of the multivariate analysis.
Discussion
In the present study, the association between the
expression levels of NER pathway-associated genes and the
recurrence and survival outcome in patients with OSCC was explored
by examining the expression levels of XRCC1, ERCC1 and ERCC2 in a
series of oral cavity epithelial cells, revealing that ERCC2, but
not ERCC1 or XRCC1, exhibited a trend of positive association with
the neoplastic development from normal epithelium to dysplasia and
OSCC. However, some cancer cells expressed lower expression levels
of XRCC1, ERCC1 and ERCC2 than HOK normal oral keratinocytes and
DOK oral precancerous cells, and this requires further
investigation. In addition, high expression levels of the three
proteins in OSCC tissues were significantly associated with worse
OS and DFS rates compared with low expression levels. Notably, the
multivariate analysis revealed that high ERCC2 expression was an
independent prognostic marker for OSCC recurrence.
High ERCC1 expression in head and neck squamous cell
carcinoma (HNSCC) has been extensively studied as a potential
prognostic biomarker for the chemoradiotherapy response and
survival prognosis of patients with HNSCC (38–42).
Although the genetic variations of ERCC2 and XRCC1 have been
reported to be associated with increased risk and worse clinical
outcome of oral cancer (43–47), there is very little data on ERCC2 and
XRCC1 expression in oral cancer. A systematic review and
case-control study has revealed that ERCC2 expression is
significantly increased in HNSCC tissues compared with in adjacent
normal tissues, and that it is positively associated with tumor
stage and grade (48). The present
study indicated that ERCC1, ERCC2 and XRCC1 expression was
associated with the survival rate of patients with OSCC. To the
best of our knowledge, the current study is the first to suggest
ERCC2 as a prognostic marker for OSCC recurrence.
The association between the synthesis of DNA repair
proteins in tumor cells and the patient response to
chemo-radiotherapy has been reported in different types of cancer.
In patients with non-small cell lung cancer, low ERCC1 expression
indicates an improved prognosis after treatment with multidrug
chemotherapy (22). High XRCC1 and
ERCC1 expression is associated with a poor prognosis in patients
with HER2+ breast cancer (47), while high ERCC1 expression has been
associated with resistance to platinum-based chemotherapy in
patients with ovarian cancer (49).
In addition, ectopic ERCC1 expression in ovarian cancer cells
increases the resistance to cisplatin-mediated growth inhibition
(50). NER capacity serves a major
role in normal tissue tolerance and drug resistance; moreover,
ERCC2 and ERCC1 act as rate-limiting enzymes in NER (51). During NER, ERCC2 participates in DNA
unwinding, and this function may alter the platinum-based
chemotherapy effect (52). In a
previous study, the genotypes of XRCC1 rs1799782 and XRCC2
rs2040639 DNA repair genes were significantly associated with oral
cancer in Taiwan (44). Several
studies have suggested that cancer cells with upregulation of DNA
repair proteins are more resistant to chemoradiotherapy (53–55).
Therefore, according to the present study, further investigation on
the mechanism of the biological process of ERCC2 may provide
potential targets for pharmacological modulation that helps improve
the efficiency of chemo-radiotherapy.
Cigarette smoking, alcohol consumption and betel
quid chewing are well-known risk factors for oral cancer. Most oral
cancer cases (~90%) in South-East Asia are associated with smoking
(56), while the proportions of
cases associated with alcohol drinking and betel quid chewing are
80 and 75%, respectively. These agents may act synergistically. A
working group of the International Agency for Research on Cancer
concluded that there was adequate evidence of an association
between chewing betel quid together with tobacco use (chewing or
smoking) as a combined risk factor (57). In areas where the habit of betel quid
chewing is widespread, these risk factors should be taken into
consideration and require further investigation. However, the
influence of smoking on clinical outcome of patients with oral
cancer is in debate, since while smoking has a negative impact on
survival, its effect is influenced by other confounding factors,
such as treatment method, age, tumor size, smoking status or
dose-response relationship (58–62).
When patients stop smoking, survival benefits have been observed in
several types of cancer, including head and neck cancer (59,63). In
the current study, cigarette smoking, alcohol consumption and betel
quid chewing were not associated with recurrence in univariate or
multivariate analysis, which may be due to fewer non-users. Further
larger scale studies are required to solve this contradiction.
According to GLOBOCAN 2020, the ratio of oral cancer
incidence in male and female is 2:1 in Asia and 2.5:1 worldwide;
however, male is the predominant sex for oral cancer in Taiwan
(2). According to the Taiwan cancer
registry annual report 2017, the proportion of female patients with
oral cancer was 10.7%, and females represented 6% of the total
sample size in the present study, which is similar to the general
oral cancer population in Taiwan (3).
