Introduction
Tongue cancer is a common malignant tumor type of
the oral and maxillofacial cavity (1). The incidence of tongue carcinoma in
males is higher than that of females and its most common
histopathological feature is squamous cell carcinoma, usually
located in the anterior two thirds of the tongue (2). Tongue adenocarcinoma is a rare distinct
clinicopathological entity of tongue cancer and is mostly located
in the root of the tongue (1).
Furthermore, lymphatic epithelial cancer and undifferentiated
carcinoma may sometimes occur in the root of the tongue (1). Tongue squamous cell carcinoma (TSCC)
accounts for ~50–60% of oral malignancies and 0.8–2% of systemic
malignancies (3,4). TSCC is highly malignant, invasive and
prone to lymph node metastasis (5).
The etiology and pathogenesis of TSCC are complicated (6,7).
Standard treatment for TSCC includes surgical resection; assisted
positive surgical margin and vein detection; lymphatic or nerve
dissection; postoperative radiotherapy and chemotherapy (8). Although substantial progress has been
made in the diagnosis and treatment, the 5-year survival rate for
patients with TSCC remains low due to an increased risk of
recurrence and lymph node metastasis (9). Therefore, TSCC poses a serious threat
to human health (10).
The etiology of TSCC is yet to be fully understood.
However, several common environmental risk factors have been
associated with TSCC, including heat, chronic damage, ultraviolet
radiation, X-rays and other radioactive carcinogenic substances
(11,12). For example, tongue and buccal mucosa
cancer may occur following chronic irritation in areas such as
residual roots, sharp cusps and tooth prosthetics (13). In addition, neuropsychiatric,
endocrine and genetic factors, as well as the immune system status
have been implicated in the development of TSCC (14). Recently, an increasing number of
studies have investigated the changes in gene expression associated
with TSCC (15–17). The occurrence of TSCC has been
associated with repeated damage, hyperemia and proliferation of
tongue mucosa cells, which are caused by several factors (18). Furthermore, tongue cancer is
triggered by a gradual increase in tongue mucosa cell metabolism,
thus resulting in repeated DNA breaks and recombination (19). Recently, several studies have
investigated the differentially expressed genes (DEGs) in patients
with TSCC, as well as their roles in various signaling pathways,
molecular functions and biological processes, using bioinformatics
analysis (20–22). Usami et al (23) found that intercellular adhesion
molecule 1 plays an important role in the development of tongue
cancer through promoting cancer cell proliferation, angiogenesis,
lymphatic vessel density and adhesion of giant cells. In addition,
Zhang et al (24)
demonstrated that galectin-3 regulates the Wnt/β-catenin signaling
pathway and Akt phosphorylation in vitro, thereby mediating
cancer cell migration and invasion, and resulting in tongue cancer
progression. Wang et al (25)
also showed that enhancer of zeste 2 polycomb repressive complex 2
subunit expression is associated with the neoplasm staging and its
overexpression increases the risk of tongue cancer. Furthermore
microarray expression datasets have been increasingly used to
identify novel microRNA (miRNA) biomarkers with diagnostic and
prognostic value in oral cancer and other types of cancer (20,26–30).
TSCC is one of the most common types of head and
neck malignant tumors and the most common cancer in the oral cavity
(31). Tongue cancer can be divided
into two types according to the anatomic location of the tumor:
Oral tongue cancer, which occurs in the anterior two thirds of the
tongue; and tongue base cancer, which involves the posterior third
of the tongue (15). The incidence
of oral tongue carcinoma is higher than that of tongue base
carcinoma (32). The majority of
tongue cancers, and especially oral tongue carcinomas, are derived
from moderately or highly differentiated squamous epithelial cells
(33). However, adenocarcinomas,
lymphatic epithelial cancer and undifferentiated carcinomas are
relatively rare and are mostly derived from the base of the tongue
(34). In addition, small salivary
gland-derived malignant tumors, such as adenoid cystic carcinoma
have also been identified as a type of tongue cancer. Treatment
strategies for tongue cancer include simple surgery; radiation
therapy, including external irradiation and inter-plant insertion
brachytherapy; systemic chemotherapy; and targeted therapy
(35,36). As a deeper understanding of the
molecular pathways involved in tongue cancer has been achieved, the
discovery of novel and promising targets for cancer treatment is
increasing (37). As such, exploring
the exact molecular mechanisms of action, as well as reliable
therapeutic targets for TSCC has attracted wide attention (15). With the development of gene
sequencing technology, a large number of DEGs have been identified
in various tumor types (38,39). DEGs play several roles in the
occurrence and development of various diseases, including
transcriptional regulation, post-transcriptional processing and
regulation of protein expression. The present study hypothesized
that DEGs could also play a key role in the development of TSCC and
in its malignant progression, thus serving as molecular markers and
therapeutic targets for TSCC.
Microarray technology allows the simultaneous
analysis of changes in the expression levels of multiple genes to
obtain gene sets that may be involved in TSCC (21). DEGs have been associated with the
tumor grade and prognosis for patients with TSCC (40). The expression of key molecular
markers may be used as independent prognostic factors; therefore,
further in-depth studies should be conducted to investigate the
potential mechanisms of action behind abnormally expressed genes.
These markers may affect the initiation and malignant progression
of TSCC and may be used as therapeutic targets (41). Therefore, there is an urgent need to
detect and analyze reliable target genes for TSCC.
Materials and methods
Microarray data
A total of three gene expression profiles, namely
GSE31056, GSE13601 and GSE78060, were retrieved from the Gene
Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo) dataset. The GSE31056
microarray data consisted of 22 TSCC and 24 normal tissue samples.
In addition, the mRNA expression profile of 31 patients with TSCC
and 26 healthy individuals was obtained from GSE13601. Similarly, a
total of 27 TSCC and three normal tissue samples were available in
the GSE78060 dataset.
Identification of DEGs
GEO2R, an online tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/), was utilized
to identify DEGs from the GEO series between TSCC and normal
tissues. Absent and duplicate probe sets were excluded. The cut-off
points were set to |log2FC|>1.5 and adjusted
P<0.05. The fold change indicated expression in TSCC tissue
samples/expression in normal tissue samples. Subsequently, DEGs
were visualized using volcano plots and heatmaps using R software
(version 3.5.3; The R Foundation) and Functional Enrichment
analysis tool (Funrich; version 3.1.3; http://funrich.org/index.html; FunRich Co. Ltd.). A
Venn diagram intersected all three datasets was constructed to
acquire the common DEGs.
Functional enrichment analysis of
DEGs
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; version 6.8; https://david.ncifcrf.gov/; DAVID Bioinformatics
Forum), an open online platform, was utilized to elucidate the
potential biological meaning of DEGs using Gene Ontology (GO) and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analyses. GO analysis of DEGs was carried out from three main
aspects of biological information, namely biological process (BP),
molecular function (MF) and cellular component (CC). P<0.05 was
considered to indicate a statistically significant difference.
Subsequently, the results were visualized using the ggplot package
of R software (version 3.5.3; The R Foundation).
