Introduction
Non-small cell lung cancer (NSCLC) is the main type
of lung cancer and has two major histological subtypes, squamous
cell carcinoma (SCC) and adenocarcinoma (AD), which accounts for
~85% of lung cancer cases (1). Due
to the late presentation of symptoms and the lack of early
diagnosis, numerous patients with NSCLC are diagnosed at advanced
stages of the disease (2), which is
a major contributing factor to the poor prognosis of NSCLC. Radical
surgery remains the most effective treatment for early-stage NSCLC
(3). The 5-year survival rate
following surgical excision is ~70% for patients with stage I
NSCLC, and only 30% for patients with stage III NSCLC (3). Therefore, effective methods of early
diagnosis are necessary to reduce the mortality rate. A large,
randomized study revealed that screening with low-dose computed
tomography (LDCT) for lung cancer results in a 20% decline in
mortality in heavy smokers compared with the findings of the chest
X-ray (4,5). However, the high false-positive error,
poor cost-effectiveness and the potential side effects associated
with LDCT screening limit its use in clinical settings (6). In addition, European research groups
have reported less significant results compared with National Lung
Screening Trial in the United States and suggested that LDCT scan
is not routinely recommended for lung cancer screening (7). Certain protein markers, such as
carcinoembryonic antigen (CEA) and cytokeratin 19 fragment 21-1
have been extensively used in clinical practice; however, these
tumor markers are not sensitive and specific to contribute to the
early detection of NSCLC (8). Thus,
the identification of novel markers that can detect the presence of
early-stage tumors and predict cancer relapse would provide useful
tools for earlier NSCLC diagnosis, with the potential to reduce
mortality.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs
that modulate gene activity and are aberrantly expressed in the
majority of cancer types (9).
Circulating miRNAs have been demonstrated to serve as biomarkers
for cancer diagnosis and prognosis prediction due to their high
stability in the bloodstream, cancer-specific regulatory role,
reproducibility and ability for non-invasive detection (10,11).
Several studies have identified various circulating miRNAs as
biomarkers for the diagnosis and prognosis of NSCLC, among numerous
differentially expressed miRNAs (12–17).
However, due to the difference in research designs, experimental
methods and study populations, the results obtained by different
groups on this topic markedly vary.
The present study performed reverse transcription-
quantitative PCR (RT-qPCR) analysis to detect a panel of different
NSCLC-relevant miRNAs in RNA isolated from the plasma of patients
with NSCLC, patients with benign lung disease (BLD) and healthy
controls. In the training set, 12 miRNAs (Table SI) that have previously been
reported to be associated with NSCLC were selected, and their
expression levels in the plasma samples were detected. The
significantly aberrant expression of these miRNAs was further
evaluated in the testing set. The present study aimed to screen out
plasma miRNA expression profiles that may act as valuable
biomarkers for the early diagnosis and prognosis of NSCLC.
Materials and methods
Study design and participants
A total of 128 patients with histopathologically
confirmed stage I–IIIA NSCLC, 70 patients with BLD and 60 healthy
controls were enrolled in the present study between January 2014
and December 2018 at the Affiliated Hospital of Jiangsu University
(Zhenjiang, China). To identify plasma miRNAs as biomarkers for
NSCLC, a prospective two-phase and case-control study was designed
to distinguish differentially expressed miRNAs between patients
with NSCLC and healthy controls. In total, 12 candidate miRNAs that
have been reported to be abnormally expressed in NSCLC tissues or
blood samples of patients with NSCLC were selected (Table SI) (16–41), and
their plasma levels were detected via RT-qPCR analysis in the
training set, which was composed of 40 patients with NSCLC and 20
healthy subjects. Next, four miRNAs with significantly different
expression levels were selected and determined in the plasma
samples of 20 patients with BLD. Subsequently, the validation of
these four significantly aberrantly expressed miRNAs in the plasma
specimens from the training set was performed in the testing set
using plasma samples from 88 NSCLC cases, 50 BLD cases and 40
healthy subjects. These detected miRNAs were compared with serum
CEA, a classical tumor marker (8).
The detailed clinical data of the study subjects are
summarized in Table I. No
significant differences were observed in the distribution of age,
sex and smoking status among patients with NSCLC or BLD and healthy
controls in the training and testing sets. The patients with NSCLC
inclued 55 cases with squamous cell carcinoma (SCC) and 70 cases
with non-SCC consisted of 67 cases with adenocarcinoma and 3 cases
with large cell carcinoma. All patients with NSCLC underwent tumor
resection, and their blood samples were collected prior to surgery,
and stored at −20°C until subsequent experimentation. In 88/128
patients with NSCLC, serial blood specimens were collected ~2 weeks
after surgery and before chemotherapy. These 88 patients were
followed up from January 2014 to December 2018 (regular outpatient
or telephone follow-up every 3 months) and their complete follow-up
data were obtained. Analysis of disease-free survival (DFS) was
performed according to high or low expression levels of the four
plasma miRNAs in the 88 patients, prior to surgery. Subgroup DFS
analysis was performed based on histological type. Patients with
SCC (n=41) and patients with non-SCC (n=47) were divided into two
groups according to high or low expression plasma levels of miR-210
and miR-150 (n=21 vs. n=20 for miR-210; and n=24 vs. n=23 for
miR-150). In addition, the 88 patients with NSCLC were divided into
two groups according to the decreased degree in the plasma levels
of the four miRNAs after surgery (>50 and ≤50%), and survival
analysis was performed to assess the impact of the decreased degree
in the four plasma miRNA levels on DFS in the patients with NSCLC
after surgery. All patients with stage IB-IIIA received
platinum-based chemotherapy after surgery. Tumor pathological
staging was classified according to the 7th edition of the
international tumor-node-metastasis system published by the
International Association for the Study of Lung Cancer (42). Blood samples were also collected from
patients with BLD and healthy controls and stored at −20°C until
subsequent experimentation. The healthy individuals were matched
not only with patients with NSCLC, but also with patients with BLD
by age, sex and smoking status (Table
I). BLD cases included pneumonia (n=24), chronic obstructive
pulmonary disease (COPD; n=20), interstitial lung disease (n=12),
asthma (n=8) and tuberculous pleurisy (n=6). The present study was
approved by the Ethics Review Board of the Affiliated Hospital of
Jiangsu University (approval no. 20140019; Zhenjiang, China) and
performed in accordance with the Declaration of Helsinki. Written
informed consent was provided by all participants prior to the
study start.
 | Table I.Demographic and clinical
characteristics of subjects in training and testing sets. |
Table I.