The current study has some limitations. First, data
regarding socioeconomic factors were not collected and analyzed,
and second, data were from a single tertiary cancer care center;
therefore, more extensive multi-center studies are required to
reinforce the present findings. Third, the median follow-up time
was 40 months, and a longer follow-up time is required for full
surveillance of cancer recurrence. Lastly, reverse
transcription-quantitative PCR may also be a useful approach to
analyze mRNA expression.
In future studies, the mechanisms of ERCC1, ERCC2
and XRCC1 participating in OSCC carcinogenesis through the NER
pathway should be analyzed both in vitro and in vivo,
and larger prospective studies to validate the possible role of
ERCC2 in OSCC outcome prediction should be performed.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was financially supported by
grants from the Ministry of Health and Welfare of Taiwan (grant no.
MOHW109-TDU-B-212-134016, Health and Welfare Surcharge of Tobacco
Products) and from the ‘Center For Intelligent Drug Systems and
Smart Bio-devices’ from The Featured Areas Research Center Program
within the framework of the Higher Education Sprout Project by the
Ministry of Education in Taiwan (grant no. IDS2B). Additionally, it
was supported by grants from the Kaohsiung Medical University
Hospital (grant nos. KMUH105-5R32, KMUH106-6R41, KMUH106-6R83,
KMUH107-7R36, KMUH108-8R42, KMUH108-8R66, KMUH-DK109001-3 and
KMUH109-9R78), Kaohsiung Medical University (Research Center Grant;
grant nos. KMU-DK108005 and KMU-DK109001), Kaohsiung Medical
University Research Center Grant (Center for Cancer Research; grant
nos. KMU-TC108A04-0 and KMU-TC108A04-1) and the Ministry of Science
and Technology of Taiwan (grant nos. MOST 107-2314-B-037-097-MY2,
MOST 108-2314-B-037-021-MY3 and MOST 108-2314-B-037-014).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on request.
Authors' contributions
YYW, PTF, CWS, YKC, JJH, MYH and SSFY conceived and
designed the experiments. YYW, YKC, MYH and SSFY performed the
experiments. YYW, PTF, JYH, MYH and SSFY analyzed and discussed the
data. YYW, PTF, MYH and SSFY contributed to
reagents/materials/analysis tools. YYW, PTF, JYH, YKC, JJH, MYH and
SSFY wrote the manuscript. MYH and SSFY are responsible for
confirming the authenticity of the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Ethics approval was obtained from the Institutional
Review Board of Kaohsiung Medical University Hospital (approval no.
20140158) and informed consent was waived due to the retrospective
nature of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hsieh TY, Chang KP, Lee SS, Chang CH, Lai
CH, Wu YC, Huang SH, Lai CS and Lin SD: Free flap reconstruction in
patients with advanced oral squamous cell carcinoma: Analysis of
patient survival and cancer recurrence. Microsurgery. 32:598–604.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. Feb 4–2021.(Epub ahead
of print). View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ministry of Health Welfare, . The cause of
death in Taiwan in 2018. https://www.hpa.gov.tw/Pages/Detail.aspx?nodeid=269&pid=13498
|
|
4
|
Rivera C and Venegas B: Histological and
molecular aspects of oral squamous cell carcinoma (Review). Oncol
Lett. 8:7–11. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liao CT, Wang HM, Ng SH, Yen TC, Lee LY,
Hsueh C, Wei FC, Chen IH, Kang CJ, Huang SF and Chang JT: Good
tumor control and survivals of squamous cell carcinoma of buccal
mucosa treated with radical surgery with or without neck dissection
in Taiwan. Oral Oncol. 42:800–809. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eich HT, Löschcke M, Scheer M, Kocher M,
Bongartz R, Wacker S, Zöller JE and Müller RP: Neoadjuvant
radiochemotherapy and radical resection for advanced squamous cell
carcinoma of the oral cavity. Outcome of 134 patients. Strahlenther
Onkol. 184:23–29. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kreppel M, Drebber U, Eich HT, Dreiseidler
T, Zöller JE, Müller RP and Scheer M: Combined-modality treatment
in advanced oral squamous cell carcinoma: Primary surgery followed
by adjuvant concomitant radiochemotherapy. Strahlenther Onkol.
187:555–560. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fan KH, Wang HM, Kang CJ, Lee LY, Huang
SF, Lin CY, Chen EY, Chen IH, Liao CT and Chang JT: Treatment
results of postoperative radiotherapy on squamous cell carcinoma of
the oral cavity: Coexistence of multiple minor risk factors results
in higher recurrence rates. Int J Radiat Oncol Biol Phys.