Construction of the protein-protein
interaction (PPI) network and module analysis
The PPI network was constructed using the Search
Tool for the Retrieval of Interacting Genes database (STRING;
http://string-db.org) and was visualized using
the Cytoscape software (version 3.7.1; http://cytoscape.org/what_is_cytoscape.html).
Molecular Complex Detection within the Cytoscape software (version
3.7.1, http://cytoscape.org/what_is_cytoscape.html) was then
applied to screen significant modules in the PPI network. The
criteria default parameters were as follows: Degree cut-off=10,
k-core=2, node score cut-off=0.2 and max. depth=100. Subsequently,
the cBioportal database (www.cbioportal.org), an online tool integrating the
International Cancer Genome Consortium (icgc.org),
the Cancer Genome Atlas (portal.gdc.cancer.gov) and other cancer genome
databases, was utilized to construct the co-expression network of
hub genes in the module. Finally, the results of the biological
process analysis and co-expression network of hub genes were
visualized using the BiNGO tool in Cytoscape software (version
3.7.1, http://cytoscape.org/what_is_cytoscape.html).
Validation and analysis of hub
genes
Published microarray data were retrieved from the
Oncomine database (http://www.oncomine.org) to validate the expression
levels of hub genes in TSCC tissues. Subsequently, survival curves
were drawn to evaluate the prognostic significance of hub genes
using the Kaplan-Meier plotter database (http://kmplot.com/analysis/). Finally, the Gene
Expression Profiling Interactive Analysis database was used to
assess the differential expression of several hub genes in each TNM
stage (42). Independent-samples T
test was applied to identify statistical differences.
Prediction and enrichment analysis of
miRNAs related to hub genes
Targetscan (www.targetscan.org), an online database that predicts
potential interactions between genes and miRNAs, was used to
predict miRNAs associated with hub genes. Subsequently, enrichment
analysis of the predicted miRNAs was performed with the DNA
Intelligent Analysis (DIANA)-miRPath software (version 3.0;
http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mirpath/index;
DIANA LAB, University of Thessaly), a handy online tool for
enrichment analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay
A total of eight individuals were recruited,
including four healthy controls and four patients with TSCC.
Following surgery, four TSCC tissue samples from four patients with
TSCC and four para-carcinoma tissues were obtained. The research
conformed to the standards set by the Declaration of Helsinki and
was authorized by the Human Ethics and Research Ethics Committees
of the Fourth Hospital of the Hebei Medical University. Written
informed consents were obtained from all participants.
Total RNA was extracted from the tissue samples
using the TRIzol® (Beijing Biolab Technology Co., Ltd.)
and reverse transcribed into cDNA with the Servicebio®RT
First Strand cDNA Synthesis kit (cat. no. G3330; Wuhan Servicebio
Biotechnology Co., Ltd.) for 60 min at 42°C. Terminate the reaction
by heating at 70°C for 5 min. RT-qPCR was performed in a Light
Cycler® 4800 System (Roche Diagnostics) with a specific
set of primers for the amplification of secreted phosphoprotein 1
(SPP1) and fibronectin 1 (FN1) genes. Primers used were as follows:
SPP1 forward, 5′-CTAAACCCTGACCCATCT-3′, reverse,
5′-CAATGCCTTCTTTCATCT-3′; GAPDH forward,
5′-ATCCGATTACCGATACCTAGACC-3′, reverse,
5′-ATGGACTATATCCGACGACGA-3′; and FN1 forward,
5′-CCAACTACCAGTAGCGAAAA −3′, reverse, 5′-GCAGGGAAAGGAAAGAAA-3′. The
thermocycling conditions used were as follows: 95°C for 15 sec
followed by 30 cycles of 60°C for 60 sec. The relative
quantification units (relative quantification=2−ΔΔCq,
where Cq represents quantification cycle values) of each sample
were calculated (43) and presented
as fold change of gene expression relative to the control group.
GAPDH was used as an endogenous control.
Statistics
The data was expressed as percentage of the total
and the mean ± SD. When two groups were compared, the paired
Student's t-test were used to determine statistical significance.
For the stage analysis, one-way ANOVA was used to compare DEG
expression levels, using the pathological stage as a variable for
calculating differential expression. All statistical analyses were
conducted using SPSS software, version 23.0 (IBM Corp.). A P-value
<0.05 was considered statistically significant.
Results
Identification of DEGs in TSCC
A total of 1,147, 731 and 1,356 DEGs were obtained
from the GSE31056, GSE13601 and GSE78060 datasets, respectively.
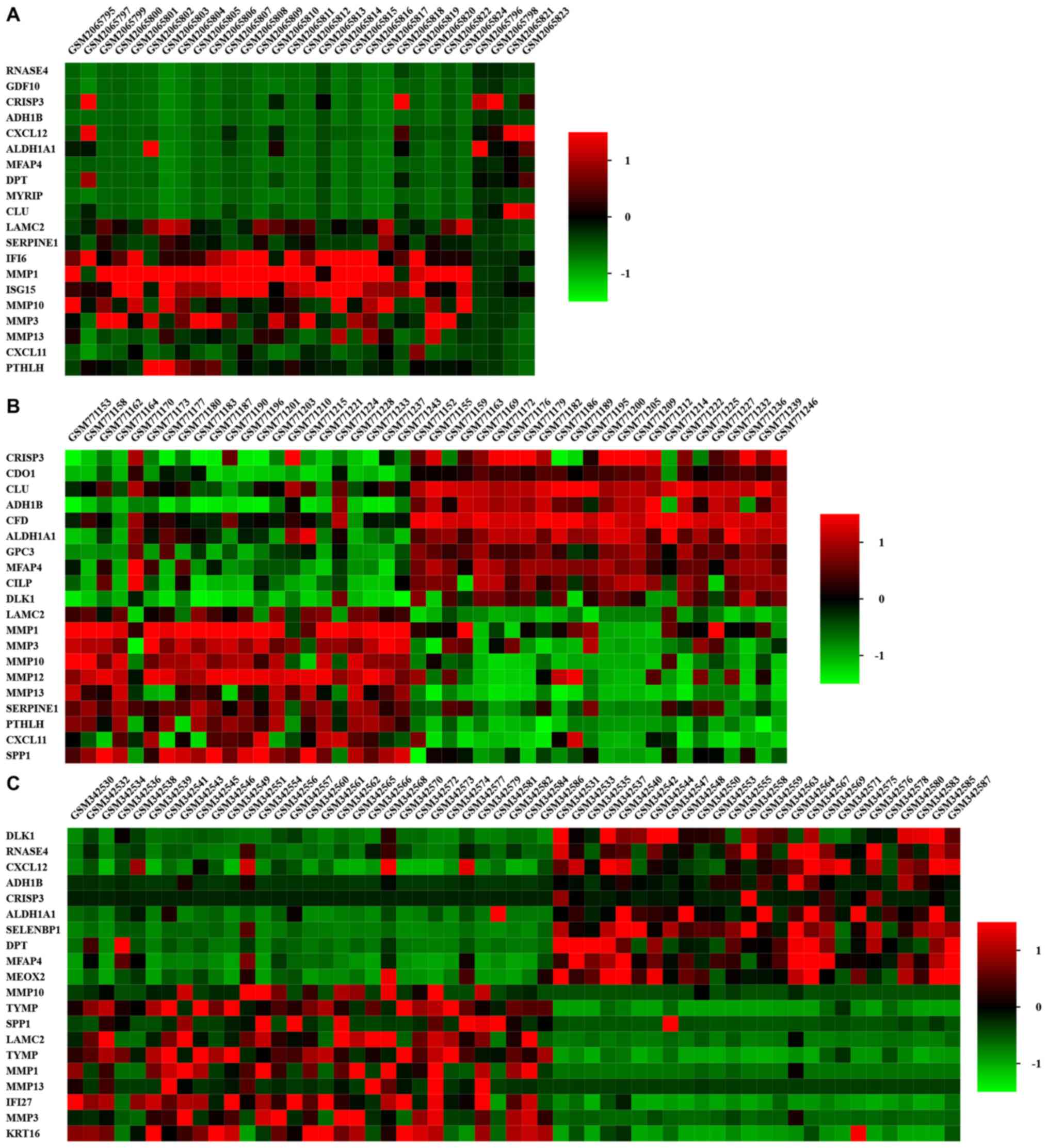
All datasets were downloaded from the GEO database (Table I). Identified DEGs were visualized
using volcano plots and heatmaps (Figs.
