Introduction
Glioblastoma is one of the most prevalent and
aggressive primary type of brain tumor and is associated with high
morbidity and mortality (1). At
present, surgery, chemotherapy, and radiation are the most common
therapeutic approaches for cancer treatment. However, the prognosis
of malignant glioma remains poor, with recurrence and low survival
times (2,3). Recently, it has been reported that a
sub-population of cells, namely cancer stem-like cells (CSCs),
contributes to poor prognosis by inducing chemoresistance,
radioresistance and recurrence (4,5).
CD133, CD44 and CD24 are stem cell surface markers expressed in
different types of cancer, but their expression and distribution
differ between cancer types and tumor cell lines (6). Therefore, the identification of CSCs
through these biomarkers is of great interest.
Nogo-A functions as a growth-inhibitory,
antiadhesive, and growth cone-collapsing factor in neurons and has
a high molecular weight (7). In
the adult central nervous system (CNS), Nogo-A exerts repulsive and
guidance functions for neurite development (8), inhibits the movement of cells in the
early stage of development (9),
and functions as an inhibitor for axonal regeneration and
plasticity after development (10). Despite its functions in the CNS,
Nogo-A is also known to exert regulatory roles in tumors. In
oligodendroglial tumors, Nogo-A has been negatively associated with
the malignancy grade (11). Schwab
et al (12) suggested that,
in neuroepithelial tumors, the expression levels of Nogo-A were
positively associated with poor prognosis. A contrasting report has
also shown that different expression levels of Nogo-A have no
independent prognostic impact in glioblastoma, despite age and
clinical status (13). Thus, the
exact role of Nogo-A in glioblastoma and CSCs remains largely
unclear.
In the present study, Nogo-A was upregulated in CSCs
derived from the human Uppsala 87 malignant glioma cell line
(U87MG-CSCs) compared with their parental cells. In contrast to its
effect in neurons, Nogo-A promoted the proliferation, invasion and
tumor formation of U87MG-CSCs. These results suggested that Nogo-A
may serve multiple functions and could represent a promising
therapeutic target.
Materials and methods
Cell culture
U87MG glioblastoma cells of unknown origin
(accession number, CVCL_0022) were obtained from the Cell Bank of
Chinese Academy of Sciences (Shanghai, China) and maintained in
Dulbecco's Modified Eagle Medium (DMEM; HyClone Laboratories, Inc.)
with 10% fetal bovine serum (FBS Gibco; Thermo Fisher Scientific,
Inc.) at 37°C in a humidified atmosphere of 5% CO2.
The CSCs were derived from U87MG cells by culturing
parental cells in a serum-free DMEM/F12 medium (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with B-27 (20 mg/ml; Thermo
Fisher Scientific, Inc.) which is an optimized serum substitute, 20
ng/ml basic fibroblast growth factor (bFGF; MilliporeSigma), 20
ng/ml epidermal growth factor (EGF; PeproTech, Inc.) at 37°C as
previously reported (14). The
medium was half-refreshed every 3 days. The primary tumor spheres
were detected within 10–14 days and subsequently dissociated and
passaged in fresh medium every 2–3 days.
For the hypoxia experiments, the cells were
initially maintained in 20% O2 and 5% CO2
(normoxia) at 37°C, then placed in an incubator (H35 Hypoxystation,
Don Whitley Scientific) with a gas mixture containing 1%
O2 and 5% CO2 (hypoxia) at 37°C. Culture
media were kept in hypoxystation for 24 h before use.
GEPIA analysis of gene expression
GEPIA (gepia.cancer-pku.cn/index.html) is based on
RNA sequences from Genotype Tissue Expression (GTEx) data and The
Cancer Genome Atlas (TCGA) programs, including 9,736 cancer and
8,587 normal samples. In the present study, GEPIA was used to
explore the mRNA expression level of Nogo-A in different kinds of
cancer compared to adjacent tissues.
Colony formation assay
For colony formation, cells were plated at final
concentration of 1×103 cells/well and cultured in
serum-free DMEM/F12 medium supplemented with 2% B-27, 20 ng/ml EGF
and 20 ng/ml bFGF. The medium was half-replaced every three days.
After 14 days, cells were then washed with PBS, fixed with 4%
paraformaldehyde in PBS at room temperature for 10 min, stained
with 0.1% crystal violet (MilliporeSigma) at room temperature for
10 min, washed with PBS, and the colonies >50 µm in diameter
were counted under a X71 (U-RFL-T) fluorescence microscope (Olympus
Corporation).
To block the Nogo-A/Nogo-A receptor (NgR)
interaction, NEP1-40 (Cat. No.: HY-P1242, MedChemExpress, United
States), a competitive antagonist of Nogo/NgR signaling pathway,
was added into culture medium at a final concentration of 100 µg/ml
at 37°C for 24 h, while saline was used as a mock control.