77:1024–1029. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Woolgar JA, Rogers S, West CR, Errington
RD, Brown JS and Vaughan ED: Survival and patterns of recurrence in
200 oral cancer patients treated by radical surgery and neck
dissection. Oral Oncol. 35:257–265. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu CH, Chen HJ, Wang PC, Chen HS and
Chang YL: Patterns of recurrence and second primary tumors in oral
squamous cell carcinoma treated with surgery alone. Kaohsiung J Med
Sci. 29:554–559. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang B, Zhang S, Yue K and Wang XD: The
recurrence and survival of oral squamous cell carcinoma: A report
of 275 cases. Chin J Cancer. 32:614–618. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bernier J, Domenge C, Ozsahin M,
Matuszewska K, Lefèbvre JL, Greiner RH, Giralt J, Maingon P,
Rolland F, Bolla M, et al: Postoperative irradiation with or
without concomitant chemotherapy for locally advanced head and neck
cancer. N Engl J Med. 350:1945–1952. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cooper JS, Pajak TF, Forastiere AA, Jacobs
J, Campbell BH, Saxman SB, Kish JA, Kim HE, Cmelak AJ, Rotman M, et
al: Postoperative concurrent radiotherapy and chemotherapy for
high-risk squamous-cell carcinoma of the head and neck. N Engl J
Med. 350:1937–1944. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fan S, Tang QL, Lin YJ, Chen WL, Li JS,
Huang ZQ, Yang ZH, Wang YY, Zhang DM, Wang HJ, et al: A review of
clinical and histological parameters associated with contralateral
neck metastases in oral squamous cell carcinoma. Int J Oral Sci.
3:180–191. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pignon JP, le Maître A, Maillard E and
Bourhis J; MACH-NC Collaborative Group, : Meta-analysis of
chemotherapy in head and neck cancer (MACH-NC): An update on 93
randomised trials and 17,346 patients. Radiother Oncol. 92:4–14.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Winquist E, Agbassi C, Meyers BM, Yoo J
and Chan KKW; Head and Neck Disease Site Group, : Systemic therapy
in the curative treatment of head and neck squamous cell cancer: A
systematic review. J Otolaryngol Head Neck Surg. 46:292017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dabholkar M, Vionnet J, Bostick-Bruton F,
Yu JJ and Reed E: Messenger RNA levels of XPAC and ERCC1 in ovarian
cancer tissue correlate with response to platinum-based
chemotherapy. J Clin Invest. 94:703–708. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Murray D and Rosenberg E: The importance
of the ERCC1/ERCC4[XPF] complex for hypoxic-cell radioresistance
does not appear to derive from its participation in the nucleotide
excision repair pathway. Mutat Res. 364:217–226. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Manic S, Gatti L, Carenini N, Fumagalli G,
Zunino F and Perego P: Mechanisms controlling sensitivity to
platinum complexes: Role of p53 and DNA mismatch repair. Curr
Cancer Drug Targets. 3:21–29. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sancar A: DNA excision repair. Annu Rev
Biochem. 65:43–81. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Petit C and Sancar A: Nucleotide excision
repair: From E. coli to man. Biochimie. 81:15–25. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Olaussen KA, Dunant A, Fouret P, Brambilla
E, André F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper HH,
et al: DNA repair by ERCC1 in non-small-cell lung cancer and
cisplatin-based adjuvant chemotherapy. N Engl J Med. 355:983–991.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Felip E and Rosell R: Testing for excision
repair cross-complementing 1 in patients with non-small-cell lung
cancer for chemotherapy response. Expert Rev Mol Diagn. 7:261–268.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Olaussen KA, Mountzios G and Soria JC:
ERCC1 as a risk stratifier in platinum-based chemotherapy for
nonsmall-cell lung cancer. Curr Opin Pulm Med. 13:284–289. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Altaha R, Liang X, Yu JJ and Reed E:
Excision repair cross complementing-group 1: Gene expression and
platinum resistance. Int J Mol Med. 14:959–970. 2004.PubMed/NCBI
|
|
26
|
Weaver DA, Crawford EL, Warner KA,
Elkhairi F, Khuder SA and Willey JC: ABCC5, ERCC2, XPA and XRCC1
transcript abundance levels correlate with cisplatin
chemoresistance in non-small cell lung cancer cell lines. Mol
Cancer. 4:182005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang MY, Tsai HL, Lin CH, Huang CW, Ma
CJ, Huang CM, Chai CY and Wang JY: Predictive value of ERCC1,
ERCC2, and XRCC1 overexpression for stage III colorectal cancer
patients receiving FOLFOX-4 adjuvant chemotherapy. J Surg Oncol.