1A-C and 2). DEGs were selected
with |log2FC|>1.5 and adj. P-value <0.05 among the mRNA
expression profiling sets in GSE13601 (Fig. 1A), GSE31056 (Fig. 1B), and GSE78060 (Fig. 1C). The Venn diagram demonstrated that
83 common DEGs were obtained from the three datasets, including 48
upregulated and 35 downregulated genes (Fig. 1D; Table
II).
 | Table I.Summary of tongue squamous cell
carcinoma microarray datasets. |
Table I.
Summary of tongue squamous cell
carcinoma microarray datasets.
| Series | Platform | GeneChip | Samples |
|---|
| GSE31056 | GPL10526 | Affymetrix Human
Genome U133 Plus 2 Array | 96 |
| GSE13601 | GPL8300 | Affymetrix Human
Genome U95 Version 2 Array | 58 |
| GSE78060 | GPL570 | Affymetrix Human
Genome U133 Plus 2 Array | 30 |
 | Table II.Screening of differentially expressed
genes in oral squamous cell carcinoma samples. |
Table II.
Screening of differentially expressed
genes in oral squamous cell carcinoma samples.
| DEGs | List of gene
symbols |
|---|
| Upregulated
DEGs | MMP1, TYMP, MMP10,
KRT16, MMP13, SPP1, MMP3, LAMC2, IFI27, PTHLH, RBP1, ISG15, MMP12,
TNC, TGFBI, CXCL11, FSCN1, MYO1B, SERPINE1, STAT1, CDH3, ITGA6,
POSTN, SNAI2, PLAU, LAMA3, FAP, XAF1, DUSP14, APOL1, COL5A2, RSAD2,
TP63, MYO10, F2RL1, PTK7, ACTN1, LAMB3, EXT1, IFIT3, FN1, IGFBP3,
ITGA3, DFNA5, IFI44, COL4A1, LOXL2, MICAL2 |
| Downregulated
DEGs | PDPN, RECK, DPT,
CFD, IFI6, MEOX2, CXCL12, ABCA6, NR3C2, ITM2A, BEX4, PBX1, GDF10,
CBX7, MYRIP, LIFR, CLU, SLITRK5, LPIN1, GPRASP1, KAT2B, CDO1, GATM,
GPC3, SORBS2, FRZB, METTL7A, CILP, RNASE4, DLK1, CRISP3, MFAP4,
ALDH1A1, SELENBP1, ADH1B |
Enrichment analysis for DEGs
The enrichment analysis indicated that DEGs were
mainly enriched in BPs associated with cell adhesion, ECM
organization, ECM disassembly and proteolysis. With respect to CC,
DEGs were primarily enriched in the extracellular space,
extracellular region, extracellular exosome and ECM. DEGs were
mainly associated with integrin binding, serine-type endopeptidase
activity and metalloendopeptidase activity in the MF category. KEGG
pathway enrichment analysis found that DEGs were remarkably
enriched in the PI3K-Akt signaling pathway, focal adhesion and
ECM-receptor interaction (Fig.
3).
PPI network construction and module
analysis
To further understand the association among DEGs, a
PPI interaction network was conducted, including 63 nodes and 218
edges (Fig. 4A). Subsequently, the
most significant module in the PPI network was selected. The top 11
candidate hub genes were also selected, namely plasminogen
activator urokinase (PLAU), signal transducer and activator of
transcription 1 (STAT1), C-X-C motif chemokine ligand 12 (CXCL12),
matrix metallopeptidase (MMP) 13, SPP1, periostin, MMP1, MMP3, FN1,
serpin family E member 1 (SERPINE1) and snail family
transcriptional repressor 2 (SNAI2). The most significant module
was obtained from PPI network of DEGs (Fig. 4B).
Hub genes and their co-expression genes were
analyzed using cBioPortal. Nodes with bold black outline represent
hub genes. Nodes with thin black outline represent the
co-expression genes. Subsequently, the co-expression network of the
11 hub genes was constructed using the cBioportal online platform
to reveal genes sharing common expression patterns with hub genes
and to further study their associations (Fig. 5). The biological process analysis of
hub genes was constructed using BiNGO. The color depth of nodes
refers to the corrected P-value of ontologies. The size of nodes
refers to the numbers of genes that are involved in the ontologies.
Finally, the potential biological characteristics of the
co-expression network were visualized (Fig. 6).
Validation and analysis of hub
genes
Τo validate the differences in the expression levels
of the hub genes, the Oncomine database was utilized. Heat map of
DEGs identified from the Oncomine database. The color depth
represents the significance of the difference. TSCC vs. normal.
References are as follows: 1, Tongue squamous cell carcinoma vs.
Normal. Estilo Head-Neck, BMC Cancer, 2009; 2, Tongue squamous cell
carcinoma vs. Normal. Kuriakose Head-Neck, Cell Mol Life Sci, 2004;
3, Tongue squamous cell carcinoma vs. Normal. Talbot Lung, Cancer
Res, 2005; 4, Tongue squamous cell carcinoma vs. Normal. Ye
Head-Neck. BMC Genomics, 2008. (Fig.
