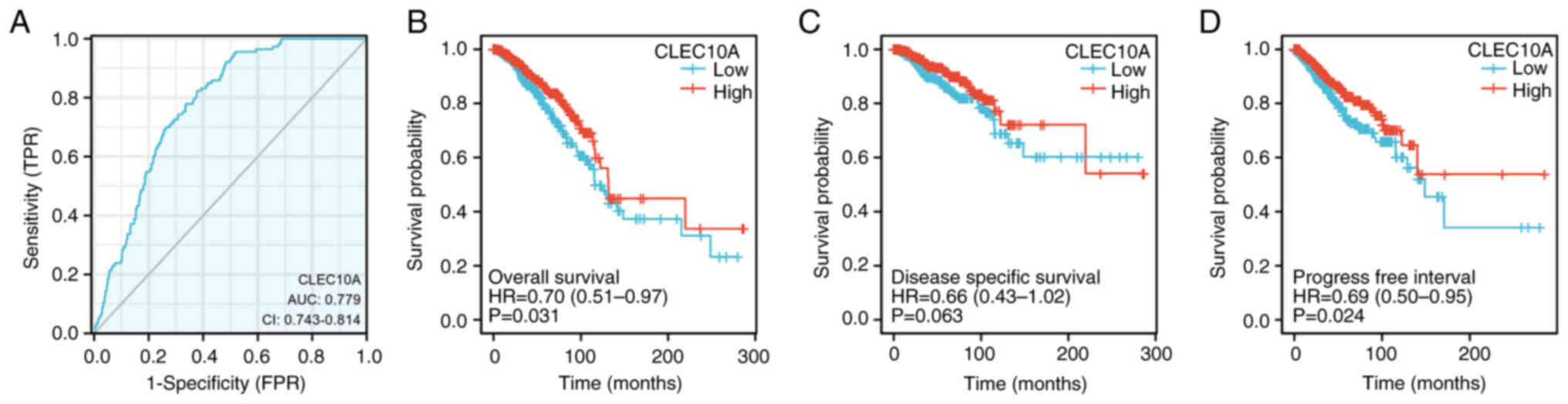
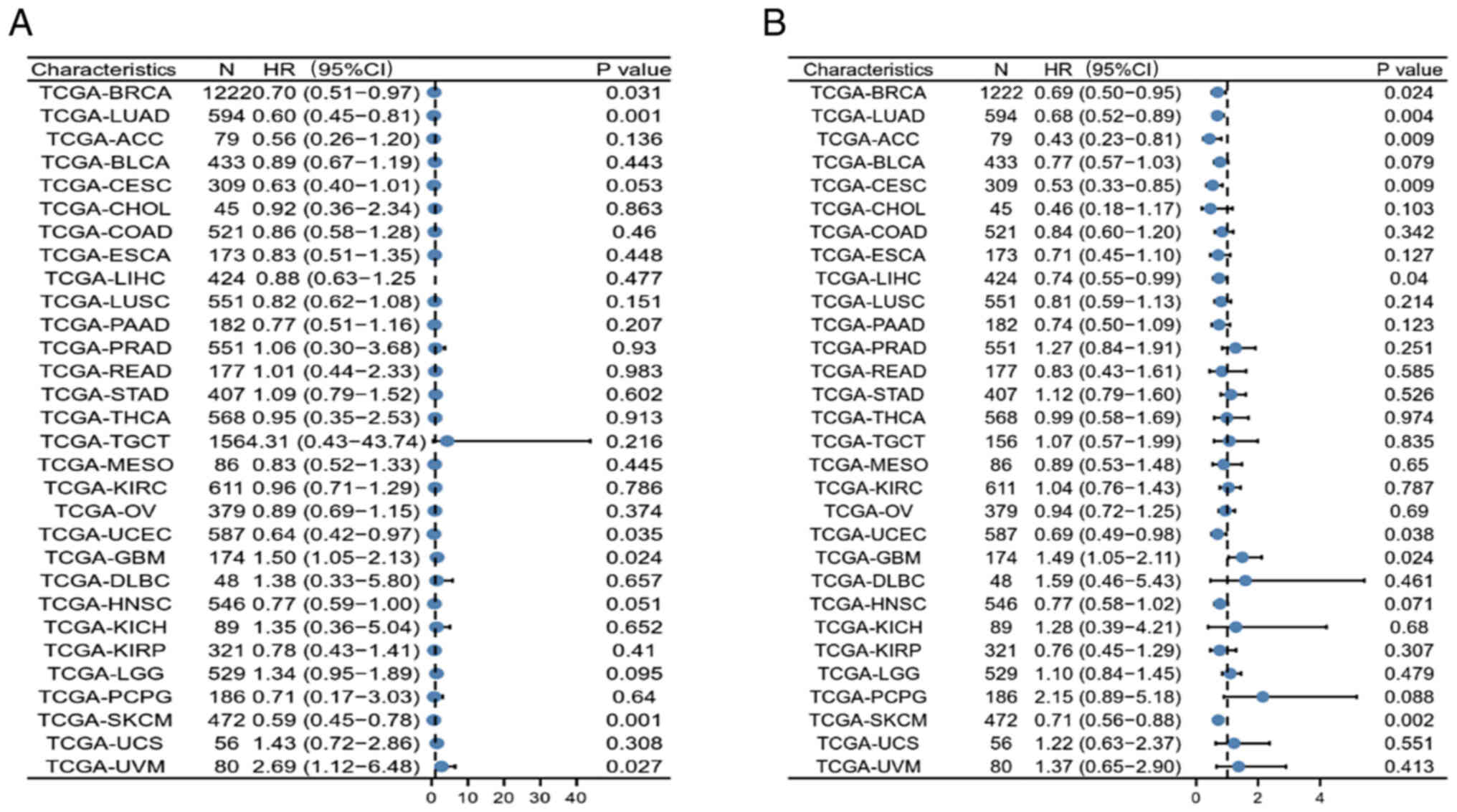
|
1
|
Aushev VN, Lee E, Zhu J, Gopalakrishnan K,
Li Q, Teitelbaum SL, Wetmur J, Degli Esposti D, Hernandez-Vargas H,
Herceg Z, et al: Novel predictors of breast cancer survival derived
from miRNA activity analysis. Clin Cancer Res. 24:581–591. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saad ED, Squifflet P, Burzykowski T,
Quinaux E, Delaloge S, Mavroudis D, Perez E, Piccart-Gebhart M,
Schneider BP, Slamon D, et al: Disease-free survival as a surrogate
for overall survival in patients with HER2-positive, early breast
cancer in trials of adjuvant trastuzumab for up to 1 year: A
systematic review and meta-analysis. Lancet Oncol. 20:361–370.
2019. View Article : Google Scholar
|
|
4
|
Shachar SS, Deal AM, Weinberg M, Williams
GR, Nyrop KA, Popuri K, Choi SK and Muss HB: Body composition as a
predictor of toxicity in patients receiving anthracycline and
taxane-based chemotherapy for early-stage breast cancer. Clin
Cancer Res. 23:3537–3543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lischka A, Doberstein N, Freitag-Wolf S,
Kocak A, Gemoll T, Heselmeyer-Haddad K, Ried T, Auer G and
Habermann JK: Genome instability profiles predict disease outcome
in a cohort of 4,003 patients with breast cancer. Clin Cancer Res.
26:4606–4615. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abu-Thuraia A, Goyette MA, Boulais J,
Delliaux C, Apcher C, Schott C, Chidiac R, Bagci H, Thibault MP,
Davidson D, et al: AXL confers cell migration and invasion by
