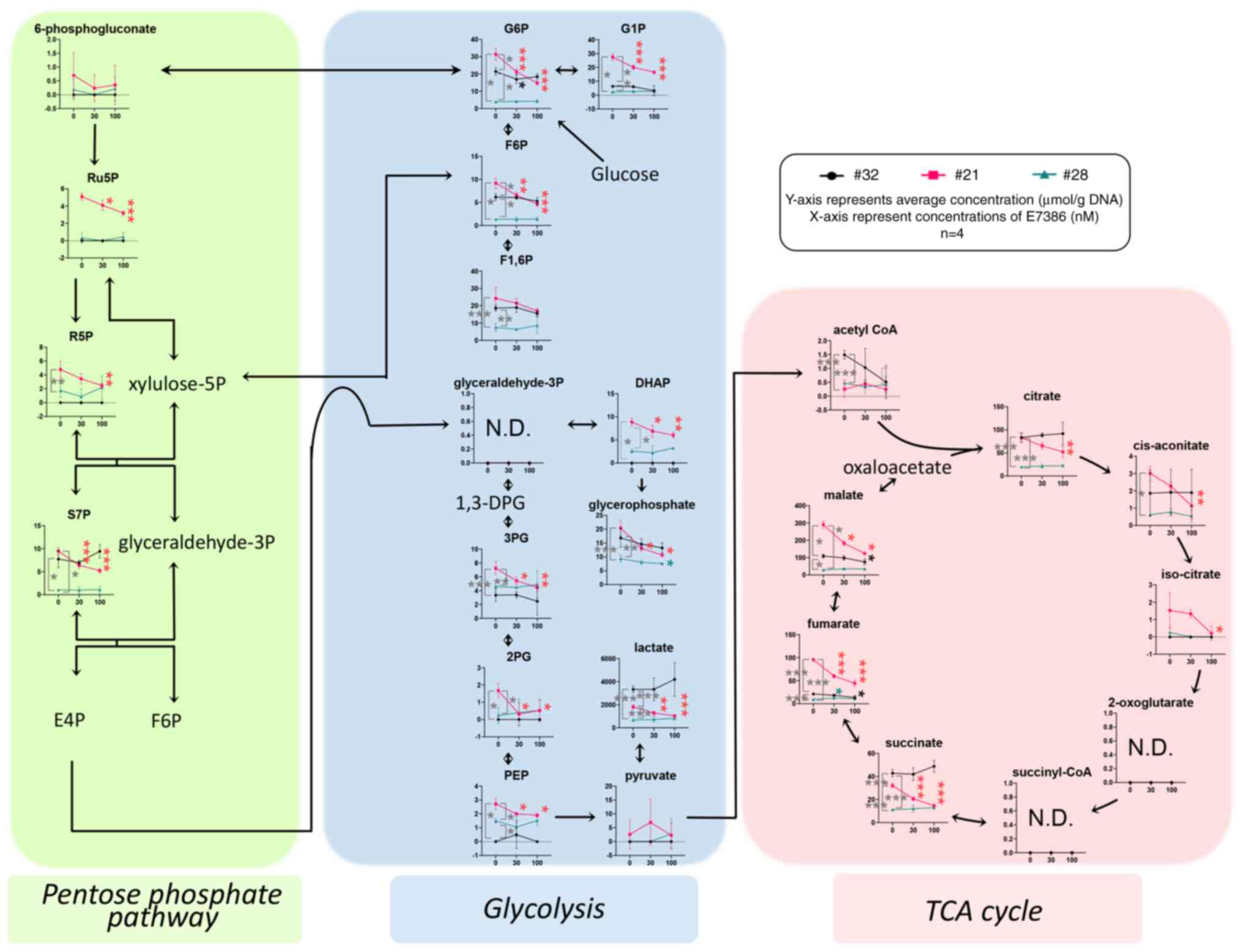
|
1
|
Dekker E, Tanis PJ, Vleugels JL, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD, Goding Sauer A,
Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA and Jemal
A: Colorectal cancer statistics, 2020. CA Cancer J Clin.
70:145–164. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar
|
|
4
|
Burrell RA, McGranahan N, Bartek J and
Swanton C: The causes and consequences of genetic heterogeneity in
cancer evolution. Nature. 501:338–345. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Normanno N, Rachiglio AM, Lambiase M,
Martinelli E, Fenizia F, Esposito C, Roma C, Troiani T, Rizzi D,
Tatangelo F, et al: Heterogeneity of KRAS, NRAS, BRAF and PIK3CA
mutations in metastatic colorectal cancer and potential effects on
therapy in the CAPRI GOIM trial. Ann Oncol. 26:1710–1714. 2015.
View Article : Google Scholar
|
|
6
|
Kim TM, Lee SH and Chung YJ: Clinical
applications of next-generation sequencing in colorectal cancers.
World J Gastroenterol. 19:6784–6793. 2013. View Article : Google Scholar
|
|
7
|
Cancer Genome Atlas Network, .
Comprehensive molecular characterization of human colon and rectal
cancer. Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Berdiel-Acer M, Sanz-Pamplona R, Calon A,
Cuadras D, Berenguer A, Sanjuan X, Paules MJ, Salazar R, Moreno V,
Batlle E, et al: Differences between CAFs and their paired NCF from
adjacent colonic mucosa reveal functional heterogeneity of CAFs,
providing prognostic information. Mol Oncol. 8:1290–1305. 2014.