7). Furthermore, the overall survival time expression analysis
of 11 hub genes was performed using the Kaplan-Meier plotter
database. The analysis results revealed that seven hub genes
exhibited a remarkable association with survival time (Figs. 8 and 9). In patients with TSCC, increased
expression levels of MMP1, MMP3, FN1, SERPINE1, SPP1, MMP13 and
SNAI2 were associated with a worse overall survival rate. In
addition, CXCL12, MMP3, FN1, SPP1 and STAT1 were found to be
differentially expressed in the various tumor stages. The
expression of CXCL12, FN1, MMP3, SPP1 and STAT1 were significantly
related with the tumor stage (P<0.05). However, the other hub
genes were not significantly related with the tumor stage
(P>0.05; Fig. 10).
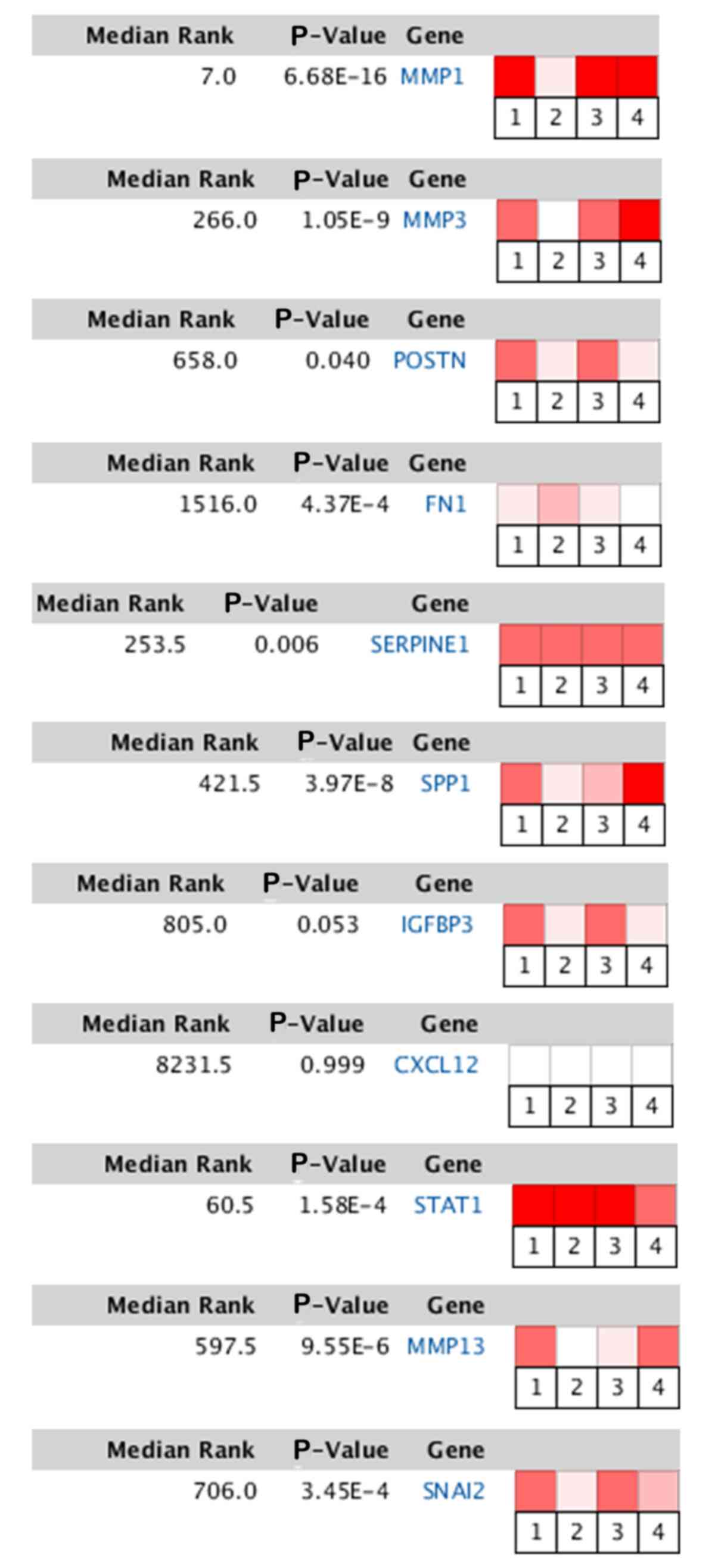 | Figure 7.Heat map of DEGs identified from the
Oncomine database. The color depth represents the significance of
the difference. TSCC vs. normal. MMP1, matrix metallopeptidase 1;
MMP3, matrix metallopeptidase 3; POSTN, periostin; FN1, fibronectin
1; SERPINE1, serpin family E member 1; SPP1, secreted
phosphoprotein 1; IGFBP3, insulin like growth factor binding
protein 3; CXCL12, C-X-C motif chemokine ligand 12; STAT1, signal
transducer and activator of transcription 1; MMP13, matrix
metallopeptidase 13; SNAI2, snail family transcriptional repressor
2. |
 | Figure 10.Hub genes expressed differentially in
each tumor stage. The expression of CXCL12, FN1, MMP3, SPP1 and
STAT1 were significantly different in different tumor stages.
However, MMP1, MMP13, PLAU, POSTN, SERPINE1 and SNAI2 were not
significantly related with the tumor stage (P>0.05). CXCL, C-X-C
motif chemokine ligand; FN, fibronectin; MMP, matrix
metallopeptidase; PLAU, plasminogen activator urokinase; POSTN,
periostin; SERPINE, serpin family E member; SNAI, snail family
transcriptional repressor; SPP, secreted phosphoprotein; STAT,
signal transducer and activator of transcription. |
Prediction and enrichment analysis of
miRNAs related to hub genes
To further elucidate the mechanisms of action and
regulatory networks of the hub genes, miRNAs associated with hub
genes were predicted (Table III).
Enrichment analysis of the predicted miRNAs was subsequently
performed. GO analysis indicated that miRNAs were significantly
enriched in the toll-like receptor (TLR)1:TLR2 signaling pathway,
nucleic acid binding transcription factor activity and cellular
protein modification process. Furthermore, KEGG pathway enrichment
analysis revealed that pathways associated with the TGF-β, Ras,
prolactin and MAPK signaling pathways were the most enriched
(Fig. 11).
 | Table III.The potential microRNAs associated
with the hub genes. |
Table III.
The potential microRNAs associated
with the hub genes.
| Gene | Predicted
microRNAs |
|---|
| MMP1 | hsa-miR-558,
hsa-miR-202-3p and hsa-miR-520g-5p |
| MMP3 | hsa-miR-365b-3p,
hsa-miR-365a-3p and hsa-miR-550b-2-5p |
| POSTN | hsa-miR-19b-3p,
hsa-miR-19a-3p and hsa-miR-5590-3p |
| FN1 | hsa-miR-613,
hsa-miR-1271-5p and hsa-miR-96-5p |
| SERPINE1 | hsa-miR-6088,
hsa-miR-148b-3p and hsa-miR-152-3p |
| SPP1 | hsa-miR-181c-5p,
hsa-miR-181a-5p and hsa-miR-181d-5p |
| IGFBP3 | hsa-miR-19a-3p,
hsa-miR-19b-3p and hsa-miR-212-5p |
| CXCL12 | hsa-miR-137,
hsa-miR-135a-5p and hsa-miR-135b-5p |
| STAT1 | hsa-miR-216a-3p,
hsa-miR-3681-3p and hsa-miR-128-3p |
| MMP13 | hsa-miR-27a-3p,
hsa-miR-27b-3p and hsa-miR-1267 |
| SNAI2 | hsa-miR-124-3p.1,
hsa-miR-206 and hsa-miR-1-3p |
Results of RT-qPCR analysis
The expression levels of SPP1 and FN1 were further
measured using RT-qPCR. The results indicated that the relative
expression levels of SPP1 and FN1 were significantly increased in
the TSCC tissue samples compared with those in the control group
(Fig. 12). This finding suggested
that SPP1 and FN1 may be considered as biomarkers for TSCC.