hijacking a PEAK1-regulated focal adhesion protein network. Nat
Commun. 11:35862020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pirro M, Mohammed Y, van Vliet SJ,
Rombouts Y, Sciacca A, de Ru AH, Janssen GMC, Tjokrodirijo RTN,
Wuhrer M, van Veelen PA and Hensbergen PJ: N-Glycoproteins have a
major role in MGL binding to colorectal cancer cell lines:
Associations with overall proteome diversity. Int J Mol Sci.
21:55222020. View Article : Google Scholar
|
|
8
|
Eggink LL, Roby KF, Cote R and Kenneth
Hoober J: An innovative immunotherapeutic strategy for ovarian
cancer: CLEC10A and glycomimetic peptides. J Immunother Cancer.
6:282018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Messenheimer DJ, Jensen SM, Afentoulis ME,
Wegmann KW, Feng Z, Friedman DJ, Gough MJ, Urba WJ and Fox BA:
Timing of PD-1 blockade is critical to effective combination
immunotherapy with Anti-OX40. Clin Cancer Res. 23:6165–6177. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Heger L, Balk S, Luhr JJ, Heidkamp GF,
Lehmann CH, Hatscher L, Purbojo A, Hartmann A, Garcia-Martin F,
Nishimura SI, et al: CLEC10A is a specific marker for human
CD1c+ dendritic cells and enhances their toll-like
receptor 7/8-induced cytokine secretion. Front Immunol. 9:7442018.
View Article : Google Scholar
|
|
11
|
Kurze AK, Buhs S, Eggert D,
Oliveira-Ferrer L, Muller V, Niendorf A, Wagener C and Nollau P:
Immature O-glycans recognized by the macrophage glycoreceptor
CLEC10A (MGL) are induced by 4-hydroxy-tamoxifen, oxidative stress
and DNA-damage in breast cancer cells. Cell Commun Signal.