Western blotting
Total protein extraction from U87MG and U87MG-CSCs
was performed using the SoniConvert® Tissue-Cell
Convertor (DocSense) and Animal Tissue/cells/bacteria total protein
isolation kit (cat. no. PP001; DocSense, Chengdu, China) following
the manufacturer's instruction. The protein concentration was
measured using the Pierce BCA protein assay kit (Thermo Fisher
Scientific, Inc.). Total protein samples (10 µg) were separated
using 10% SDS-PAGE, then transferred onto a nitrocellulose
membrane. After transferring, the membrane was blocked with PBS
supplemented with 0.25% Tween 20 (PBS-T) and 5% skimmed milk for 1
h at room temperature. Following blocking, membranes were incubated
for 1 h at room temperature with the following primary antibodies:
Anti-CD24 antibody diluted 1:1,000 (cat. no. ab202073), anti-CD44
antibody diluted 1:2,500 (cat. no. ab157107), anti-CD133 antibody
diluted 1:1,000 (cat. no. ab226355), anti-microtubule associated
protein 2 (MAP2) antibody diluted 1:1,000 (cat. no. ab183830),
anti-β III tubulin antibody diluted 1:1,000 (cat. no. ab18207),
anti-GFAP antibody diluted 1:1,500 (cat. no. ab7260), anti-nestin
antibody diluted 1:1,000 (cat. no. ab105389) and anti-β-actin
(loading control) antibody diluted 1:5,000 (cat. no. ab8226). All
primary antibodies and secondary antibody were bought from Abcam.
The membranes were washed three times with PBS-T, then incubated
with HRP-labeled goat anti-mouse IgG antibody diluted in 1:5,000
(cat. no. ab97040) or goat anti-rabbit IgG antibody diluted in
1:5,000 (cat. no. ab7090) for 1 h at room temperature. The protein
bands were then detected using SuperSignal Western Femto Maximum
Sensitivity Substrate (Thermo Fisher Scientific, Inc.) and analyzed
using Quantity One (version 4.6.9, Bio-Rad Laboratories, Inc.).
Cell counting kit 8 (CCK-8)
assays
Proliferation was evaluated using Cell Counting Kit
8 (CCK-8) assays. Cells (5×103 cells/well) were plated
into 24-well culture plates containing 500 µl DMEM/F12 medium
supplemented with 2% B-27, 20 ng/ml EGF and 20 ng/ml bFGF. Cells
were incubated for 1–2 h at 37°C and 5% CO2 CCK-8
reagent (10 µl/well; MilliporeSigma) following the manufacturer's
instructions. The cell growth curves represent normalized mean
optical density at 450 nm of three independent samples recorded on
days 1 to 5.
Transwell assays
Cell culture matrix Matrigel (MilliporeSigma) was
diluted 1:5 in DMEM/F12. An 80-µl volume of diluted Matrigel was
added to the upper chambers of Transwell inserts for pre-coating at
37°C in a 5% CO2 incubator for 4 h. CSCs were made into
single-cell suspension, resuspended in DMEM/F12, and plated into
the upper chamber at a density of 5×103 cells/well and
cultured at 37°C. DMEM/F12 (600 µl) supplemented with 20 ng/ml EGF,
20 ng/ml bFGF and 2% B-27 was added to the lower chamber. After
24-h incubation, cells on the lower side were fixed in 4%
paraformaldehyde at room temperature for 10 min and stained with
0.25% crystal violet at room temperature for 15 min. The cells were
counted in five random fields of view under a light microscope.
Immunofluorescence staining
Cells were fixed and permeabilized with 4%
paraformaldehyde in 1X PBS containing 0.1% Triton X-100 for 10 min
at room temperature and non-specific binding was blocked with 10%
normal goat serum (Sigma) in 1X PBS for 1 h at room temperature.
The cells were incubated with primary antibodies against anti-β III
tubulin antibody diluted 1:1,000 (cat. no. ab18207), anti-GFAP
antibody diluted 1:1,500 (cat. no. ab7260), anti-nestin antibody
diluted 1:1,000 (cat. no. ab105389) at room temperature for 1 h.
The Alexa Fluor 488-conjugated secondary antibody (Cat. No.:
ab150077) was then added at dilution of 1:1,000 and incubated for 1
h at room temperature. All primary antibodies and secondary
antibody were bought from Abcam. The cells were mounted in
Vectashield® (Vector Laboratories, Ltd.) with Hoechst
33342 and imaged under a X71 (U-RFL-T) fluorescence microscope
(Olympus Corporation).
Transduction
The shRNA targeting Nogo-A mRNA (shCCDC88A;
targeting sequence: 5′-AATGATTCCGAGGCAGATTAT-3′) and the scrambled
negative control shRNA (shScrambled; scrambled sequence:
5′-AACGAACGAGTACCGTACACT-3′) were designed and bought from
Guangzhou RiboBio Co., Ltd. Both shRNAs were inserted into GV248
lentiviral vectors. For lentiviral packaging, the shNogo-A or
shScrambled vector was co-transfected into 293T cells (Shanghai
Institute of Biochemistry and Cell Biology) with packing vectors
(pVSVG and psPAX2, Addgene) at equal amount (1.2 µg each plasmid)
using Lipofectamine® 2000 (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol at 37°C. The medium
was replaced 4 h later. After 72 h of transfection, the medium were
centrifuged at 12.000 g, 4°C for 15 min, and supernatant was
collected and filtered. To silence Nogo-A, CSCs derived from U87MG
glioblastoma cells were transduced with lentivirus with a
multiplicity of infection of 10 at 37°C for 4 h, followed by the
replacement of supernatant with regular medium. GFP-expressing
cells were imaged using a fluorescence microscope (Olympus
Corporation). Cells stably expressing shRNA were established by
culture in the presence of 10 µg/ml puromycin for selection and 5
µg/ml puromycin was used for maintenance.