Discussion
The results of the present study demonstrated that
SPP1 and FN1 were highly expressed in TSCC compared with normal
tissues. Furthermore, the SPP1 and FN1 expression levels were
gradually elevated with the increase of TSCC pathological staging.
Patients with high SPP1 and FN1 expression levels exhibited poorer
overall survival compared with those with decreased ones.
SPP1, also known as osteopontin, is a widely
expressed viscous glycoprotein with several biological activities,
which is secreted by various cell types, including osteoclasts and
T cells (44). SPP1 may be detected
in normal tissues, body fluids and cells and is involved in various
physiological processes, such as development, differentiation,
inflammation and wound healing (45). It has been reported that increased
expression of SPP1 plays an important role in the occurrence and
metastasis of various malignant tumor types (46). When tumor cells invade the ECM, SPP1
promotes the expression of MMPs in tumor cells through the
NF-κB-dependent signal transduction pathway (47). Subsequently, MMPs degrade the cell
basement membrane and ECM, resulting in tumor invasion and
metastasis, as well as a poor prognosis for patients with cutaneous
melanoma (47,48). SPP1 is also overexpressed in lung
adenocarcinomas and serves as an independent prognostic biomarker,
especially for T1, T2 and N0 tumor stages (44). However, the predictive value of SPP1
in lung squamous cell carcinoma needs further investigation
(49). In addition, SPP1 is
significantly upregulated in patients with colorectal cancer
compared with healthy individuals (50). Therefore, it has been previously
demonstrated that SPP1 promotes colorectal cancer metastasis by
activating uncommon interstitial transformation, thus, SPP1 is
considered as a potential therapeutic target for patients with
colorectal cancer (51).
Overexpression of SPP1 may also be involved in the development of
nasopharyngeal carcinoma (52,53),
while a previous study has revealed that polymorphisms in the SPP1
gene are associated with other malignant tumor types such as
gliomas and lung cancer (54). Zou
et al (55) performed
bioinformatics analysis to identify the key DEGs between oral
squamous cell carcinoma (OSCC) and normal tissues. The analysis
revealed that SPP1, integrin subunit α3 and PLAU could be the key
candidate genes of OSCC (55).
Furthermore, SPP1 was upregulated in OSCC, indicating that it could
serve as an underlying predictor of OSCC (56). A previous study demonstrated that
SPP1 was significantly associated with esophageal squamous cell
carcinoma and could; therefore, be a potential target for therapy
(57). Several studies have also
suggested that SPP1 may be involved in the occurrence and
development of OSCC (58–61). These studies indicated that SPP1 may
act as an underlying biomarker of TSCC, a subtype of OSCC. The
present study provided direct evidence that SPP1 expression was
associated with TSCC. Therefore, the present data revealed that
SPP1 was highly expressed in TSCC compared with normal tissues,
while SPP1 and FN1 levels were gradually elevated with the increase
in TSCC pathological staging. Compared with individuals exhibiting
decreased SPP1 expression levels, upregulation of SPP1 and FN1 in
patients with TSCC was associated with a poor overall survival
rate. However, to the best of our knowledge, no studies have been
performed to investigate the role of SPP1 in TSCC. The molecular
and biological functions of SPP1 and its cancer-promoting effects
in other types of cancer (62), led
to the hypothesis that SPP1 also played an important role in the
occurrence of TSCC. Therefore, the clinical detection of SPP1 may
be used as a diagnostic biomarker for TSCC.
FN1 is composed of a variety of homologous repeating
units and participates in cell movement, growth and differentiation
as well as matrix formation, while it also plays an important role
in the mechanisms behind cell adhesion (63,64). It
has been reported that FN1 affects cell migration by mediating
cell-to-cell and cell matrix adhesion (65). Overexpression of FN1 promotes
adhesion and aggregation of tumor cells by affecting the movement,
differentiation and growth of tumor cells (66). Furthermore, FN1 inhibits migration of
tumor cells and triggers tumor metastasis through mediating
intercellular and cell matrix adhesion (67). In addition, FN1, serves as a ligand
for 12 members of the integrin receptor family, an important family
of ECM-associated adhesion receptors (68). Morita et al (69) demonstrated that FN1 overexpression
accelerates the progress and lymph node metastasis of OSCC through
promoting the expression of vascular endothelial growth factor-C. A
number of previous studies have provided insights into the role of
FN1 as a novel biomarker for OSCC (70–72),
indicating that FN1 may also be used as an underlying biomarker of
TSCC. This hypothesis was further verified in the present study.
FN1 affects the fourth stage of local invasion and spread of
tumors, namely the migration of tumor cells (67). Tumor cells migrate through the
basement membrane with an amoeboid form of movement, following the
binding of the integrin transmembrane receptor to FN1 (73). FN1 also participates in matrix
remodeling and affects cell movement by regulating actin
aggregation (74). Therefore,
investigating the interactions and mechanisms through which FN1
functions may improve the understanding of its role in TSCC,
provide novel approaches for investigating its underlying
mechanisms of action and develop more effective treatments.
Studying the molecular mechanism of action of FN1 behind the
occurrence and progression of TSCC may further the current
understanding of the potential targeting of FN1 for the treatment
of TSCC (70).
It should be noted that the present study has some
limitations. Firstly, this was an observational study based on the
bioinformatics analysis of 11 key genes. Therefore, the results of
the present study provided novel clues that could be used for
subsequent, in-depth studies investigating the developmental
mechanisms of action behind TSCC. In addition, future studies
involving more samples should be conducted to verify the results of
the present study.
In conclusion, a total of 83 DEGs and 11 hub genes,
especially SPP1 and FN1, were obtained from the bioinformatics and
microarray assays between TSCC and normal tissues. These genes
could be used as diagnostic and therapeutic biomarkers for TSCC.
Furthermore, SPP1 and FN1 may be associated with the occurrence,
lymph node metastasis and malignant progression of TSCC. Therefore,
both molecules were considered as potential biomarkers for
monitoring TSCC progression.