Demographic and clinical
characteristics of subjects in training and testing sets.
|
| Training set | Testing set |
|---|
|
|
|
|
|---|
| Characteristic | Healthy controls
(n=20) | BLD (n=20) | NSCLC (n=40) | Healthy controls
(n=40) | BLD (n=50) | NSCLC (n=88) |
|---|
| Age, years (mean ±
SD) | 58±9.3 | 60±7.8 | 62±11.5 | 59±10.4 | 56±11.5 | 61±12.1 |
| Sex, n (%) |
|
|
|
|
|
|
|
Male | 10 (50.0) | 9
(45.0) | 19 (47.5) | 20 (50.0) | 26 (52.0) | 47 (53.4) |
|
Female | 10 (50.0) | 11 (55.0) | 21 (52.5) | 20 (50.0) | 24 (48.0) | 41 (46.6) |
| Smoking status, n
(%) |
|
|
|
|
|
|
| Ever
and current | 8
(40.0) | 8
(40.0) | 18 (45.0) | 15 (37.5) | 21 (42.0) | 39 (44.3) |
|
Never | 12 (60.0) | 12 (60.0) | 22 (55.0) | 25 (62.5) | 29 (58.0) | 49 (55.7) |
| Histology, n
(%) |
|
|
|
|
|
|
|
SCC | NA | NA | 17 (42.5) | NA | NA | 41 (46.6) |
|
aNon-SCC | NA | NA | 23 (57.5) | NA | NA | 47 (53.4) |
| TNM stage, n
(%) |
|
|
|
|
|
|
| I | NA | NA | 12 (30.0) | NA | NA | 23 (26.1) |
| II | NA | NA | 15 (37.5) | NA | NA | 40 (45.5) |
|
IIIA | NA | NA | 13 (32.5) | NA | NA | 25 (28.4) |
Plasma preparation and RNA
extraction
For plasma preparation, 4 ml venous blood was
collected and placed in EDTA-containing tubes (Invitrogen; Thermo
Fisher Scientific, Inc.). Whole blood was separated into plasma via
centrifugation at 3,000 × g for 10 min at 4°C, within 2 h of
collection. Subsequently, 1 ml aliquots of the plasma specimens
were transferred into 1.5 ml tubes and centrifuged at 4,000 × g for
10 min at room temperature to remove any remaining cellular debris.
The supernatant was subsequently transferred into clean tubes and
stored at −80°C until subsequent experimentation.
Total RNA was isolated from plasma samples using the
mirVana PARIS miRNA Isolated kit (Ambion; Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions. The
concentration and purity of the extracted RNA were analyzed using a
NanoDrop 1000 spectrophotometer (NanoDrop Technologies; Thermo
Fisher Scientific, Inc.), and RNA with a concentration >10 ng/µl
was considered acceptable.
RT-qPCR
RT-qPCR analysis was performed using the TaqMan
MicroRNA RT kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions. Briefly, 5 µl
(5–10 ng/µl) total RNA was reverse transcribed into cDNA using
Avian Myeloblastosis Virus Reverse Transcriptase (Takara
Biotechnology Co., Ltd.) and stem-loop RT primers (Takara
Biotechnology Co., Ltd.). qPCR was subsequently performed on an
Applied Biosystems 7900HT Fast Real-time PCR System (Thermo Fisher
Scientific, Inc.) at 90°C for 10 min, followed by 40 cycles of 90°C
for 15 sec and 60°C for 1 min. All reactions were performed in
triplicate. U6 snRNA was used as the reference gene due to its
stability and reproducibly among patients and healthy controls
(9–11). The fold change in each miRNA
expression relative to U6 snRNA was calculated using the
2−∆∆Cq method (43). The
expression levels of each target miRNA relative to miRNA expressed
in healthy controls were calculated using the 2−∆∆Cq
method (43). The fold-change of
relative miRNA expression was log2 transformed.
Serum CEA levels were determined using the
chemiluminescent microparticle immunoassay ARCHITECT i2000SR
(Abbott Pharmaceutical Co. Ltd.), according to the manufacturer's
instructions.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 5 (GraphPad Software, Inc.) and SPSS 20.0 software (IBM
Corp.). Data are presented as the mean ± SD from at least three
separate experiments. Baseline characteristics of patients with
NSCLC or BLD and controls were compared using the Kruskal-Wallis
test (age) and Pearson's χ2 or Fisher's exact tests (sex
and smoking status). For multiple comparison of the values of miRNA
expression levels, the Kruskal-Wallis test was initially used to
determine whether the samples originated from the same distribution
followed by Steel's-Dwass post hoc test to compare multiple groups
with a control. Wilcoxon's test was used to compare miRNA values in
paired plasma samples obtained before and 2 weeks after tumor
resection.
Receiver operating characteristic (ROC) curve
analysis was performed to obtain area under the curve (AUC) values
for evaluating diagnostic performance of each plasma miRNA for
NSCLC. According to the optimal cut-off values provided by ROC
curve analysis, the sensitivity, specificity, and positive and
negative predictive values were calculated. Risk scores were
assigned to all patients according to a linear combination of the
expression levels of the miRNAs in plasma, weighted according to
the regression coefficient. Stepwise logistic regression analysis
was performed to construct diagnostic miRNA panels based on RT-qPCR
results in training and testing setting (44). The prediction probability of being
diagnosed with NSCLC from healthy control and BLD or stage I NSCLC
from stage II and IIIA NSCLC was used as an index to construct the
ROC curve. AUC was used as an index for the diagnostic performance
evaluation of the miRNA panels.
To further determine whether the altered miRNAs and
other clinical characteristics were independent powerful diagnostic
markers for NSCLC, forward stepwise univariate and multivariate
logistic regression analyses were performed using the healthy
subjects as the reference category. The median value was used as
the cut-off to categorize the expression of each miRNA as high or
low when assessing their association with disease-free survival
(DFS). DFS was defined from the day of surgery to the time of
recurrence, mortality or end of follow-up, and was analyzed using
the Kaplan-Meier method and log-rank test. P<0.05 was considered
to indicate a statistically significant difference.
Results
Plasma levels of selected miRNAs in
patients with NSCLC, patients with BLD and healthy controls
In the initial candidate miRNA selection, a panel of
12 candidate miRNAs that have previously been reported to be
aberrantly expressed in NSCLC tissues or blood samples of patients
with NSCLC (Table SI) (16–41) were
analyzed via RT-qPCR analysis in plasma specimens from 40 patients
with NSCLC and 20 healthy controls. For the training set, when
comparing patients with NSCLC vs. healthy controls, only those
miRNAs with a mean fold-change ≥2 and P<0.05 were selected for
further analysis in the testing set. The results demonstrated that
the expression levels of 4/12 plasma miRNAs, including miR-210,
miR-1290, miR-150 and miR-21-5p, were significantly higher in
patients with NSCLC compared with those found in the healthy
controls (P=0.042 NSCLC compared with healthy control in miR-210;
P<0.001 NSCLC compared with healthy control in miR-1290, miR-150
and miR-215p; Table SI and Fig. 1A-D). The expression levels of these
four miRNAs were detected via RT-qPCR analysis in 20 patients with
BLD, and no significant differences in the expression levels of
these miRNAs were observed between patients with BLD and healthy
controls (Fig. 1A-D).
These four miRNAs were further evaluated via RT-qPCR
analysis in another independent sample set consisting of 88
patients with NSCLC, 50 patients with BLD and 40 healthy controls
(testing set). Similar to the results from the training set, the
plasma levels of these four miRNAs in patients with NSCLC were
significantly higher compared with the healthy controls and
patients with BLD (P=0.058, NSCLC compared with BLD and healthy
control; P<0.001, NSCLC compared with BLD or healthy control in
miR-1290, miR-150 and miR-21-5p; Fig.
2A-D), in the testing set. Similar results were observed for
serum CEA levels across the three group in both the training and
testing sets (Figs. 1E and 2E).