Cell cycle analysis
Cell cycle distribution was analyzed by quantifying
DNA content. CSCs with Nogo-A knockdown were harvested, washed with
ice-cold PBS, and fixed overnight at 4°C with ice-cold 70% ethanol.
The fixed cells were washed with PBS for three times and incubated
with final concentration of 100 µg/ml RNase A and 40 µg/ml
propidium iodide (PI; Beyotime) for 30 min in the dark. Cells were
analyzed using the three-laser Navios cytometer (Beckman Coulter
Inc.).
Tumor formation in soft agar
Cells (5×103) were seeded in 1 ml of 0.3%
melted agar in DMEM/F12 containing 2% B27, 20 ng/ml of EGF, and 20
ng/ml of bFGF and plated in 6-well plate with 0.6% agar in the same
medium at 37°C. Two weeks later, colonies were stained with 10
mg/ml of nitro blue tetrazolium (Sigma-Aldrich; Merck KGaA) at 37°C
and scanned 4 h later with an Epson Perfection 3200 scanner.
ATP production
Cells (1×106) cells were plated in 6-well
plates and allowed to adhere overnight. ATP Lite assay kit (cat.
no. 6016943; PerkinElmer, Inc.) was used to measure ATP production
following the manufacturer's instruction.
Reactive oxygen species (ROS)
detection
Total intracellular ROS was measured by flow
cytometry after dichlorofluorescein (DCF) oxidation assays (cat.
no. D399; Thermofisher Scientific, Inc.). The intracellular ROS
oxidizes the cleaved dichlorodihydrofluorescein diacetate (DCFH-DA)
which enters into the cells. Target cells (5×105) were
incubated with DCFH-DA (10 µM) for 1 h at 37°C, followed by three
washes with ice-cold PBS and ROS fluorescence was analyzed using a
microplate reader (Sinergy H1; BioTek). The signal of DCFH in
shScrambled group at 24 h time point was used for
normalization.
Statistical analysis
All data are presented as mean ± SD. Statistical
differences were assessed using unpaired Student's t-tests. One-way
analysis of variance (ANOVA) was performed to compare two groups
with one variable followed by Tukey's post hoc test using Prism 6
(GraphPad Software). P<0.05 was considered to indicate a
statistically significant difference. All experiments were repeated
three times.
Results
Enrichment and identification of CSCs
from glioblastoma cells
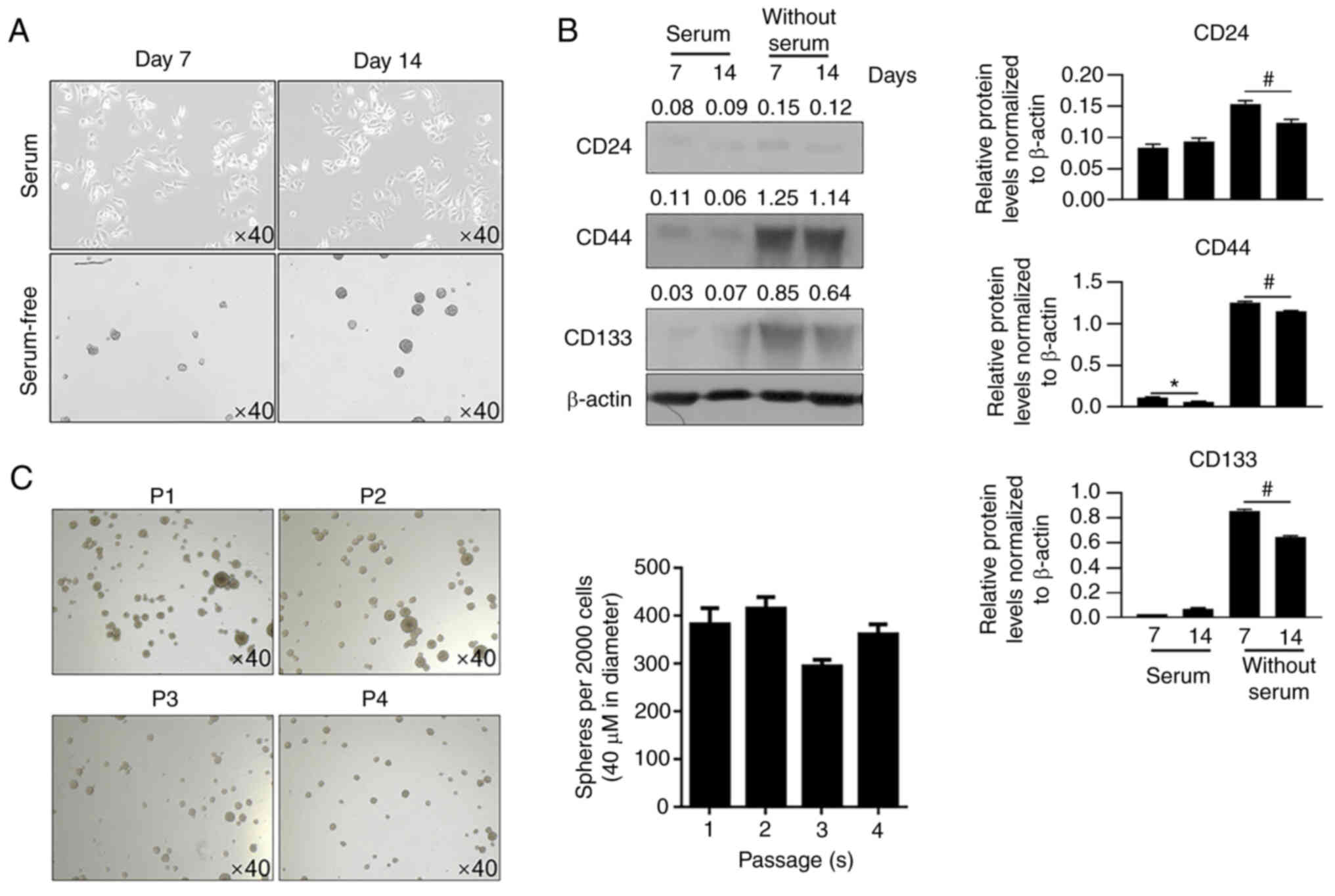
To obtain CSCs from U87MG glioblastoma cells, U87MG
cells were cultured in serum-free medium. After 14 days, spheres
were imaged (Fig. 1A). Spheres
formed and suspended in medium, whereas parental cells attached to
bottom of well. Western blotting was then performed to detect CD44,
CD24, and CD133, which are three CSC markers (15,16).