108:457–464. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chiu TJ, Chen CH, Chien CY, Li SH, Tsai HT
and Chen YJ: High ERCC1 expression predicts cisplatin-based
chemotherapy resistance and poor outcome in unresectable squamous
cell carcinoma of head and neck in a betel-chewing area. J Transl
Med. 9:312011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Caldecott KW: XRCC1 and DNA strand break
repair. DNA Repair (Amst). 2:955–969. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nix P, Greenman J, Stafford N and Cawkwell
L: Expression of XRCC 1 and ERCC 1 proteins in radioresistant and
radiosensitive laryngeal cancer. Cancer Ther. 2:47–53. 2004.
|
|
31
|
Estilo CL, O-charoenrat P, Talbot S, Socci
ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y,
Boyle JO, et al: Oral tongue cancer gene expression profiling:
Identification of novel potential prognosticators by
oligonucleotide microarray analysis. BMC Cancer. 9:112009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Peng CH, Liao CT, Peng SC, Chen YJ, Cheng
AJ, Juang JL, Tsai CY, Chen TC, Chuang YJ, Tang CY, et al: A novel
molecular signature identified by systems genetics approach
predicts prognosis in oral squamous cell carcinoma. PLoS One.
6:e234522011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cromer A, Carles A, Millon R, Ganguli G,
Chalmel F, Lemaire F, Young J, Dembélé D, Thibault C, Muller D, et
al: Identification of genes associated with tumorigenesis and
metastatic potential of hypopharyngeal cancer by microarray
analysis. Oncogene. 23:2484–2498. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ginos MA, Page GP, Michalowicz BS, Patel
KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL and Gaffney PM:
Identification of a gene expression signature associated with
recurrent disease in squamous cell carcinoma of the head and neck.
Cancer Res. 64:55–63. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Verschuur HP, Irish JC, O'Sullivan B, Goh
C, Gullane PJ and Pintilie M: A matched control study of treatment
outcome in young patients with squamous cell carcinoma of the head
and neck. Laryngoscope. 109:249–258. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Boccard SG, Marand SV, Geraci S, Pycroft
L, Berger FR and Pelletier LA: Inhibition of DNA-repair genes Ercc1
and Mgmt enhances temozolomide efficacy in gliomas treatment: A
pre-clinical study. Oncotarget. 6:29456–29468. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang PY, Li Y, Mai HQ, Luo RZ, Cai YC and
Zhang L: Expression of ERCC1 predicts clinical outcome in
locoregionally advanced nasopharyngeal carcinoma treated with
cisplatin-based induction chemotherapy. Oral Oncol. 48:964–968.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gao Y and Liu D: The roles of excision
repair cross-complementation group1 in objective response after
cisplatin-based concurrent chemoradiotherapy and survival in head
and neck cancers: A systematic review and meta-analysis. Oral
Oncol. 51:570–577. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bauman JE, Austin MC, Schmidt R, Kurland
BF, Vaezi A, Hayes DN, Mendez E, Parvathaneni U, Chai X, Sampath S
and Martins RG: ERCC1 is a prognostic biomarker in locally advanced
head and neck cancer: Results from a randomised, phase II trial. Br
J Cancer. 109:2096–2105. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
An HJ, Jo H, Jung CK, Kang JH, Kim MS, Sun
DI, Cho KJ, Cho JH, Won HS, Sun S and Ko YH: Prognostic implication
of ERCC1 protein expression in resected oropharynx and oral cavity
cancer. Pathol Res Pract. 213:949–955. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang L, Wei W, Zhou L, Wang J and Hu G:
High/positive expression of ERCC1 predicts poor treatment response
and survival prognosis in nasopharyngeal carcinoma: A systematic
meta-analysis from 21 studies. Medicine (Baltimore). 98:e156412019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Y, Wang Y, Wu J and Li LJ: XRCC1
Arg194Trp polymorphism is associated with oral cancer risk:
Evidence from a meta-analysis. Tumour Biol. 34:2321–2327. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang CH, Lin YD, Yen CY, Chuang LY and
Chang HW: A systematic gene-gene and gene-environment interaction
analysis of DNA repair genes XRCC1, XRCC2, XRCC3, XRCC4, and oral
cancer risk. Omics. 19:238–247. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Avci H, Ergen A, Bireller ES, Ertugrul B
and Cakmakoglu B: A strong relationship between oral squamous cell
carcinoma and DNA repair genes. Biochem Genet. 55:378–386. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang E, Cui Z, Xu Z, Duan W, Huang S, Tan
X, Yin Z, Sun C and Lu L: Association between polymorphisms in
ERCC2 gene and oral cancer risk: Evidence from a meta-analysis. BMC
Cancer. 13:5942013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Vaezi A, Feldman CH and Niedernhofer LJ:
ERCC1 and XRCC1 as biomarkers for lung and head and neck cancer.