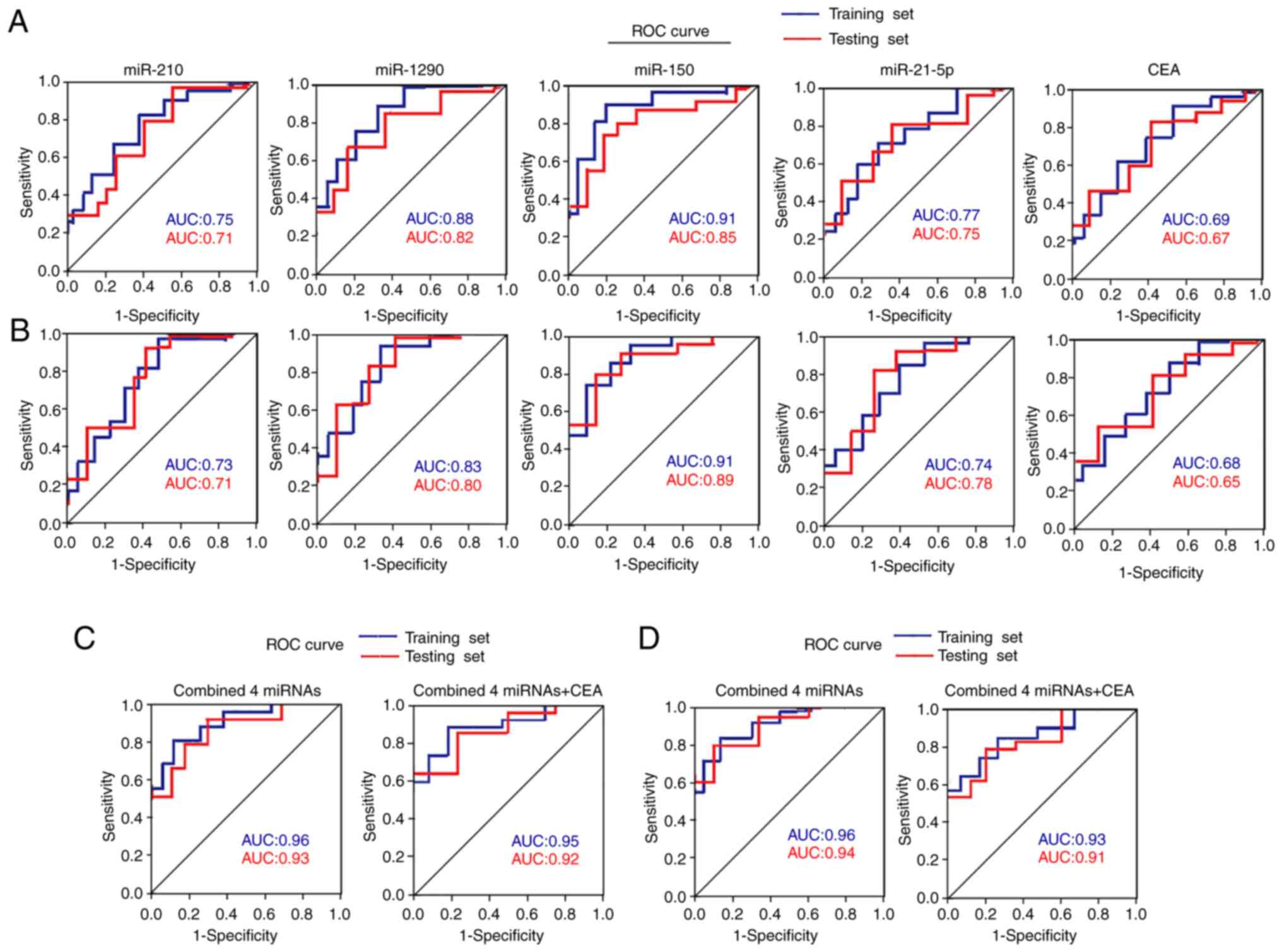
Diagnostic performance of four plasma
miRNAs by ROC curve analysis
ROC curve analyses were performed on the four plasma
miRNAs to determine their diagnostic performance for
differentiating between patients with NSCLC and healthy controls in
the training and testing sets. The results demonstrated that the
four miRNAs exhibited similar performance between the two sets,
with AUC values of 0.71–0.91 (Fig.
3A and Table SII). When the
optimal diagnostic cut-off value of each miRNA was determined via
ROC curve analysis, these miRNAs yielded a sensitivity of 70–90%, a
specificity of 70–85% and an accuracy of 70–88% (Table SII).
ROC curve analyses were performed in the two sets to
distinguish between patients with NSCLC and BLD. The AUC values for
these four miRNAs ranged from 0.71–0.91, with a sensitivity of
72–90%, a specificity of 70–86% and an accuracy of 72–88% (Fig. 3B and Table SIII). The AUC values for serum CEA
in the two sets were 0.69, 0.67, 0.68 and 0.65, respectively, which
were inferior to the four aforementioned miRNAs (miR-210, mtR-1290,
miR-150 and miR-21-5p) (Fig. 3A and
B, and Tables SII and SIII).
To further determine whether the four altered miRNAs
and other clinical characteristics are independent powerful
diagnostic markers for NSCLC, forward stepwise univariate logistic
regression analysis was performed using the healthy controls as the
reference category. The results demonstrated that the odds ratios
for each of the four miRNAs were statistically significant in the
two sets combined (Table SIV).
Multivariate logistic regression analysis demonstrated that
miR-1290, miR-150 and miR-21-5p were independently associated with
NSCLC after adjusting for age, sex and smoking status (Table SIV).
When the four miRNAs were merged as a panel, they
displayed a higher diagnostic performance than that of any
individual miRNA alone in distinguishing patients with NSCLC from
healthy controls in the training and testing sets, with AUC values
of 0.96 and 0.93, respectively (Fig.
3C and Table SII). The
diagnostic performance of the four-miRNA panel was also assessed to
distinguish patients with NSCLC from patients with BLD, and the AUC
values were 0.96 and 0.94, respectively, in the two sets (Fig. 3D and Table SIII). However, combining the four
miRNAs with CEA failed to further improve the diagnostic
performance in any of the two sets (Fig.
3C and D, and Tables SII and
SIII).