On days 7 and 14, spheres cultured in serum-free medium presented a
marked increase in CD44 and CD133 levels, but not in CD24 levels,
compared with cells cultured in serum-containing medium (Fig. 1B). Passages 1 to 4 presented
consistent self-renewal capacity, confirming the stemness of the
U87MG-derived CSCs (Fig. 1C).
CCK-8 assays were performed to detect cell
proliferation from day 1 to day 4. U87MG-CSCs presented increased
proliferation compared with their parental cells (Fig. 2A). Transwell invasion and tumor
formation assays in soft agar were performed. U87MG CSCs presented
enhanced invasion and tumor formation compared to U87MG cells
(Fig. 2B and C). To further
confirm CSC differentiation, CSCs were cultured in medium
supplemented with FBS for 5 or 10 days to promote differentiation.
MAP2, β-III tubulin, GFAP and nestin, which are markers of CSC
differentiation (6), were then
detected using western blot analysis. As shown in Fig. 2D and E, MAP2, β-III tubulin, and
GFAP levels were upregulated in CSCs after 5 and 10-day culture in
medium supplemented with FBS, and these proteins were also imaged
through immunofluorescence staining. These data indicated that CSCs
were successfully enriched from U87MG cells. Furthermore, these
results indicated that enriched CSCs from U87MG presented increased
viability, invasion and tumor formation compared with the parental
cells.
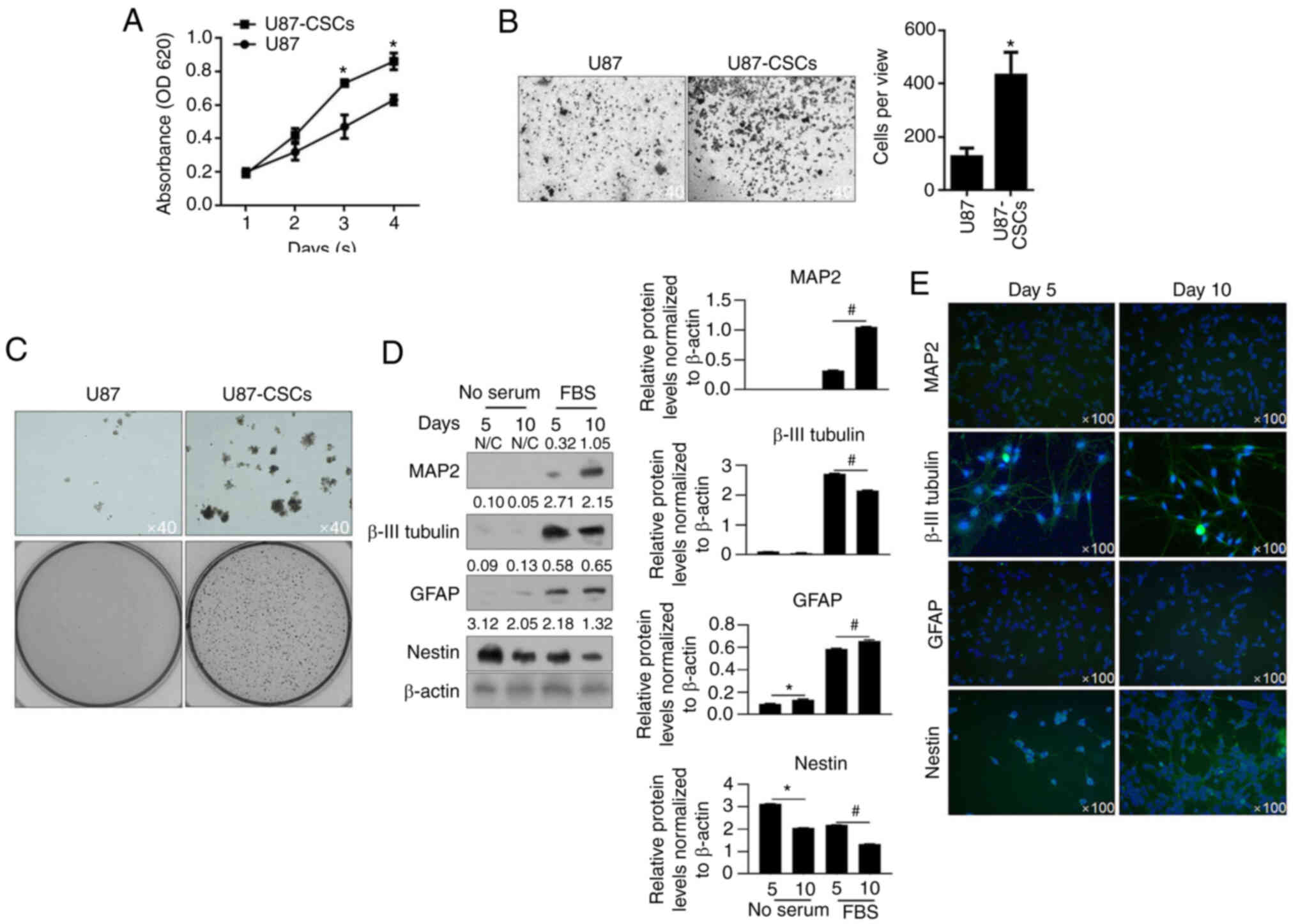 | Figure 2.Proliferation, invasion and colony
formation of U87 CSCs and its parental cells. (A) By performing
CCK-8 assays, the proliferation of U87MG-CSCs from day 1 to 4 was
measured. *P<0.05 vs. U87MG parental cells. (B) Transwell assays
were performed to detect invasiveness of U87MG-CSCs. *P<0.05 vs.
U87MG parental cells. (C) Tumor formation in soft agar was employed
to detect the tumor formation in U87MG-CSCs compared to their
parental cells. Upper lane, Magnification, ×40; lower lane, imaged
by digital camera. (D) To analyze differentiation, CSCs were
cultured in FBS-supplemented medium for 5 and 10 days, then
analyzed by western blotting. *P<0.05 vs. 5 days without serum;
#P<0.05 vs. 5 days with serum. (E) Immunofluorescence
staining of the neuronal markers, MAP2, β-III tubulin, GFAP and
nestin. CCK-8, Cell Counting Kit-8; CSCs, cancer stem cells; MAP2,
microtubule-associated protein 2; GFAP, glial fibrillary acidic
protein; CSCs, cancer stem cells. |
Nogo-A is upregulated in
U87MG-CSCs
By considering that Nogo-A is critically involved in