Acknowledgements
The authors would like to thank Dr. Peng Guo (The
Fourth Hospital of Hebei Medical University, Shijiazhuang, China)
for his statistical assistance and suggestions during the
submitting process.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XLX and HL performed the experiments and were major
contributors in writing the manuscript and submitting the
manuscript. TKL made substantial contributions to research
conception and designed the draft of the research process. YZ and
SXZ were involved in revising manuscript critically for important
intellectual content and made substantial contributions to
conception and design, acquisition of data, and analysis and
interpretation of all data. ZC and YB analyzed the data. YZ and SXZ
confirmed the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The data of this research was downloaded from the
GEO database, a public database. All institutional and national
guidelines for the care and use of participates were followed. The
research conformed to the standards set by the Declaration of
Helsinki and was authorized by the Human Ethics and Research Ethics
Committees of the Fourth Hospital of the Hebei Medical University.
Written informed consents were obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Paderno A, Morello R and Piazza C: Tongue
carcinoma in young adults: A review of the literature. Acta
Otorhinolaryngol Ital. 38:175–180. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mannelli G, Arcuri F, Agostini T,
Innocenti M, Raffaini M and Spinelli G: Classification of tongue
cancer resection and treatment algorithm. J Surg Oncol.
117:1092–1099. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sun L, Liang J, Wang Q, Li Z, Du Y and Xu
X: MicroRNA-137 suppresses tongue squamous carcinoma cell
proliferation, migration and invasion. Cell Prolif. 49:628–635.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Semsettin B, Sinan E and Nigar V:
Comparison of the effects of topical cyclosporine a 0.05%,
cyclosporine a 2%, epinastine hydrochloride 0.05%, and prednisolone
acetate 1% on allergic inflammation in an experimental allergic
conjunctivitis model. Cornea. 32:1465–1469. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bello IO, Soini Y and Salo T: Prognostic
evaluation of oral tongue cancer: Means, markers and perspectives
(II). Oral Oncol. 46:636–643. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ahmadi N, Chan M, Huo YR, Sritharan N and
Chin RY: Survival outcome of tonsillar squamous cell carcinoma
(TSCC) in the context of human papillomavirus (HPV): A systematic
review and meta-analysis. Surgeon. 17:6–14. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ramqvist T, Grün N and Dalianis T: Human
papillomavirus and tonsillar and base of tongue cancer. Viruses.
7:1332–1343. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lozev I, Ruseva S, Pidakev I, Cardoso JC,
Wollina U, Lotti T, Maximov GK, Terziev I and Tchernev G:
Mucoepidermoid carcinoma (MEC) of parotid gland with massive
cutaneous involvement: Bilateral pedicle advancement flap
(U-Plasty) as adequate surgical approach. Open Access Maced J Med
Sci. 6:134–136. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu L, Wang Y, Li R, Liu A, Zhang X, Zuo C
and Xu X: Surgical treatment of early tongue squamous cell
carcinoma and patient survival. Oncol Lett. 17:5681–5685.
2019.PubMed/NCBI
|
|
10
|
Ng JH, Iyer NG, Tan MH and Edgren G:
Changing epidemiology of oral squamous cell carcinoma of the
tongue: A global study. Head Neck. 39:297–304. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wade MH and Plotnick H: Xeroderma
pigmentosum and squamous cell carcinoma of the tongue.
Identification of two black patients as members of complementation
group C. J Am Acad Dermatol. 12:515–521. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang P, Zhang L, Liu H, Zhao L, Li Y,
Shen JX, Liu Q, Liu MZ and Xi M: Clinicopathologic characteristics
and prognosis of tongue squamous cell carcinoma in patients with
and without a history of radiation for nasopharyngeal carcinoma: A
matched case-control study. Cancer Res Treat. 49:695–705. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Velly AM, Franco EL, Schlecht N, Pintos J,
Kowalski LP, Oliveira BV and Curado MP: Relationship between dental
factors and risk of upper aerodigestive tract cancer. Oral Oncol.
34:284–291. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Troiano G, Rubini C, Togni L, Caponio VC,
Zhurakivska K, Santarelli A, Cirillo N, Lo Muzio L and Mascitti M:
The immune phenotype of tongue squamous cell carcinoma predicts
early relapse and poor prognosis. Cancer Med. 9:8333–8344. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hussein AA, Forouzanfar T, Bloemena E, de
Visscher J, Brakenhoff RH, Leemans CR and Helder MN: A review of
the most promising biomarkers for early diagnosis and prognosis
prediction of tongue squamous cell carcinoma. Br J Cancer.
119:724–736. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakamura K, Akiba J, Ogasawara S, Naito Y,
Nakayama M, Abe Y, Kusukawa J and Yano H: SUOX is negatively
associated with multistep carcinogenesis and proliferation in oral
squamous cell carcinoma. Med Mol Morphol. 51:102–110. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng G, Zhang Z, Liu H, Xiong Y, Luo L,
Jia X, Peng C, Zhang Q, Li N, Gu Y, et al: HSP27-mediated
extracellular and intracellular signaling pathways synergistically
confer chemoresistance in squamous cell carcinoma of tongue. Clin
Cancer Res. 24:1163–1175. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen H and Dai J: miR-409-3p suppresses
the proliferation, invasion and migration of tongue squamous cell
carcinoma via targeting RDX. Oncol Lett. 16:543–551.
2018.PubMed/NCBI
|
|
19
|
Solomon B, Young RJ and Rischin D: Head
and neck squamous cell carcinoma: Genomics and emerging biomarkers
for immunomodulatory cancer treatments. Semin Cancer Biol.
52:228–240. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Falzone L, Lupo G, La Rosa G, Crimi S,
Anfuso CD, Salemi R, Rapisarda E, Libra M and Candido S:
Identification of novel MicroRNAs and their diagnostic and
prognostic significance in oral cancer. Cancers (Basel).
11:6102019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu M, Wu G, Chen Y, Wang H, Gao Y and Wang
A: Bioinformatic screening and experimental analysis identify SFRP1
as a prognostic biomarker for tongue squamous cell carcinomas. Arch
Oral Biol. 110:1045872020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang R, Zhou X, Wang H, Zhou B, Dong S,
Ding Q, Peng M, Sheng X, Yao J, Huang R, et al: Integrative
analysis of gene expression profiles reveals distinct molecular
characteristics in oral tongue squamous cell carcinoma. Oncol Lett.