Subgroup analyses of four plasma
miRNAs in patients with NSCLC
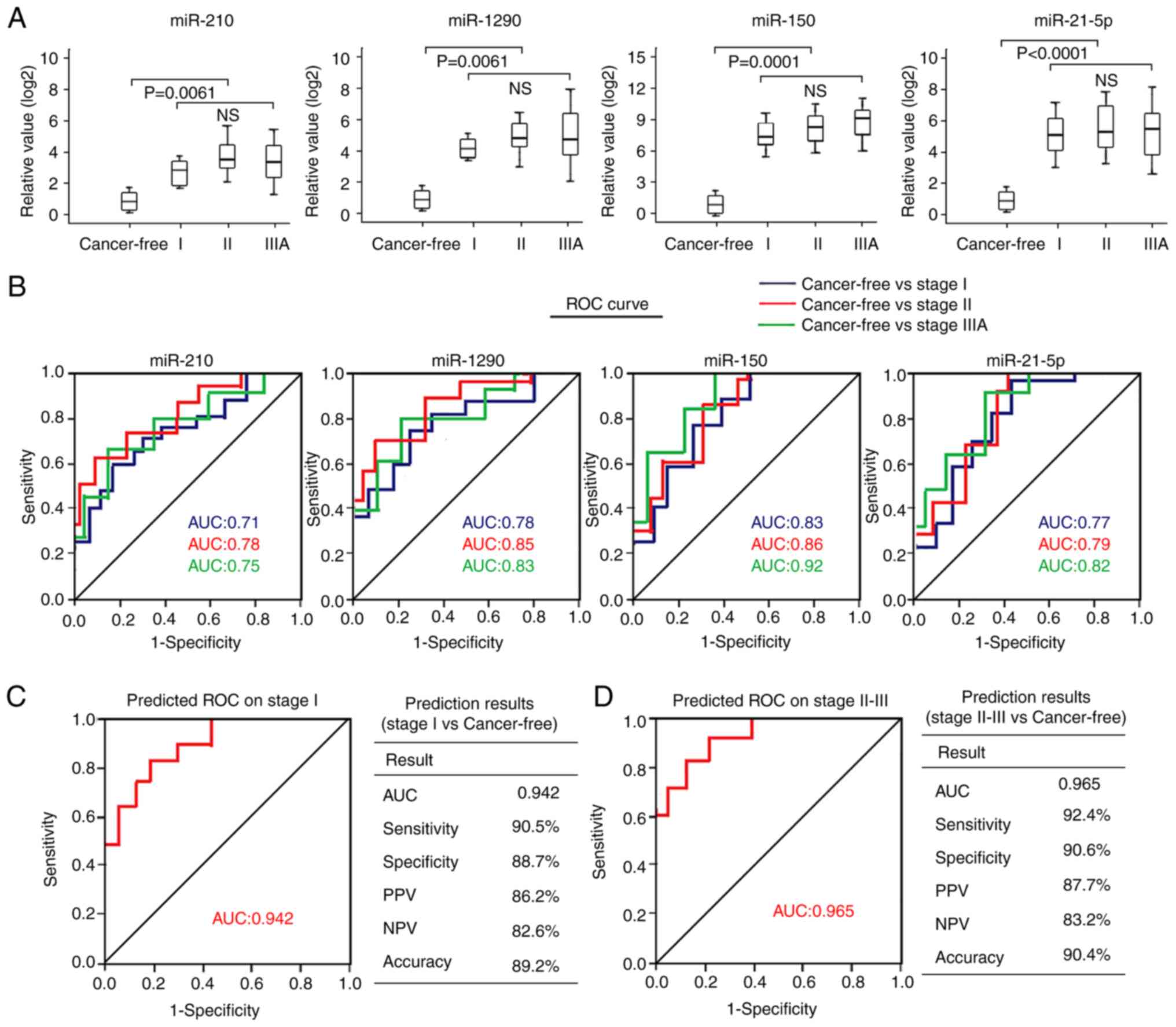
The present study assessed the association between
the four plasma miRNAs and tumor pathological stage of NSCLC. The
results demonstrated that none of the four plasma miRNAs exhibited
significantly different expression in patients with stage I NSCLC
than those with stage II or IIIA disease (Fig. 4A). The AUC values of the four
individual miRNAs in patients with stage I NSCLC (AUCs, 0.71–0.83)
were relatively lower than those with stage II NSCLC (AUCs,
0.78–0.86) or stage IIIA NSCLC (AUCs, 0.75–0.92) (Fig. 4B). When combining the four miRNAs as
a panel in stage I patients with another panel, including patients
with stage II–IIIA disease, the AUC value of the four-miRNA panel
was 0.942 to distinguish patients with stage I NSCLC from healthy
individuals, with a sensitivity of 90.5%, a specificity of 88.7%
and an accuracy of 89.2% (Fig. 4C).
The AUC value of the four-miRNA panel in differentiating patients
with stage II–IIIA NSCLC from healthy individuals was 0.965, with a
sensitivity of 92.4%, a specificity of 90.6% and an accuracy of
90.4% (Fig. 4D). Taken together,
these results suggest that the four-miRNA signature may possess
similar diagnostic performance for patients with stage I and stage
II–IIIA NSCLC.
The plasma levels of the four miRNAs in patients
with different histological types were analyzed. No significant
differences in any of the four miRNAs were observed between
patients with SCC and patients with non-SCC (AD and large cell lung
cancer) (Kruskal-Wallis test; Fig.
5A). The AUC values of the four miRNAs in patients with SCC
were similar to those found in patients without SCC when using the
healthy subjects as the control (Fig.
5B).
Association between the four miRNAs
and DFS time of patients with NSCLC
To determine whether the plasma levels of the four
miRNAs were associated with DFS in patients with NSCLC, 88 patients
in the training and testing sets were followed up and the follow-up
data were analyzed. The median follow-up time was 42.5 months. The
cut-off value for high or low miRNA expression was defined using
the median levels of each plasma miRNA. Kaplan-Meier survival
analysis demonstrated that high plasma levels of miR-210 and
miR-150 were significantly associated with a short DFS time in
patients with NSCLC (Fig. 6A and C).
However, no significant associations between miR-1290 or miR-21-5p
levels and DFS were observed (Fig. 6B
and D).
Given the potential impact of histological type on
the DFS of patients with NSCLC who exhibited different plasma
levels of miR-210 and miR-150, subgroup analysis was performed to
further assess the association between these two miRNAs and DFS in
patients with NSCLC. For patients with histologically confirmed
non-SCC lung cancer, high plasma levels of miR-210 and miR-150 were
associated with shorter DFS times (Fig.
6F and H), whereas the DFS time of patients with SCC was not
affected by the plasma levels of miR-210 or miR-150 (Fig. 6E and G).