regulating physiological processes in glioblastoma cancer (17,18),
the expression of Nogo-A was analyzed in several types of cancer
using GEPIA (Gene Expression Profiling Interactive Analysis), a web
server for cancer and normal gene expression profiling and
interactive analyses (19).
Despite the upregulation of Nogo-A in pancreatic adenocarcinoma, no
evident differences were observed in Nogo-A expression between
tumor and adjacent tissue, including in bladder urothelial
carcinoma (BLCA), breast invasive carcinoma (BRCA), colon
adenocarcinoma (COAD), kidney renal clear cell carcinoma (KIRC),
lung adenocarcinoma (LUAD) and pancreatic adenocarcinoma (PAAD)
(Fig. 3A).
Furthermore, Nogo-A was detected in U87MG and
U87MG-CSCs from passage 1 to 4 by western blot analysis. As shown
in Fig. 3B, stable expression
levels of Nogo-A at all passages were observed in U87MG-CSCs, which
were significantly higher than those in U87MG cells. After 5 and 10
days of differentiation, Nogo-A expression levels were
significantly decreased, indicating that Nogo-A promotes stemness
of CSCs derived from U87MG (Fig.
3C).
Nogo-A regulates malignant behaviors
in U87MG-CSCs via interaction with NgR
Aiming to evaluate the effect of Nogo-A on malignant
behaviors in U87MG-CSCs, Nogo-A was knocked down by transfecting
Nogo-A targeting shRNA (shNogo-A), compared to negative control
(shScrambled, Fig. 4A).
Considering that the role of Nogo-A mainly is exerted by binding to
NgR (20), NEP1-40, a competitive
antagonist of the Nogo/NgR signaling pathway, was used to block
Nogo-A activity (21). As shown in
Fig. 4B, cell viability after days
4 and 5 was significantly inhibited by both Nogo-A knockdown and
Nogo/NgR pathway inhibition by NEP1-40. Cell cycle distribution was
then measured by performing PI staining followed by flow cytometry
analysis. Moreover, the frequency of cells in the G1 to
G0 phase of the cell cycle was significantly increased
both by Nogo-A knockdown and Nogo/NgR signaling pathway inhibition
using NEP1-40, indicating that cell proliferation was stimulated by
Nogo-A via Nogo-A/NgR signaling pathway by promoting cell cycle
entry (Fig. 4C). Using colony
formation assays, it was observed that Nogo-A knockdown
significantly decreased sphere formation, which is similar with the
effect of addition of NEP1-40 in U87MG-CSCs (Fig. 4D). Other malignant behaviors were
also assessed in U87MG-CSCs, including tumor formation and invasion
in soft agar. As it is shown in Fig.