17:2377–2387. 2019.PubMed/NCBI
|
|
23
|
Usami Y, Ishida K, Sato S, Kishino M,
Kiryu M, Ogawa Y, Okura M, Fukuda Y and Toyosawa S: Intercellular
adhesion molecule-1 (ICAM-1) expression correlates with oral cancer
progression and induces macrophage/cancer cell adhesion. Int J
Cancer. 133:568–578. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang D, Chen ZG, Liu SH, Dong ZQ, Dalin
M, Bao SS, Hu YW and Wei FC: Galectin-3 gene silencing inhibits
migration and invasion of human tongue cancer cells in vitro via
downregulating β-catenin. Acta Pharmacol Sin. 34:176–184. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang C, Liu X, Chen Z, Huang H, Jin Y,
Kolokythas A, Wang A, Dai Y, Wong DT and Zhou X: Polycomb group
protein EZH2-mediated E-cadherin repression promotes metastasis of
oral tongue squamous cell carcinoma. Mol Carcinog. 52:229–236.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Candido S, Lupo G, Pennisi M, Basile MS,
Anfuso CD, Petralia MC, Gattuso G, Vivarelli S, Spandidos DA, Libra
M and Falzone L: The analysis of miRNA expression profiling
datasets reveals inverse microRNA patterns in glioblastoma and
Alzheimer's disease. Oncol Rep. 42:911–922. 2019.PubMed/NCBI
|
|
27
|
Falzone L, Romano GL, Salemi R, Bucolo C,
Tomasello B, Lupo G, Anfuso CD, Spandidos DA, Libra M and Candido
S: Prognostic significance of deregulated microRNAs in uveal
melanomas. Mol Med Rep. 19:2599–2610. 2019.PubMed/NCBI
|
|
28
|
Falzone L, Scola L, Zanghì A, Biondi A, Di
Cataldo A, Libra M and Candido S: Integrated analysis of colorectal
cancer microRNA datasets: Identification of microRNAs associated
with tumor development. Aging (Albany NY). 10:1000–1014. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Falzone L, Candido S, Salemi R, Basile MS,
Scalisi A, McCubrey JA, Torino F, Signorelli SS, Montella M and
Libra M: Computational identification of microRNAs associated to
both epithelial to mesenchymal transition and NGAL/MMP-9 pathways
in bladder cancer. Oncotarget. 7:72758–72766. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hafsi S, Candido S, Maestro R, Falzone L,
Soua Z, Bonavida B, Spandidos DA and Libra M: Correlation between
the overexpression of Yin Yang 1 and the expression levels of
miRNAs in Burkitt's lymphoma: A computational study. Oncol Lett.
11:1021–1025. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li R, Faden DL, Fakhry C, Langelier C,
Jiao Y, Wang Y, Wilkerson MD, Pedamallu CS, Old M, Lang J, et al:
Clinical, genomic, and metagenomic characterization of oral tongue
squamous cell carcinoma in patients who do not smoke. Head Neck.
37:1642–1649. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li B, Li CH, Guo H, Chen J and Wang SX:
Analysis of 27 cases of defect restoration using infrahyoid
myocutaneous flap after intraoral cancer surgery. Zhonghua Er Bi
Yan Hou Tou Jing Wai Ke Za Zhi. 43:826–829. 2008.(In Chinese).
PubMed/NCBI
|
|
33
|
Liu X, Qiao B, Zhao T, Hu F, Lam AK and
Tao Q: Sox2 promotes tumor aggressiveness and
epithelial-mesenchymal transition in tongue squamous cell
carcinoma. Int J Mol Med. 42:1418–1426. 2018.PubMed/NCBI
|
|
34
|
Tang Q, Cheng B, Xie M, Chen Y, Zhao J,
Zhou X and Chen L: Circadian clock gene bmal1 inhibits
tumorigenesis and increases paclitaxel sensitivity in tongue
squamous cell carcinoma. Cancer Res. 77:532–544. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tanaka Y, Araki K, Tanaka S, Miyagawa Y,
Suzuki H, Kamide D, Tomifuji M, Uno K, Kimura E, Yamashita T, et
al: Sentinel lymph node-targeted therapy by oncolytic sendai virus
suppresses micrometastasis of head and neck squamous cell carcinoma
in an orthotopic nude mouse model. Mol Cancer Ther. 18:1430–1438.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xiong J, Feng J, Qiu L, Gao Z, Li P, Pang
L and Zhang Z: SDF-1-loaded PLGA nanoparticles for the targeted
photoacoustic imaging and photothermal therapy of metastatic lymph
nodes in tongue squamous cell carcinoma. Int J Pharm. 554:93–104.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McCubrey JA, Lertpiriyapong K, Steelman
LS, Abrams SL, Yang LV, Murata RM, Rosalen PL, Scalisi A, Neri LM,
Cocco L, et al: Effects of resveratrol, curcumin, berberine and
other nutraceuticals on aging, cancer development, cancer stem
cells and microRNAs. Aging (Albany NY). 9:1477–1536. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qiu Y, Meng LB, Di CY, Huo YH, Yao BC,
Zhang TJ and Hua Z: Exploration of the differentially expressed
long noncoding RNAs and genes of morphine tolerance via
bioinformatic analysis. J Comput Biol. 26:1379–1393. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zou YF, Meng LB, Wang QQ, He ZK, Hu CH,
Shan MJ, Wang DY and Yu X: Identification and functional enrichment
analysis of potential diagnostic and therapeutic targets in
adamantinomatous craniopharyngioma. J Comput Biol. 27:55–68. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang H, Liu J, Fu X and Yang A:
Identification of key genes and pathways in tongue squamous cell
carcinoma using bioinformatics analysis. Med Sci Monit.
23:5924–5932. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li T, Wu Q, Liu D and Wang X: miR-27b
suppresses tongue squamous cell carcinoma epithelial-mesenchymal
transition by targeting ITGA5. Onco Targets Ther. 13:11855–11867.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen Y, Guo Y and Yan W: lncRNA
RP5-916L7.2 correlates with advanced tumor stage, and promotes
cells proliferation while inhibits cells apoptosis through
targeting miR-328 and miR-939 in tongue squamous cell carcinoma.
Clin Biochem. 67:24–32. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Meng LB, Shan MJ, Qiu Y, Qi R, Yu ZM, Guo
P, Di CY and Gong T: TPM2 as a potential predictive biomarker for
atherosclerosis. Aging (Albany NY). 11:6960–6982. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang Y, Du W, Chen Z and Xiang C:
Upregulation of PD-L1 by SPP1 mediates macrophage polarization and
facilitates immune escape in lung adenocarcinoma. Exp Cell Res.
359:449–457. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kramerova I, Kumagai-Cresse C, Ermolova N,
Mokhonova E, Marinov M, Capote J, Becerra D, Quattrocelli M,
Crosbie RH, Welch E, et al: Spp1 (osteopontin) promotes TGFβ
processing in fibroblasts of dystrophin-deficient muscles through
matrix metalloproteinases. Hum Mol Genet. 28:3431–3442. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Morse C, Tabib T, Sembrat J, Buschur KL,
Bittar HT, Valenzi E, Jiang Y, Kass DJ, Gibson K, Chen W, et al:
Proliferating SPP1/MERTK-expressing macrophages in idiopathic
pulmonary fibrosis. Eur Respir J. 54:18024412019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Guarneri C, Bevelacqua V, Polesel J,
Falzone L, Cannavò PS, Spandidos DA, Malaponte G and Libra M: NF-κB
inhibition is associated with OPN/MMP-9 downregulation in cutaneous
melanoma. Oncol Rep. 37:737–746. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shevde LA and Samant RS: Role of
osteopontin in the pathophysiology of cancer. Matrix Biol.