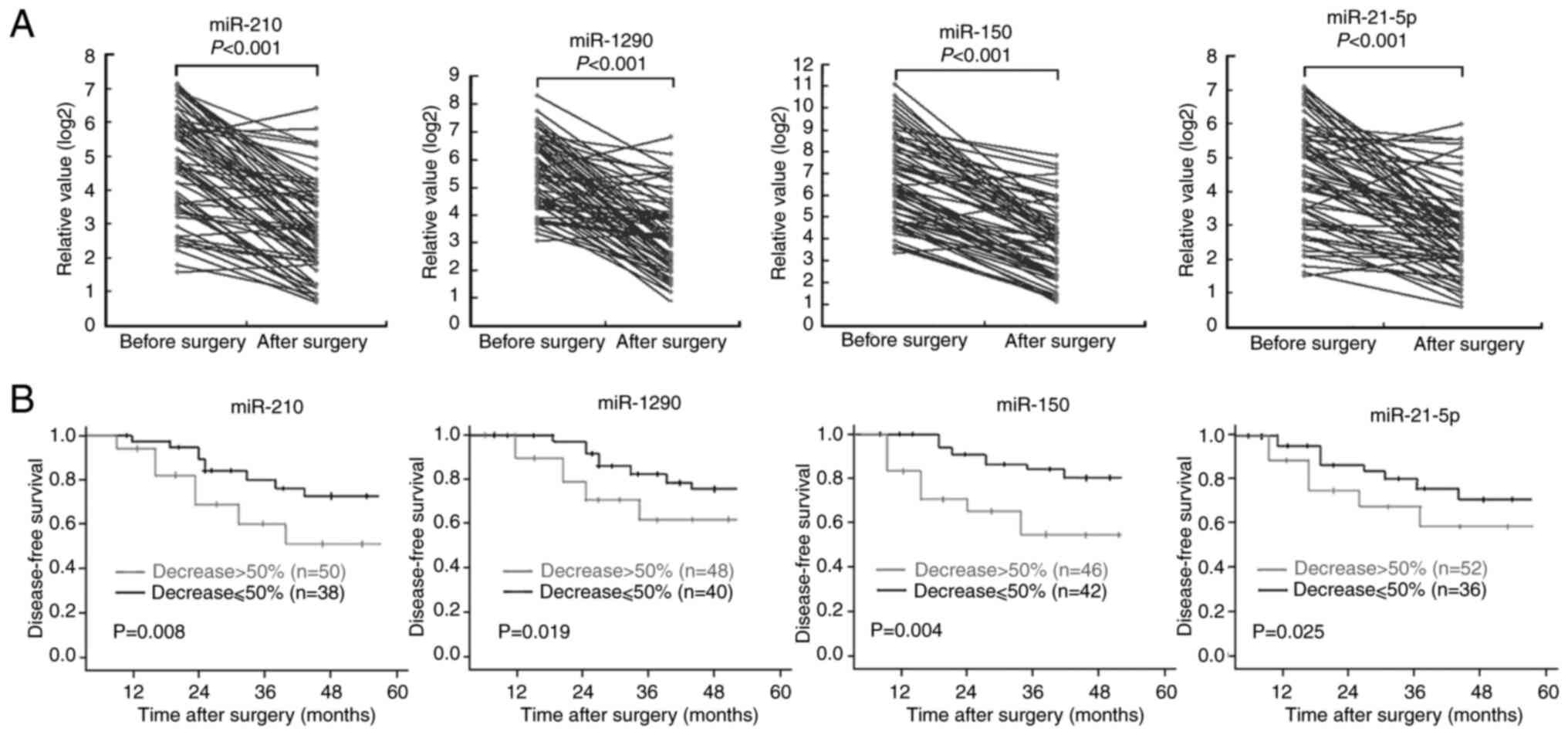
In the 88 patients with NSCLC, additional plasma
samples were collected ~2 weeks after surgery and before
chemotherapy. The results demonstrated that the high pre-operative
plasma levels of the four miRNAs significantly decreased after
surgery, despite a slight increase in the levels of these four
miRNAs in a small number of patients with NSCLC (Fig. 7A). Patients with NSCLC were divided
into two groups (>50 and ≤50%) according to the degree of
post-operative decrease in the expression levels of the four
miRNAs. Survival analysis demonstrated that the DFS time of
patients whose plasma levels of the four individual miRNAs
decreased by >50% after surgery was significantly longer than
those with <50% reduction in the plasma levels of the four
miRNAs (Fig. 7B).
Discussion
Several circulating miRNAs have been reported to act
as diagnostic markers in NSCLC; however, the miRNAs identified by
different researchers vary from study to study (12–17). In
addition to differences in populations and specimens, variations in
the methods used and project design may also result in
inconsistencies between these studies. The results of the present
study demonstrated that miR-210, miR-1290, miR-150 and miR-21-5P
were significantly upregulated in the plasma of patients with NSCLC
compared with their expression levels in patients with BLD and
healthy controls. The four miRNAs were selected by RT-qPCR analysis
from 12 candidate miRNAs that have previously been reported to be
aberrantly expressed in NSCLC tissues or blood samples of patients
with NSCLC (16–41). The diagnostic value of the four
miRNAs for NSCLC was verified via ROC curve analyses in two
independent cohorts of age and sex-matched plasma samples (training
and testing sets). Univariate and multivariate logistic regression
analyses demonstrated the reliability of the diagnostic efficiency
of these four miRNAs. Notably, the results of the present study
demonstrated that combining the four miRNAs had a higher diagnostic
power than that of single miRNAs, and it accurately distinguished
patients with NSCLC from patients with BLD and healthy
controls.
The diverse and complex molecular events involved in
the initiation and development of a malignancy limits the utility
of an individual miRNA as a tumor biomarker (17,19). A
panel of miRNAs can represent several aspects of carcinogenesis,
and the use of these miRNAs in combination can constitute a more
complex indicator for NSCLC diagnosis and prognosis than single
miRNAs (45). Some miRNAs, such as
miR-25 and miR-214, that have been reported in previous studies
(17,28), were markedly elevated in the plasma
of patients with NSCLC in the training set in the present study.
However, these miRNAs were not assessed in the testing set as they
did not meet the selection criteria for further analyses.
The main cause of BLD in the present study included
infectious and non-infectious inflammation. Benign disease can lead
to various substance changes at the molecular level (46), whereby the microenvironment of the
lung is likely to be affected. Abnormal expression of miRNAs occurs
in BLD (47–49). Previous studies have reported that
several circulating miRNAs are aberrantly expressed in patients
with COPD or pneumonia. For example, miR-23a, miR-25, miR-145 and
miR-224 are downregulated (47),
while miR-29 and miR-126 are upregulated in patients with COPD
(48), and miR-193a-5p, miR-542-3p
and miR-1246 are markedly elevated in patients with pneumonia,
which is associated with disease severity (49). However, no significant differences
were observed in the plasma levels of the four miRNAs between
patients with BLD and healthy controls in the present study,
suggesting that the four plasma miRNAs can be used to distinguish
patients with NSCLC from patients with BLD.
The results of the present study demonstrated that
the AUC values of the four miRNAs were higher than that of serum
CEA levels. Notably, combination of the four miRNAs with CEA did
not improve the AUCs of the miRNA-based biomarker for
distinguishing patients with NSCLC from the healthy controls,
suggesting that the diagnostic performance of the four miRNAs is
superior to that of CEA alone, a tumor biomarker widely used in the
clinic (50). Notably, the plasma
levels of the four miRNAs in patients with stage I NSCLC were
markedly elevated compared with the healthy controls and patients
with BLD. ROC curve analysis demonstrated that this panel displayed
similar diagnostic performance in stage I and stage II–IIIA
patients, supporting the four-miRNA panel as a diagnostic marker
for early detection of NSCLC.
Given that patients with NSCLC diagnosed at an early
stage can undergo radical surgery of tumors (3), the results of the present study suggest
that the use of the four-miRNA panel as a marker for defining early
events of NSCLC may be an effective approach to improve prognosis.
Histopathological analysis of biopsy tissue is the gold standard
for NSCLC diagnosis, which requires invasive methods, such as
transthoracic needle puncture or bronchoscopy (12). However, for early-stage NSCLC, it is
relatively difficult for a physician to obtain tissue samples by
biopsy due to the small lesion size depicted in lung imaging
(45). Therefore, it is important to
develop novel non-invasive methods and markers with high
specificity and sensitivity for the detection of NSCLC.