4E and F, Nogo-A knockdown or addition of NEP1-40 significantly
decreased invasion and tumor formation capacities in
U87MG-CSCs.
Nogo-A regulates ATP synthesis and ROS
accumulation and exerts protective effect against hypoxia-induced
cell death
It has been reported that
hypoxia/reoxygenation-induced mitochondria-dependent apoptosis is
tightly regulated by Nogo-A/NgR signaling pathway (22). This prompted us to determine
whether Nogo-A/NgR signaling pathway is involved in mitochondrial
energy metabolism. To achieve this goal, mitochondrial ATP
synthesis, ROS accumulation and apoptosis were assessed. Firstly,
ATP synthesis was measured following Nogo-A knockdown or Nogo-A/NgR
signaling pathway inhibition. Both approaches resulted in decreased
intracellular ATP levels (Fig.
5A), indicating that Nogo-A/NgR signaling could promote
metabolism. After 24-, 48- and 72-h inhibition of the Nogo-A/NgR
signaling pathway, ROS accumulation was significantly decreased.
This observation could be explained by a decrease in ATP synthesis
and utilization with a concomitant decrease in ROS generation and
accumulation (Fig. 5B). These
results suggested that Nogo-A/NgR signaling could sensitize cells
to hypoxia, which could induce ROS accumulation.
Moreover, cell death was detected in CSCs following
hypoxia exposure. Hypoxia treatment notably increased apoptosis
compared with normoxia group (Fig.
5C). Following Nogo-A knockdown or addition of NEP1-40, both
non-apoptotic and apoptotic cell death were significantly
increased. Interestingly, non-apoptotic cell death was increased by
NEP1-40, whereas apoptotic cell death was not affected by NEP1-40.
Taken together, these results indicated that Nogo-A/NgR signaling
pathway tightly regulates hypoxia/reoxygenation-induced
mitochondria-dependent apoptosis.
Discussion
In the present study, high levels of Nogo-A protein
expression were observed in U87MG-CSCs, which decreased after CSC
differentiation. Upregulated Nogo-A promoted the proliferation,
entry into the cell cycle and tumor formation in soft agar in CSCs
derived from U87MG cells. Although it is difficult to rule out the
confounding effect of cell proliferation on cell invasion,
invasiveness was also potentially enhanced in U87-CSCs compared to
parental cells. Moreover, the effects of Nogo-A on these was
dependent on its receptor, NgR. Knockdown of Nogo-A and inhibition
of Nogo-A/NgR signaling pathway inhibited their roles in regulating
CSCs. However, according to GEPIA database, no differences in
Nogo-A expression levels were observed between glioblastoma tumor
and adjacent tissues. These results indicated that Nogo-A could be
expressed differently and serve different functions in CSCs,
compared with parental glioblastoma cells.
Glioblastoma is the most common and fatal type of
primary brain tumor due to the occurrence of chemoresistance and
radioresistance (23), resulting
in tumor growth, metastasis, and relapse (24,25).
CSCs in glioblastoma are a sub-population emerging from the
increased self-renewing division of glioblastoma cells or from the
reprogramming of differentiated glioblastoma cells to
undifferentiated forms (26).
Thus, improving the understanding of the differences between CSCs
derived from glioblastoma and their parental cells is of utmost
importance to provide insights into strategies to overcome
chemoresistance and radioresistance. Therefore, in the present
study, CSCs were enriched from U87MG and their stemness was
confirmed by detecting markers of stemness, including CD44 and
CD133 (6). Self-renewal capacity
was also assessed by performing serial replating assays, which is
considered the gold standard of cell stemness detection. However,
one limitation of this study is that additional experiments
required to verify the CSC phenotype, such as RNA sequencing, were
not performed.
Nogo-A was initially liked to the regulation of
neurons in CNS (7,9,17)
and, in recent years, accumulating evidence has emerged presenting
its regulatory effects on the malignancy of glioblastoma cells
(13,18,25).
However, conflicting roles of Nogo-A in glioblastoma cells
indicated that Nogo-A could exert complex functions under different
conditions. The GEPIA server was used to determine the Nogo-A
expression levels in different types of cancers and it was found
that, except for pancreatic adenocarcinoma, no clear difference in
Nogo-A expression was observed between tumor tissues and adjacent
tissues, in agreement with the previous report (13). Instead of using clinical samples,
CSCs were derived from U87MG glioblastoma cells and Nogo-A was
upregulated compared with the parental cells. Moreover, after
differentiation induced by culturing in serum-containing medium, a
marked decrease in Nogo-A protein expression was observed in
differentiated CSCs, further confirming the positive association
between Nogo-A and stemness in CSCs derived from U87MG cells. As a
limitation, however, the effect of the Nogo-A overexpression on the
properties of CSCs could not be evaluated due to the relatively
high endogenous Nogo-A expression level. In future studies, Nogo-A
in U87MG glioblastoma cells overexpression may help confirm the
alterations in the stemness of U87MG cells observed in the present
study. The U87MG is widely used for investigating stemness of
glioblastoma. However, as a limitation of the present study, only
one source of CSCs was obtained from U87MG and more sources of CSCs
are needed for further investigation.