37:131–141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li S, Yang R, Sun X, Miao S, Lu T, Wang Y,
Wo Y and Jiao W: Identification of SPP1 as a promising biomarker to
predict clinical outcome of lung adenocarcinoma individuals. Gene.
679:398–404. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Choe EK, Yi JW, Chai YJ and Park KJ:
Upregulation of the adipokine genes ADIPOR1 and SPP1 is related to
poor survival outcomes in colorectal cancer. J Surg Oncol.
117:1833–1840. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xu C, Sun L, Jiang C, Zhou H, Gu L, Liu Y
and Xu Q: SPP1, analyzed by bioinformatics methods, promotes the
metastasis in colorectal cancer by activating EMT pathway. Biomed
Pharmacother. 91:1167–1177. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang HH, Wang XW and Tang CE: Osteopontin
expression in nasopharyngeal carcinoma: Its relevance to the
clinical stage of the disease. J Cancer Res Ther. 7:138–142. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ma R, Luo X, Feng S, Li J, Fan Y, Wen W
and Li H: Osteopontin promotes EZH2 expression and tumor
progression in nasopharyngeal carcinoma. ORL J Otorhinolaryngol
Relat Spec. 76:273–281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang G, Peng X, Guo P and Yang G:
Association of osteopontin polymorphism with cancer risk: A
meta-analysis. Int J Clin Exp Med. 8:20911–20917. 2015.PubMed/NCBI
|
|
55
|
Zou B, Li J, Xu K, Liu JL, Yuan DY, Meng Z
and Zhang B: Identification of key candidate genes and pathways in
oral squamous cell carcinoma by integrated Bioinformatics analysis.
Exp Ther Med. 17:4089–4099. 2019.PubMed/NCBI
|
|
56
|
Zhang C, Man DP, Ma SM, Cao SW and Li DW:
Expressions and significances of CD147, OPN and MMP-2 in oral
squamous cell carcinoma. Sichuan Da Xue Xue Bao Yi Xue Ban.
43:683–686. 2012.(In Chinese). PubMed/NCBI
|
|
57
|
Ito T, Hashimoto Y, Tanaka E, Kan T,
Tsunoda S, Sato F, Higashiyama M, Okumura T and Shimada Y: An
inducible short-hairpin RNA vector against osteopontin reduces
metastatic potential of human esophageal squamous cell carcinoma in
vitro and in vivo. Clin Cancer Res. 12:1308–1316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hu Q, Peng J, Chen X, Li H, Song M, Cheng
B and Wu T: Obesity and genes related to lipid metabolism predict
poor survival in oral squamous cell carcinoma. Oral Oncol.
89:14–22. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang X, Zhang L, Tan X, Lin Y, Han X,
Wang H, Ming H, Li Q, Liu K and Feng G: Systematic analysis of
genes involved in oral cancer metastasis to lymph nodes. Cell Mol
Biol Lett. 23:532018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
D'Addazio G, Artese L, Traini T, Rubini C,
Caputi S and Sinjari B: Immunohistochemical study of osteopontin in
oral squamous cell carcinoma allied to fractal dimension. J Biol
Regul Homeost Agents. 32:1033–1038. 2018.PubMed/NCBI
|
|
61
|
Huang CF, Yu GT, Wang WM, Liu B and Sun
ZJ: Prognostic and predictive values of SPP1, PAI and caveolin-1 in
patients with oral squamous cell carcinoma. Int J Clin Exp Pathol.
7:6032–6039. 2014.PubMed/NCBI
|
|
62
|
Wang Y, Su J, Wang Y, Fu D, Ideozu JE,
Geng H, Cui Q, Wang C, Chen R, Yu Y, et al: The interaction of YBX1
with G3BP1 promotes renal cell carcinoma cell metastasis via
YBX1/G3BP1-SPP1- NF-κB signaling axis. J Exp Clin Cancer Res.
38:3862019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Aota Y, An HS, Homandberg G, Thonar EJ,
Andersson GB, Pichika R and Masuda K: Differential effects of
fibronectin fragment on proteoglycan metabolism by intervertebral
disc cells: A comparison with articular chondrocytes. Spine (Phila
Pa 1976). 30:722–728. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Beumer S, Heijnen HF, IJsseldijk MJ,
Orlando E, de Groot PG and Sixma JJ: Platelet adhesion to
fibronectin in flow: The importance of von Willebrand factor and
glycoprotein Ib. Blood. 86:3452–3460. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Filenius S, Tervo T and Virtanen I:
Production of fibronectin and tenascin isoforms and their role in
the adhesion of human immortalized corneal epithelial cells. Invest
Ophthalmol Vis Sci. 44:3317–3325. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Amary F, Perez-Casanova L, Ye H, Cottone
L, Strobl AC, Cool P, Miranda E, Berisha F, Aston W, Rocha M, et
al: Synovial chondromatosis and soft tissue chondroma: Extraosseous
cartilaginous tumor defined by FN1 gene rearrangement. Mod Pathol.
32:1762–1771. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Cai X, Liu C, Zhang TN, Zhu YW, Dong X and
Xue P: Down-regulation of FN1 inhibits colorectal carcinogenesis by
suppressing proliferation, migration, and invasion. J Cell Biochem.
119:4717–4728. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Pankov R and Yamada KM: Fibronectin at a
glance. J Cell Sci. 115:3861–3863. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Morita Y, Hata K, Nakanishi M, Omata T,
Morita N, Yura Y, Nishimura R and Yoneda T: Cellular fibronectin 1
promotes VEGF-C expression, lymphangiogenesis and lymph node
metastasis associated with human oral squamous cell carcinoma. Clin
Exp Metastasis. 32:739–753. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yen CY, Huang CY, Hou MF, Yang YH, Chang
CH, Huang HW, Chen CH and Chang HW: Evaluating the performance of
fibronectin 1 (FN1), integrin α4β1 (ITGA4), syndecan-2 (SDC2), and
glycoprotein CD44 as the potential biomarkers of oral squamous cell
carcinoma (OSCC). Biomarkers. 18:63–72. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Suresh A, Vannan M, Kumaran D, Gümüs ZH,
Sivadas P, Murugaian EE, Kekatpure V, Iyer S, Thangaraj K and
Kuriakose MA: Resistance/response molecular signature for oral
tongue squamous cell carcinoma. Dis Markers. 32:51–64. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang Z, Pan J, Li L, Wang Z, Xiao W and
Li N: Survey of risk factors contributed to lymphatic metastasis in
patients with oral tongue cancer by immunohistochemistry. J Oral
Pathol Med. 40:127–134. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Brinkhof B, Zhang B, Cui Z, Ye H and Wang
H: ALCAM (CD166) as a gene expression marker for human mesenchymal
stromal cell characterisation. Gene X. 5:1000312020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhan S, Li J, Wang T and Ge W:
Quantitative proteomics analysis of sporadic medullary thyroid
cancer reveals FN1 as a potential novel candidate prognostic
biomarker. Oncologist. 23:1415–1425. 2018. View Article : Google Scholar : PubMed/NCBI
|