Pharmgenomics Pers Med. 4:47–63. 2011.PubMed/NCBI
|
|
48
|
Zafeer M, Mahjabeen I and Kayani MA:
Increased expression of ERCC2 gene in head and neck cancer is
associated with aggressive tumors: A systematic review and
case-control study. Int J Biol Markers. 31:e17–e25. 2016.
View Article : Google Scholar
|
|
49
|
Du P, Wang Y, Chen L, Gan Y and Wu Q: High
ERCC1 expression is associated with platinum-resistance, but not
survival in patients with epithelial ovarian cancer. Oncol Lett.
12:857–862. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu J, Zhang L, Mao P, Jiang G, Liu L,
Wang J, Yang W, Owusu L and Li W: Functional characterization of a
novel transcript of ERCC1 in chemotherapy resistance of ovarian
cancer. Oncotarget. 8:85759–85771. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gossage L and Madhusudan S: Current status
of excision repair cross complementing-group 1 (ERCC1) in cancer.
Cancer Treat Rev. 33:565–577. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lai JI, Tzeng CH, Chen PM, Lin JK, Lin TC,
Chen WS, Jiang JK, Wang HS and Wang WS: Very low prevalence of XPD
K751Q polymorphism and its association with XPD expression and
outcomes of FOLFOX-4 treatment in Asian patients with colorectal
carcinoma. Cancer Sci. 100:1261–1266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Curtin NJ: DNA repair dysregulation from
cancer driver to therapeutic target. Nat Rev Cancer. 12:801–817.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Schulz A, Meyer F, Dubrovska A and
Borgmann K: Cancer stem cells and radioresistance: DNA repair and
beyond. Cancers (Basel). 11:8622019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Huang RX and Zhou PK: DNA damage response
signaling pathways and targets for radiotherapy sensitization in
cancer. Signal Transduct Target Ther. 5:602020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Johnson N: Tobacco use and oral cancer: A
global perspective. J Dent Educ. 65:328–339. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ko YC, Huang YL, Lee CH, Chen MJ, Lin LM
and Tsai CC: Betel quid chewing, cigarette smoking and alcohol
consumption related to oral cancer in Taiwan. J Oral Pathol Med.
24:450–453. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Abrahão R, Anantharaman D, Gaborieau V,
Abedi-Ardekani B, Lagiou P, Lagiou A, Ahrens W, Holcatova I, Betka
J, Merletti F, et al: The influence of smoking, age and stage at
diagnosis on the survival after larynx, hypopharynx and oral cavity
cancers in Europe: The ARCAGE study. Int J Cancer. 143:32–44. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cao W, Liu Z, Gokavarapu S, Chen Y, Yang R
and Ji T: Reformed smokers have survival benefits after head and
neck cancer. Br J Oral Maxillofac Surg. 54:818–825. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Colares N, Souza Rodrigues DF, Freitas MO,
Dantas TS, Cunha MDPSS, Sousa FB and Barros Silva PG: Smoking
history decreases survival in patients with squamous cell carcinoma
of the mouth: A retrospective study with 15 years of follow-up.
Asian Pac J Cancer Prev. 20:1781–1787. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kawakita D, Hosono S, Ito H, Oze I,
Watanabe M, Hanai N, Hasegawa Y, Tajima K, Murakami S, Tanaka H and
Matsuo K: Impact of smoking status on clinical outcome in oral
cavity cancer patients. Oral Oncol. 48:186–191. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lee SU, Moon SH, Choi SW, Cho KH, Park JY,
Jung YS, Ryu J, Ryu CH, Yun T, Kim TH, et al: Prognostic
significance of smoking and alcohol history in young age oral
cavity cancer. Oral Dis. 26:1440–1448. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Koshiaris C, Aveyard P, Oke J, Ryan R,
Szatkowski L, Stevens R and Farley A: Smoking cessation and
survival in lung, upper aero-digestive tract and bladder cancer:
Cohort study. Br J Cancer. 117:1224–1232. 2017. View Article : Google Scholar : PubMed/NCBI
|

