CSCs derived from glioblastoma cells act
differently, frequently promoting malignancies (4). The aim of this study was to determine
whether Nogo-A was associated with malignancies in CSCs derived
from glioblastoma cells. Nogo-A was knocked down by introducing
shRNA targeting Nogo-A mRNA and the Nogo-A/NgR signaling pathway
was blocked by adding NEP1-40, an antagonist of the Nogo-A/NgR
signaling pathway (21). As
expected, knockdown of Nogo-A and inhibition of the Nogo-A/NgR
signaling pathway decreased malignant behavior, including cell
proliferation, invasion and tumor formation in soft agar.
Nogo-A/NgR signaling pathway was associated with the inhibition of
the proliferation and differentiation in glioblastoma stem cells
(27); however, in this study,
following Nogo-A knockdown and Nogo-A/NgR pathway inhibition, the
stemness of CSCs derived from U87MG glioblastoma cells were
inhibited, which is in disagreement with the previous report
(27). This could be due to the
use of different CSCs enrichment methods, which could result in
different sub-populations of CSCs.
It has been reported that, under hypoxic conditions,
Nogo-A binds to Apg-1, a member of the stress-induced heat-shock
protein of 110 kDa (Hsp110), and thus exerts a protective effect
against hypoxia-induced cell death (28). Considering the regulatory role
exerted by Nogo-A under hypoxic conditions, CSCs were also exposed
to hypoxic conditions for 2 h to find out that a decrease in Nogo-A
expression and the Nogo-A/NgR signaling pathway inhibition were
associated with a decrease in mitochondrial ATP synthesis and ROS
accumulation. Furthermore, non-apoptotic and apoptotic cell death
were also increased in hypoxic conditions, indicating that in CSCs,
Nogo-A could exert protective effects against hypoxia and oxidative
stress. It was also hypothesized that altered mitochondrial
functions, including ATP synthesis, could also affect malignant
behaviors of CSCs in a Nogo-A/NgR signaling pathway related
manner.
Nogo-A was upregulated in CSCs derived from
glioblastoma and functioned as a key factor in promoting malignant
behaviors and protecting cells from exposure to hypoxic conditions.
Moreover, it was found that Nogo-A critically regulated
mitochondrial function, ATP synthesis, and maintenance of stemness
via interacting with NgR. These data highlight Nogo-A as a
potential therapeutic target for glioblastoma.
Acknowledgements
The authors would like to thank Dr Yin Li (Chongqing
University) for language editing.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CA, YuZ and YiZ designed the experiments and
performed most of the experiments included in this study. CA, YuZ
and KP performed cell culture, RNA isolation and protein extraction
experiments. CA, YY and YiZ are responsible for data collection and
performed statistical analysis. All authors read and approved the
final manuscript. All authors confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mitre AO, Florian AI, Buruiana A, Boer A,
Moldovan I, Soritau O, Florian SI and Susman S: Ferroptosis
involvement in glioblastoma treatment. Medicina (Kaunas).
58:3192022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang Y and Jiang T: Understanding high
grade glioma: Molecular mechanism, therapy and comprehensive
management. Cancer Lett. 331:139–146. 2013. View Article : Google Scholar
|
|
3
|
Ramaswamy V and Taylor MD: The amazing and
deadly glioma race. Cancer Cell. 28:275–277. 2015. View Article : Google Scholar
|
|
4
|
Alcantara Llaguno S and Parada LF: Cancer
stem cells in gliomas: Evolving concepts and therapeutic
implications. Curr Opin Neurol. 34:868–874. 2021. View Article : Google Scholar
|
|
5
|
Lathia JD, Mack SC, Mulkearns-Hubert EE,
Valentim CL and Rich JN: Cancer stem cells in glioblastoma. Genes
Dev. 29:1203–1217. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Seifert C, Balz E, Herzog S, Korolev A,
Gaßmann S, Paland H, Fink MA, Grube M, Marx S, Jedlitschky G, et
al: PIM1 inhibition affects glioblastoma stem cell behavior and
kills glioblastoma stem-like cells. Int J Mol Sci. 22:111262021.
View Article : Google Scholar
|
|
7
|
GrandPré T, Nakamura F, Vartanian T and
Strittmatter SM: Identification of the Nogo inhibitor of axon
regeneration as a reticulon protein. Nature. 403:439–444. 2000.
View Article : Google Scholar
|
|
8
|
Petrinovic MM, Duncan CS, Bourikas D,
Weinman O, Montani L, Schroeter A, Maerki D, Sommer L, Stoeckli ET
and Schwab ME: Neuronal Nogo-A regulates neurite fasciculation,
branching and extension in the developing nervous system.
Development. 137:2539–2550. 2010. View Article : Google Scholar
|
|
9
|
Mathis C, Schröter A, Thallmair M and
Schwab ME: Nogo-a regulates neural precursor migration in the
embryonic mouse cortex. Cereb Cortex. 20:2380–2390. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schwab ME: Functions of Nogo proteins and
their receptors in the nervous system. Nat Rev Neurosci.