The present study also assessed the individual role
of the four miRNAs in predicting DFS in 88 patients with NSCLC who
received surgery and follow-up. The results demonstrated that high
plasma levels of miR-210 and miR-150 were associated with shorter
DFS time. Notably, high plasma levels of these two miRNAs were
significantly associated with a shorter DFS time in patients
without SCC lung cancer, but not in patients with SCC. However, no
significant differences in the plasma levels of miR-210 or miR-150
were observed between patients with SCC and patients without SCC
lung cancer. Furthermore, the results demonstrated an association
between DFS and changes in the plasma levels of the four miRNAs
before and after surgery. Patients whose plasma levels of the four
miRNAs were reduced by >50% after surgery had a longer DFS time,
suggesting that changes in the pre- and post-operative plasma
levels of these four miRNAs may predict prognosis in patients with
NSCLC.
Circulating miRNAs can derive from various cell
types, including cancer cells, which passively leak or actively
transport them into the bloodstream as a way of cell-to-cell
communication or an alternative source of circulating miRNAs
(51,52). Thus, the deregulation profile, and
the diagnostic and prognostic roles of circulating miRNAs may be
independent from tissue samples. As circulating miRNAs originate
from all cancer cells within an individual, analyzing circulating
miRNAs can reduce the impact of the wide heterogeneity of a whole
solid tumor compared with miRNAs isolated from a small piece of
tissue (9,10). The four aberrantly expressed miRNAs
identified in plasma from patients with NSCLC in the present study
are well documented NSCLC-related miRNAs. miR-210 is a reliable
biomarker for the early diagnosis of NSCLC, which is found in blood
samples and sputum and bronchoalveolar lavage fluid samples
(53–55). miR-1290 is a tumor-initiating
cell-specific miRNA, and together with miR-1246, plays a crucial
role in tumor initiation and cancer progression in human NSCLC
(56), and it was identified as a
potential prognostic biomarker for NSCLC (24). Zhang et al (57) reported that circulating miR-150 can
predict prognosis in early-stage NSCLC, and can facilitate cancer
cell proliferation by suppressing the tumor suppressor gene, SRC
kinase signaling inhibitor 1. Li et al (58) demonstrated that miR-150-caused
autophagy inhibition triggered endoplasmic reticulum stress,
increased cellular reactive oxygen species levels, activated the
DNA damage response and facilitated NSCLC cell proliferation and
tumor growth. miR-21-5p upregulation in tumor samples has been
observed in patients with NSCLC, and has been confirmed as an
independent prognostic predictor for overall survival (59). Another study reported that miR-21-5p
expression in NSCLC tissue is associated with histological subtype,
tumor volume, regional lymph node and distal metastasis, and that
miR-21-5p promotes the progression of NSCLC by modulating SMAD7
expression (60).
The four miRNAs in blood samples of other types of
cancer have also been assessed. For example, circulating miR-210 is
significantly increased in patients with breast cancer and
hepatocellular carcinoma (61,62).
Furthermore, serum miR-1290 is markedly overexpressed in patients
with pancreatic cancer and ovarian cancer (63,64).
Circulating miR-150 is upregulated in patients with colorectal
cancer (65), and serum miR-21-5p is
significantly elevated in patients with advanced papillary renal
cell carcinoma (66). Taken
together, these results suggest that the four miRNAs are involved
in the occurrence and development of other cancers, and may be used
as biomarkers for these cancers.
The present study is not without limitations. First,
the total sample size was relatively small, which may have resulted
in bias. Furthermore, the follow-up time of patients after surgery
was not long enough, and only a small number of patients were
included. Furthermore, the impact of hemolysis on the expression of
miRNAs was not evaluated by measuring the hemolysis grade of the
plasma samples, which may influence the diagnostic accuracy of the
four plasma miRNAs for NSCLC.
In conclusion, the results of the present study
demonstrated the potential of a four-plasma miRNA signature in the
detection of early-stage NSCLC. The results also indicated that the
plasma expression profiles of miR-210 and miR-150 can act as
prognostic biomarkers for patients with NSCLC, mainly for those
with the lung AD subtype. In addition, a significant decrease in
the levels of these four plasma miRNAs (>50%) after surgery is a
predictive factor of a longer DFS time. However, prospective
cohorts are required to validate the results presented here.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by research grants
from the Medical Research Program of Jiangsu Health Committee in
China (grant no. ZDB2020022) and the Social Development Foundation
of Zhenjiang in China (grant nos. SH2014076 and SH2015063).
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL, HGJ and CHD conceived and designed the present
study. YPX, XBX, HGJ, CHD, QJ and YS performed the experiments, and
analyzed and interpreted the data. QJ and YS confirm the
authenticity of all the raw data. HGJ and JL were involved in
project development, data analysis and editing the manuscript. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics Review
Board of the Affiliated Hospital of Jiangsu University (approval
no. 20140019; Zhenjiang, China) and performed in accordance with
International Ethical Guidelines for Biomedical Research Involving
Human Subjects (CIOMS) (67) and the
Declaration of Helsinki of 1964 (68) and a later version (69). Written informed consent was provided
by all participants prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JHM, Beasly MB, Chirieac LC, Dacic S, Duhig E,
Flieder DB, et al: The 2015 world health organization
classification of lung tumors: Impact of genetic, clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He L: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mominioni L, Imperatori A, Rovera F,
Ochetti A, Torrigiotti G and Paolucci M: Stage I nonsmall cell lung
carcinoma: Analysis of survival and implications for screening.
Cancer. 89 (11 Suppl):S2334–S2344. 2000. View Article : Google Scholar
|
|
4
|
National Lung Screening Trial Research
Team, ; Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD,
Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM and Sicks JD:
Reduced lung-cancer mortality with low-dose computed tomographic
screening. N Engl J Med. 365:395–409. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
National Lung Screening Trial Research
Team, ; Church TR, Black WC, Aberle DR, Berg CD, Clingan KL, Duan
F, Fagerstrom RM, Gareen IF, Gierada DS, et al: Results of initial
low-dose computed tomographic screening for lung cancer. N Engl J
Med. 368:1980–1991. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Snowsill T, Yang H, Griffin E, Long L,
Varley-Campbell J, Coelho H, Robinson S and Hyde C: Low-dose
computed tomography for lung cancer screening in high risk
populations: A systematic review and economic evaluation. Health
Technol Assess. 22:1–276. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Veronesi G: Lung cancer screening: The
European perspective. Thorac Sury Clin. 25:161–174. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Okarnura K, Takayama K, Izumi M, Harada T,
Furuyama K and Nakanish Y: Diagostic value of CEA and CYFRA 21-1
tumor markers in primary lung cancer. Lung Cancer. 80:45–49. 2013.
View Article : Google Scholar
|
|
9
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnosis, monitoring and therapeutics-A
comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pardini B, Sabo AA, Birolo G and Calin GA:
Noncoding RNAs in extracellular fluids as cancer biomarkers: The
new frontier of liquid biopsies. Cancers (Basel). 11:11702019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Larrea E, Sole C, Manterola L, Goicoechea
I, Armesto M, Arestin M, Caffarel MM, Araujo AM, Araiz M,
Fernandez-Mercado M and Lawrie CH: New concepts in cancer
biomarkers: Curculating miRNA in liquied biopsies. Int J Mol Sci.