17:1072019. View Article : Google Scholar
|
|
12
|
He M, Han Y, Cai C, Liu P, Chen Y, Shen H,
Xu X and Zeng S: CLEC10A is a prognostic biomarker and correlated
with clinical pathologic features and immune infiltrates in lung
adenocarcinoma. J Cell Mol Med. 25:3391–3399. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Z, Li J, He T and Ding J:
Bioinformatics identified 17 immune genes as prognostic biomarkers
for breast cancer: Application study based on artificial
intelligence algorithms. Front Oncol. 10:3302020. View Article : Google Scholar
|
|
14
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar
|
|
16
|
Nagy A, Lanczky A, Menyhart O and Gyorffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep. 8:92272018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Svetnik V, Liaw A, Tong C, Culberson JC,
Sheridan RP and Feuston BP: Random forest: A classification and
regression tool for compound classification and QSAR modeling. J
Chem Inf Comput Sci. 43:1947–1958. 2003. View Article : Google Scholar
|
|
18
|
Bindea G, Mlecnik B, Tosolini M,
Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T,
Lafontaine L, Berger A, et al: Spatiotemporal dynamics of
intratumoral immune cells reveal the immune landscape in human
cancer. Immunity. 39:782–795. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shang Y, Zhang Y, Liu J, Chen L, Yang X,
Zhu Z, Li D, Deng Y, Zhou Z, Lu B and Fu CG: Decreased E2F2
expression correlates with poor prognosis and immune infiltrates in
patients with colorectal cancer. J Cancer. 13:653–668. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ru B, Wong CN, Tong Y, Zhong JY, Zhong
SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, et al: TISIDB: An
integrated repository portal for tumor-immune system interactions.
Bioinformatics. 35:4200–4202. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sameni M, Tovar EA, Essenburg CJ,
Chalasani A, Linklater ES, Borgman A, Cherba DM, Anbalagan A, Winn
ME, Graveel CR and Sloane BF: Cabozantinib (XL184) inhibits growth
and invasion of preclinical TNBC models. Clin Cancer Res.
22:923–934. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin W, Noel P, Borazanci EH, Lee J, Amini
A, Han IW, Heo JS, Jameson GS, Fraser C, Steinbach M, et al:
Single-cell transcriptome analysis of tumor and stromal
compartments of pancreatic ductal adenocarcinoma primary tumors and
metastatic lesions. Genome Med. 12:802020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Narayanan K, Kumar S, Padmanabhan P,
Gulyas B, Wan ACA and Rajendran VM: Lineage-specific exosomes could
override extracellular matrix mediated human mesenchymal stem cell
differentiation. Biomaterials. 182:312–322. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang G, Lu X, Dey P, Deng P, Wu CC, Jiang
S, Fang Z, Zhao K, Konaparthi R, Hua S, et al: Targeting
YAP-Dependent MDSC infiltration impairs tumor progression. Cancer
Discov. 6:80–95. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nollau P, Wolters-Eisfeld G, Mortezai N,
Kurze AK, Klampe B, Debus A, Bockhorn M, Niendorf A and Wagener C:
Protein domain histochemistry (PDH): Binding of the carbohydrate
recognition domain (CRD) of recombinant human glycoreceptor CLEC10A
(CD301) to formalin-fixed, paraffin-embedded breast cancer tissues.
J Histochem Cytochem. 61:199–205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zizzari IG, Napoletano C, Battisti F,
Rahimi H, Caponnetto S, Pierelli L, Nuti M and Rughetti A: MGL
Receptor and immunity: When the ligand can make the difference. J
Immunol Res. 2015.4506952015.
|
|
28
|
Hoober JK: ASGR1 and its enigmatic
relative, CLEC10A. Int J Mol Sci. 21:48182020. View Article : Google Scholar
|
|
29
|
Heger L, Hofer TP, Bigley V, de Vries IJM,
Dalod M, Dudziak D and Ziegler-Heitbrock L: Subsets of CD1c(+) DCs:
Dendritic cell versus monocyte lineage. Front Immunol.
11:5591662020. View Article : Google Scholar
|
|
30
|
Zelensky AN and Gready JE: The C-type
lectin-like domain superfamily. FEBS J. 272:6179–6217. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sawant DV, Yano H, Chikina M, Zhang Q,
Liao M, Liu C, Callahan DJ, Sun Z, Sun T, Tabib T, et al: Adaptive
plasticity of IL-10+ and IL-35+
Treg cells cooperatively promotes tumor T cell
exhaustion. Nat Immunol. 20:724–735. 2019. View Article : Google Scholar
|
|
32
|
Plitas G, Konopacki C, Wu K, Bos PD,
Morrow M, Putintseva EV, Chudakov DM and Rudensky AY: Regulatory T
cells exhibit distinct features in human breast cancer. Immunity.
45:1122–1134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aguilera TA and Giaccia AJ: Molecular
pathways: Oncologic pathways and their role in T-cell exclusion and
immune evasion-A new role for the AXL receptor tyrosine kinase.
Clin Cancer Res. 23:2928–2933. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kajikawa M, Ose T, Fukunaga Y, Okabe Y,
Matsumoto N, Yonezawa K, Shimizu N, Kollnberger S, Kasahara M and
Maenaka K: Structure of MHC class I-like MILL2 reveals
heparan-sulfate binding and interdomain flexibility. Nat Commun.
9:43302018. View Article : Google Scholar : PubMed/NCBI
|