11:799–811. 2010. View
Article : Google Scholar
|
|
11
|
Xiong NX, Zhao HY, Zhang FC and He ZQ:
Negative correlation of Nogo-A with the malignancy of
oligodendroglial tumor. Neurosci Bull. 23:41–45. 2007. View Article : Google Scholar
|
|
12
|
Schwab DE, Lepski G, Borchers C, Trautmann
K, Paulsen F and Schittenhelm J: Immunohistochemical comparative
analysis of GFAP, MAP-2, NOGO-A, OLIG-2 and WT-1 expression in WHO
2016 classified neuroepithelial tumors and their prognostic value.
Pathol Res Pract. 214:15–24. 2018. View Article : Google Scholar
|
|
13
|
Behling F, Barrantes-Freer A, Behnes CL,
Stockhammer F, Rohde V, Adel-Horowski A, Rodríguez-Villagra OA,
Barboza MA, Brück W, Lehmann U, et al: Expression of Olig2, nestin,
NogoA and AQP4 have no impact on overall survival in IDH-wildtype
glioblastoma. PLoS One. 15:e2292742020. View Article : Google Scholar
|
|
14
|
Wang X, Zhou W, Li X, Ren J, Ji G, Du J,
Tian W, Liu Q and Hao A: Graphene oxide suppresses the growth and
malignancy of glioblastoma stem cell-like spheroids via epigenetic
mechanisms. J Transl Med. 18:2002020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shtivelman E and Bishop J: Expression of
CD44 is repressed in neuroblastoma cells. Mol Cell Biol.
11:5446–5453. 1991. View Article : Google Scholar
|
|
16
|
Nanduri LS, Maimets M, Pringle SA, van der
Zwaag M, van Os RP and Coppes RP: Regeneration of irradiated
salivary glands with stem cell marker expressing cells. Radiother
Oncol. 99:367–372. 2011. View Article : Google Scholar
|
|
17
|
Wälchli T, Pernet V, Weinmann O, Shiu JY,
Guzik-Kornacka A, Decrey G, Yüksel D, Schneider H, Vogel J, Ingber
DE, et al: Nogo-A is a negative regulator of CNS angiogenesis. Proc
Natl Acad Sci USA. 110:E1943–E1952. 2013. View Article : Google Scholar
|
|
18
|
Wirthschaft P, Bode J, Soni H, Dietrich F,
Krüwel T, Fischer B, Knobbe-Thomsen CB, Rossetti G, Hentschel A,
Mack N, et al: RhoA regulates translation of the Nogo-A decoy SPARC
in white matter-invading glioblastomas. Acta Neuropathol.
138:275–293. 2019. View Article : Google Scholar
|
|
19
|
Aldape K, Zadeh G, Mansouri S,
Reifenberger G and von Deimling A: Glioblastoma: Pathology,
molecular mechanisms and markers. Acta Neuropathol. 129:829–848.
2015. View Article : Google Scholar
|
|
20
|
Zagrebelsky M and Korte M: Maintaining
stable memory engrams: New roles for Nogo-A in the CNS.
Neuroscience. 283:17–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fang Y, Yao L, Li C, Wang J, Wang J, Chen
S, Zhou XF and Liao H: The blockage of the Nogo/NgR signal pathway
in microglia alleviates the formation of Aβ plaques and tau
phosphorylation in APP/PS1 transgenic mice. J Neuroinflammation.
13:562016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sarkey JP, Chu M, McShane M, Bovo E, Ait
Mou Y, Zima AV, de Tombe PP, Kartje GL and Martin JL: Nogo-A
knockdown inhibits hypoxia/reoxygenation-induced activation of
mitochondrial-dependent apoptosis in cardiomyocytes. J Mol Cell
Cardiol. 50:1044–1055. 2011. View Article : Google Scholar
|
|
23
|
Lima FR, Kahn SA, Soletti RC, Biasoli D,
Alves T, da Fonseca AC, Garcia C, Romão L, Brito J, Holanda-Afonso
R, et al: Glioblastoma: Therapeutic challenges, what lies ahead.
Biochim Biophys Acta. 1826:338–349. 2012.
|
|
24
|
Diaz A and Leon K: Therapeutic approaches
to target cancer stem cells. Cancers (Basel). 3:3331–3352. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ortensi B, Setti M, Osti D and Pelicci G:
Cancer stem cell contribution to glioblastoma invasiveness. Stem
Cell Res Ther. 4:182013. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Safa AR, Saadatzadeh MR, Cohen-Gadol AA,
Pollok KE and Bijangi-Vishehsaraei K: Glioblastoma stem cells
(GSCs) epigenetic plasticity and interconversion between
differentiated non-GSCs and GSCs. Genes Dis. 2:152–163. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Meng Y, Shang F and Zhu Y: miR-124
participates in the proliferation and differentiation of brain
glioma stem cells through regulating Nogo/NgR expression. Exp Ther
Med. 18:2783–2788. 2019.PubMed/NCBI
|
|
28
|
Kern F, Stanika RI, Sarg B, Offterdinger
M, Hess D, Obermair GJ, Lindner H, Bandtlow CE, Hengst L and
Schweigreiter R: Nogo-A couples with Apg-1 through interaction and
co-ordinate expression under hypoxic and oxidative stress. Biochem
J. 455:217–227. 2013. View Article : Google Scholar : PubMed/NCBI
|