17:6272016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C,
Wang C, Ren Z, Zhao Y, Wu S, et al: Identification of ten serum
microRNAs from a genome-wide serum micreRNA expression profile as
novel noninvasive biomarkers for nonsmall cell lung cancer
diagnosis. Int J Cancer. 130:1620–1628. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nadal E, Truini A, Nakata A, Lin J, Reddy
RM, Chang AC, Ramnath N, Gotoh N, Beer DG and Chen G: A novel serum
4-microRNA signature for lung cancer detection. Sci Rep.
5:124642015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lv S, Xue J, Wu C, Wang L, Wu J, Xu S,
Liang X and Lou J: Identification of a panel of serum microRNAs as
biomarkers for early detection of lung adenocarcinoma. J Cancer.
8:48–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shang AQ, Xi YN, Wang J, Sun L, Wei J, Lu
WY, Lan JY, Wang WW, Wang L and Wang LL: Predictive value of serum
microRNA-22 and microRNA-126 levels for non-small cell lung cancer
development and metastasis: A case-control study. Neoplasma.
64:453–459. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Foss KM, Sima C, Ugolini D, Neri M, Allen
KE and Weiss GJ: MiR-1254 and miR-574-5P: Serum-based microRNA
biomarkers for early-stage non-small cell lung cancer. J Thorac
Oncol. 6:482–488. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang C, Ding M, Xia M, Chen S, Van Le A,
Soto-Gil R, Shen Y, Wang N, Wang J, Gu W, et al: A Five-miRNA panel
identified from a multicentric case-control study serves as a novel
diagnostic tool for ethnically diverse non-small-cell lung cancer
patients. EBioMedicine. 2:1377–1385. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tan X, Qin W, Zhang L, Hang J, Li B, Zhang
C, Wan J, Zhou F, Shao K, Sun Y, et al: A 5-microRNA signature for
lung squamous cell carcinoma diagnosis and hsa-miR-31 for
prognosis. Clin Cancer Res. 17:6802–6811. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guan P, Yin Z, Li X, Wu W and Zhou B:
Meta-analysis of human lung cancer microRNA expression profiling
studies comparing cancer tissue with normal tissue. J Exp Clin
Cancer Res. 31:542012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang C, Sun C, Liang X, Xie S, Huang J and
Li D: Integrative analysis of microRNA and mRNA expression profiles
in non-small-cell lung cancer. Cancer Gene Ther. 23:90–97. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Eilertsen M, Andersen S, Al-Saad S,
Richardsen E, Stenvold H, Hald SM, Al-Shibli K, Donnem T, Busund LT
and Bremnes RM: Postive prognostic impact of miR-210 in non-small
cell lung cancer. Lung Cancer. 83:272–278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Duncavage E, Goodgame B, Sezhiyan A,
Govindan R and Pfeifer J: Use of microRNA expression levels to
predict outcomes in resected stage I non-small cell lung cancer. J
Thorac Oncol. 5:1755–1760. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao M, Hou D, Liang H, Gong F, Wang Y, Yan
X, Jiang X, Wang C, Zhang J, Zen K, et al: MiR-150 promotes the
proliferation and migration of lung cancer cells by targeting SRC
kinase signalling inhibitor 1. Eur J Cancer. 50:1013–1024. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mo D, Gu B, Gong X, Wu L, Wang H, Jiang Y,
Zhang B, Zhang M, Zhang Y, Xu J and Pan S: MiR-1290 is a potential
prognostic biomarker in non-small cell lung cancer. J Thorac Dis.
7:1570–1579. 2015.PubMed/NCBI
|
|
25
|
Liang B, Wang GX, Long G, Qiu JH and Hu
ZL: Tumor suppressor miR-22 suppresses lung cancer cell progression
through post-transcriptional regulation of ErbB3. J Cancer Res Clin
Oncol. 138:1355–1361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shin YM, Yun J, Lee OJ, Han HS, Lim SN, An
JY, Lee KH, Lee KM and Choe KH: Diagnostic value of circulating
extracellular miR-134, miR-183, and miR-22 levels in lung
adenocarcinoma-associated malignant pleural effusion. Cancer Res
Treat. 46:178–185. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu T, Chen W, Kong D, Li X, Lu H, Liu S,
Wang J, Du L, Kong Q, Huang X and Lu Z: MiR-25 targets the
modulator of apoptosis 1 gene in lung cancer. Carcinogenesis.
36:925–935. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang P, Yang D, Zhang H, Wei X, Ma T,
Cheng Z, Hong Q, Hu J, Zhuo H, Song Y, et al: Early detection of
lung cancer in serum by a panel of microRNA biomarkers. Clin Lung
Cancer. 16:313–319.e1. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Heegaard NH, Schetter AJ, Welsh JA, Yoneda
M, Bowman ED and Harris CC: Circulating miroRNA expression profiles
in early stage non-small cell lung cancer. Int J Cancer.
130:1378–1386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu W, He J, Chen D, Zhang B, Xu L, Ma H,
Liu X, Zhang Y and Le H: Expression of miR-29c, miR-93, and miR-429
as potential biomarkers for detection of early stage non-small lung
cancer. PLoS One. 9:e877802014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Boeri M, Verri C, Conta D, Roz L, Modena
P, Facchinetti F, Calabrò E, Croce CM, Pastorino U and Sozzi G:
MicroRNA signatures in tissues and plasma predict development and
prognosis of computed tomography detected lung cancer. Proc Natl
Acad Sci USA. 108:3713–3718. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Patnaik SK, Yendamuri S, Kannisto E,
Kucharczuk JC, Singhal S and Vachani A: MicroRNA expression
profiles of whole blood in lung adenocarcinoma. PLoS One.
7:e460452012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Long H, Wang Z, Chen J, Xiang T, Li Q,
Diao X and Zhu B: MicroRNA-214 promotes epithelial-mesenchymal
transition and metastasis in lung adenocarcinoma by targeting the
suppressor-of-fused protein (Sufu). Oncotarget. 6:38705–38718.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim G, An HJ, Lee MJ, Song JY, Jeong JY,
Lee JH and Jeong HC: Hsa-miR-1246 and hsa-miR-1290 are associated
with stemness and invasiveness of non-small cell lung cancer. Lung
Cancer. 91:15–22. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rani S, Gately K, Crown J, O'Byrne K and
O'Driscoll L: Global analysis of serum microRNAs as potential
biomarkers for lung adenocarcinoma. Cancer Biol Ther. 14:1104–1112.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu S, Kong H, Hou Y, Ge D, Huang W, Ou J,
Yang D, Zhang L, Wu G, Song Y, et al: Two plasma microRNA panels
for diagnosis and subtype discrimination of lung cancer. Lung
Cancer. 123:44–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang W, Li H and Luo R: The microRNA-1246
promotes metastasis in non-small cell lung cancer by targeting
cytoplasmic polyadenylation element-binding protein 4. Diagn
Pathol. 10:1272015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang N, Wei X and Xu L: MiR-150 promotes
the proliferation of lung cancer cells by targeting P53. FEBS Lett.