View Article : Google Scholar
|
|
9
|
Naruse M, Ochiai M, Sekine S, Taniguchi H,
Yoshida T, Ichikawa H, Sakamoto H, Kubo T, Matsumoto K, Ochiai A
and Imai T: Re-expression of REG family and DUOXs genes in CRC
organoids by co-culturing with CAFs. Sci Rep. 11:20772021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamada K, Hori Y, Inoue S, Yamamoto Y, Iso
K, Kamiyama H, Yamaguchi A, Kimura T, Uesugi M, Ito J, et al:
E7386, a selective inhibitor of the interaction between
beta-catenin and CBP, exerts antitumor activity in tumor models
with activated canonical Wnt signaling. Cancer Res. 81:1052–1062.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Soga T, Baran R, Suematsu M, Ueno Y, Ikeda
S, Sakurakawa T, Kakazu Y, Ishikawa T, Robert M, Nishioka T and
Tomita M: Differential metabolomics reveals ophthalmic acid as an
oxidative stress biomarker indicating hepatic glutathione
consumption. J Biol Chem. 281:16768–16776. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hirayama A, Kami K, Sugimoto M, Sugawara
M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, et
al: Quantitative metabolome profiling of colon and stomach cancer
microenvironment by capillary electrophoresis time-of-flight mass
spectrometry. Cancer Res. 69:4918–4925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Naruse M, Ishigamori R and Imai T: The
unique genetic and histological characteristics of DMBA-induced
mammary tumors in an organoid-based carcinogenesis model. Front
Genet. 12:7687312021. View Article : Google Scholar
|
|
16
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar
|
|
17
|
Bae JH, Kim SJ, Kim MJ, Oh SO, Chung JS,
Kim SH and Kang CD: Susceptibility to natural killer cell-mediated
lysis of colon cancer cells is enhanced by treatment with epidermal
growth factor receptor inhibitors through UL16-binding protein-1
induction. Cancer Sci. 103:7–16. 2012. View Article : Google Scholar
|
|
18
|
Vincent EE, Sergushichev A, Griss T,
Gingras MC, Samborska B, Ntimbane T, Coelho PP, Blagih J, Raissi
TC, Choiniere L, et al: Mitochondrial phosphoenolpyruvate
carboxykinase regulates metabolic adaptation and enables
glucose-independent tumor growth. Mol Cell. 60:195–207. 2015.
View Article : Google Scholar
|
|
19
|
Arce L, Pate KT and Waterman ML: Groucho
binds two conserved regions of LEF-1 for HDAC-dependent repression.
BMC Cancer. 9:1592009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen Z, Chen G and Zhao H: FDPS promotes
glioma growth and macrophage recruitment by regulating CCL20 via
Wnt/β-catenin signalling pathway. J Cell Mol Med. 24:9055–9066.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu D, Fu X, Wang Y, Wang X, Wang H, Wen J
and Kang N: Protein diaphanous homolog 1 (Diaph1) promotes
myofibroblastic activation of hepatic stellate cells by regulating
Rab5a activity and TGFβ receptor endocytosis. FASEB J.
34:7345–7359. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar
|
|
23
|
Kalluri R: The biology and function of
fibroblasts in cancer. Nat Rev Cancer. 16:582–598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lyssiotis CA and Kimmelman AC: Metabolic
interactions in the tumor microenvironment. Trends Cell Biol.
27:863–875. 2017. View Article : Google Scholar
|
|
25
|
Dixon SJ, Patel DN, Welsch M, Skouta R,
Lee ED, Hayano M, Thomas AG, Gleason CE, Tatonetti NP, Slusher BS
and Stockbell BR: Pharmacological inhibition of cystine-glutamate
exchange induces endoplasmic reticulum stress and ferroptosis.
Elife. 3:e025232014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Raulet DH, Gasser S, Gowen BG, Deng W and
Jung H: Regulation of ligands for the NKG2D activating receptor.
Annu Rev Immunol. 31:413–441. 2013. View Article : Google Scholar
|
|
27
|
Chocarro-Calvo A, Garcia-Martinez JM,
Ardila-Gonzalez S, De la Vieja A and Garcia-Jimenez C:
Glucose-induced β-catenin acetylation enhances Wnt signaling in
cancer. Mol Cell. 49:474–486. 2013. View Article : Google Scholar
|
|
28
|
Kling JC and Blumenthal A: Roles of WNT,
NOTCH, and Hedgehog signaling in the differentiation and function
of innate and innate-like lymphocytes. J Leukoc Biol. 101:827–840.
2017. View Article : Google Scholar
|
|
29
|
Kling JC, Jordan MA, Pitt LA, Meiners J,
Thanh-Tran T, Tran LS, Nguyen TT, Mittal D, Villani R, Steptoe RJ,
et al: Temporal regulation of natural killer T Cell interferon
gamma responses by beta-catenin-dependent and -independent Wnt
signaling. Front Immunol. 9:4832018. View Article : Google Scholar
|
|
30
|
Kim BK, Yoo HI, Lee AR, Choi K and Yoon
SK: Decreased expression of VLDLR is inversely correlated with
miR-200c in human colorectal cancer. Mol Carcinog. 56:1620–1629.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee K, Shin Y, Cheng R, Park K, Hu Y,
McBride J, He X, Takahashi Y and Ma JX: Receptor heterodimerization
as a novel mechanism for the regulation of Wnt/β-catenin signaling.
J Cell Sci. 127:4857–4869. 2014.
|