587:2346–2351. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yin QW, Sun XF, Yang GT, Li XB, Wu MS and
Zhao J: Increased expression of microRNA-150 is associated with
poor prognosis in non-small cell lung cancer. Int J Clin Exp
Pathol. 8:842–846. 2015.PubMed/NCBI
|
|
40
|
Tian F, Li R, Chen Z, Shen Y, Lu J, Xie X
and Ge Q: Differentially expressed miRNAs in tumor, adjacent, and
normal tissues of lung adenocarcinoma. Biomed Res Int.
2016:14282712016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ma R, Wang C, Wang J, Wang D and Xu J:
MiRNA-mRNA interaction network in non-small cell lung cancer.
Interdiscip Sci. 8:209–219. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Goldstraw P: Staging Manual in Thoracic
Oncology. Orange Pakk, FL: Editorial Rx Press; pp. 57–65. 2009
|
|
43
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
DeLong ER, DeLong DM and Clarke-Pearson
DL: Comparing the area under two or more correlated receiver
operating characteristic curve: A nonparametric approach.
Biomentrics. 44:837–845. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhou Q, Huang SX, Zhang F, Li SJ, Liu C,
Xi YY, Wang L, Wang X, He QQ, Sun CC and Li DJ: MicroRNAs: A novel
potential biomarker for diagnosis and therapy in patients with
non-small cell lung cancer. Cell Prolif. 50:e123942017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Belinsky SA: Gene-promoter
hypermrthylation as a biomaker in lung cancer. Nat Rev Cancer.
4:707–717. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu X, Qu J, Xue W, He L, Wang J, Xi X,
Liu X, Yin Y and Qu Y: Bioinformatics-based identification of
potential microRNA biomarkers in frequent and non-frequent
exacerbators of COPD. Int J Chron Obstruct Pulmon Dis.
13:1217–1228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kara M, Kirkil G and Kalemci S:
Differential expression of microRNAs in chronic obstructive
pulmonary disease. Adv Clin Exp Med. 25:21–26. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hermann S, Brandes F, Kirchner B,
Buschmann D, Borrmann M, Klein M, Kotschote S, Bonin M, Reithmair
M, Kaufmann I, et al: Diagnostic potential of circulating cell-free
microRNAs for comminity-acquired pneumonia and pneumonia-related
sepsis. J Cell Mol Med. 24:12054–12064. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang DW, Zhang Y, Hong QY, Hu J, Li C, Pan
BS, Wang Q, Ding FH, Ou JX, Liu FL, et al: Role of a serum-based
biomarker panel in the early diagnosis of lung cancer for a cohort
of high-risk patients. Cancer. 121 (Suppl):S3113–S3121. 2015.
View Article : Google Scholar
|
|
51
|
Kosaka N, Iguchi H, Yoshioka Y, Takeshita
F, Matsuki Y and Ochiya T: Secretory mechanisms and intercellular
transfer of microRNAs in living cells. J Boil Chem.
285:17442–17452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Schwarzenbach H, Nishida N, Calin GA and
Panted K: Clinical relevance of circulating cell-free microRNAs in
cancer. Nat Rev Clin Oncol. 11:145–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang X, Zhi X, Zhang Y, An G and Feng G:
Role of plasma microRNAs in the early diagnosis of non-small-cell
lung cancer: A case-control study. J Thorac Dis. 8:1645–1652. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xing L, Todd NW, Yu L, Fang H and Jiang F:
Early detection of squamous cell lung cancer in sputum by a panel
of microRNA markers. Mod Pathol. 23:1157–1164. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kim JO, Gazala S, Razzak R, Guo L, Ghosh
S, Roa WH and Béard EL: Non-small cell lung cancer detection using
microRNA expression profiling of bronchoalveolar lavage fluid and
sputum. Anticancer Res. 35:1873–1880. 2015.PubMed/NCBI
|
|
56
|
Zhang WC, Chi TM, Yang H, Nga ME, Lunny
DP, Lim EK, Sun LL, Pang YH, Leow YN, Malusay SR, et al:
Tumor-initiating cell-specific miR-1246 and miR-1290 expression
converge to promote non-small cell lung cancer progression. Nat
Commun. 7:117022016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang L, Lin J, Ye Y, Oba T, Gentile E,
Lian J, Wang J, Zhao Y, Gu J, Wistuba II, et al: Serum microRNA-150
predicts prognosis for early-stage non-small cell lung cancer and
promotes tumor cell proliferation by targeting tumor suppressor
gene SRCIN1. Clin Pharmacol Ther. 103:1061–1073. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li H, Liu J, Cao W, Xiao X, Liang L,
Liu-Smith F, Wang W, Liu H, Zhou P, Ouyang R, et al:
C-myc/miR150/EPG5 axis mediated dysfunction of autophagy promotes
development of non-small cell lung cancer. Theranostics.
9:5135–5148. 2019. View Article : Google Scholar
|
|
59
|
Li C, Yin Y, Liu X, Xi X, Xue W and Qu Y:
Non-small cell lung cancer associated microRNA expression
signature: Integrated bioinformatic analysis, valilation and
clinical significance. Oncotarget. 8:24564–24578. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li X and Wu X: MiR-21-5P promotes the
progression of non-small-cell lung cancer by regulating the
expression of SMAD7. Onco Targets Ther. 11:8445–8454. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bertol G, Cava C and Castigliohi I:
MicroRNAs: New biomarkers for diagnosis, prognosis, therapy
prediction and therapeutic tools for breast cancer. Theranostics.
5:1122–1143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ahmed EK, Fahmy SA, Effat H and Wahab AHA:
Circulating miR-210 and miR-1246 as potential biomarkers for
differentiating hepatocellular carcinoma from metastatic tumors in
the liver. J Med Biochem. 38:109–117. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wei J, Yang L, Wu YN and Xu J: Serum
miR-1290 and miR-1246 as potential diagnostic biomarkers of human
pancreatic cancer. J Cancer. 11:1325–1333. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kobayashi M, Sawada K, Nakamura K,
Yoshimura A, Miyamoto M, Shimizu A, Ishida K, Nakatzuka E, Kodama
M, Hashimoto K, et al: Exosomal miR-1290 is a potential biomarker
of high-grade serous ovarian carcinoma and can disriminate patients
from those with malignanies of other histologial types. J Ovarian
Res. 11:812018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sur D, Burz C, Sabarimurugan S and Irimie
A: Diagnostic and prognostic significance of miR-150 in colorectal
cancer: A systematic review and meta-analysis. J Pers Med.
10:992020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kalogirou C, Ellinger J, Kristianse G,
Hatzichristodoulou G, Kubler H, Kneitz B, Busch J and Fendler A:
Identification of miR-21-5p and miR-210-3p serum levels as
biomarkers for patients with papillary renal cell carcinoma: A
multicenter analysis. Transl Androl Urol. 9:1314–1322. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Council for International Organizations of
Medical Sciences, . International ethical guidelines for biomedical
research involving human subjects. Bull Med Ethics. 182:17–23.
2002.PubMed/NCBI
|
|
68
|
World Medical Association, . Declaration
of Helsinki. 1964.http://www.wma.net/wp-content/uploads/2018/07/DoH-Jun1964.pdf
|
|
69
|
World Medical Association, . Declaration
of Helsinki. 2008.https://www.wma.net/wp-content/uploads/2018/07/DoH-Oct2008.pdf
|