Introduction
Oral squamous cell carcinoma (OSCC), especially
gingival OSCC (GOSCC), frequently invades the mandible or maxilla;
jawbone destruction due to OSCC decreases the quality of life of
patients (1–4). OSCC invades subgingival connective
tissue, destroying the jawbone via vertical infiltration or
reaching the periodontal ligament (PDL) tissue via horizontal
infiltration. OSCC induces osteoclasts that produce bone resorption
in and around cancerous tissue, destroying the jawbone as they
progress deeper into the jaw (5).
The nature of OSCC bone destruction is hypothesized to be a result
of differential activation of osteoclasts by cancer cells, but the
mechanism is unknown. To date, several studies have reported
mechanisms of bone resorption in OSCC, which were relevant with the
size of OSCC and the expression level of osteoprotegerin (3,4).
However, the complete picture of bone resorption in OSCC is not yet
clear. Multiple animal models are available for the study of OSCC
bone resorption (6).
Histologically, bone destruction by OSCC is directly mediated by
osteoclasts rather than by cancer cells (7,8); In
bone invasion of OSCC, osteoclastogenesis is mediated by receptor
activator of nuclear factor κB (RANK), RANK ligand (RANKL) and
osteoprotegerin, which belong to the tumor necrosis factor (TNF)
family (9–12). OSCC cells promote expression of
RANKL in stromal cells (SCs) and osteoclasts near OSCC bone
invasion regions by secreting factors such as parathyroid
hormone-related peptide (PTHrP), interleukin (IL)-6, IL-11, TNF-α
and prostaglandin E2 (13–16). Furthermore, SCs in the tumor
microenvironment of OSCC secret IL-6 to promote RANKL production in
fibroblastic SCs (17). Therefore,
it is necessary to study the potential regulatory mechanisms of
bone invasion to improve treatment and prognosis of patients with
OSCC.
Solid tumors are composed of parenchymal and stromal
components, which are hypothesized to regulate cancer progression
by crosstalk with each other. In OSCC, an epithelial tumor, the
interaction between the tumor parenchyma and stroma also serves a
role in tumor progression (18,19).
However, previous studies on the tumor stroma have only reported
that the cancer parenchyma subordinately dominates the stroma and
contributes to the progression of cancerous tissue (18,19);
therefore, the mechanisms underlying the effect of the cancer
stroma on the parenchyma are not clear. Our previous studies
reported that the cancer stroma regulates bone invasion of human
OSCC cell lines, HSC-2 and HSC-3 cells (20–23).
Therefore, not only gingival cancer but also tongue cancer induces
bone invasion. The behavior of OSCC is hypothesized to depend on
its location; however, the mechanisms remain unknown (5). The tumor microenvironment of gingival
epithelium-derived OSCC is complex and changes depending on
direction of invasion (5). To the
best of our knowledge, however, the effect of gingival-(G-) and PDL
tissue-derived (P-)SCs in the GOSCC microenvironment on bone
invasion in OSCC of different origins have been poorly
investigated.
In the present study, the role of G-SCs and P-SCs in
bone invasion in OSCC and their potential regulatory mechanisms
were assessed. The differences in bone resorption capacity between
gingival connective tissue and the PDL with respect to osteoclasts,
MMPs and epithelial-mesenchymal transition (EMT) and RANKL and
PTHrP, which have been reported as associated with bone resorption
(22), were evaluated. The human
OSCC HSC-3 cell line was selected for use as a cancer cell model as
it is a moderately to poorly differentiated oral cancer cell line
with bone invasion ability and is widely used in bone invasion
studies (17,22). Similarly, the murine leukemia
RAW264.7 cell line was selected for use as an osteoclast cell model
based on its reported applications in bone invasion research
(22,24). Human dermal fibroblasts (HDFs) were
selected for use as a negative control for G-SCs and P-SCs as they
are normal fibroblasts that remain unaffected by the cancer cells
(23). The role and function of
G-SCs and P-SCs in bone invasion of HSC-3 cells both in
vitro and in vivo were evaluated. The present study will
provide a potential regulatory mechanism for bone invasion in
OSCC.
Materials and methods
Establishment of primary normal
SCs
The human OSCC HSC-3 cell line (cat. no. JCRB0623),
HDFs (cat. no. CC-2511) and the murine macrophage RAW264.7 cell
line (cat. no. RCB0535) were purchased from the Japanese Collection
of Research Bioresources Cell Bank, Lonza Group, Ltd. and RIKEN
BioResource Center respectively. G-SCs and P-SCs were isolated from
normal gingival tissue and the root surface of the tooth,
respectively, of a male patient (age, 50 years), which were stored
cells from the previous study (5).
HSC-3 and RAW264.7 cells, G-SCs, P-SCs and HDFs were cultured in
α-MEM (Thermo Fisher Scientific, Inc.) containing 10% fetal bovine
serum (Biowest) and 1% antimycotic antibiotic (Thermo Fisher
Scientific, Inc.) in a cell culture incubator at 37°C in a
humidified atmosphere of 5% CO2. The present study was
approved by the Ethics Committee of Okayama University (approval
no. 1703-042-001). Written informed consent was obtained from the
patient.
Tartrate-resistant acid phosphatase
(TRAP) staining of cells
When cell density reached 90%, RAW264.7 and HSC-3
cells were collected using EDTA (Thermo Fisher Scientific, Inc.)
and G-SCs, P-SCs and HDFs were collected using Accutase®
solution (Invitrogen; Thermo Fisher Scientific, Inc.) respectively.
RAW264.7, G-SCs/P-SCs/HDFs and HSC-3 were mixed at a ratio of
3:3:1. The combined cells were added to a 6-well plate containing
coverslips (22×22 mm) at a density of 3.5×105
cells/well. After 3 days in the cell culture incubator (37°C), the
slides were washed three times with Tris-buffered saline (TBS) and
fixed with 4% paraformaldehyde (PFA) for 15 min in room
temperature. The slides were stained using a TRAP staining kit
(cat. no. AK04F; Cosmo Bio Co., Ltd.) in a 37°C incubator overnight
after washing three times with TBS. The percentage of positive
osteoclast cells (defined as the percentage of TRAP-positive cells)
and proliferation of osteoclast cells were calculated from ten
images acquired using a BX51 bright-field microscope
(magnification, ×40; Olympus Corporation) and ImageJ software
(version 1.53; National Institutes of Health). Independent
experiments were performed three times.
Generation of HSC-3/SC xenograft mouse
model
Animal research was performed in accordance with the
guidelines and regulations (20)
of the Animal Care and Use Committee, Okayama University (approval
no. OKU-2022354). Anesthesia was performed as reported in
Laboratory Animal Anesthesia, 3rd edition (ketamine hydrochloride,
75 mg/kg body weight; medetomidine hydrochloride, 0.5 mg/kg body
weight) (25,26). Anesthetization of mice was
confirmed by assessing whether mice returned to a prone position
when placed on their backs. Following intraperitoneal anesthesia,
200 µl mixed cells (HSC-3, 1×106, 100 µl mixed with
G-SCs, P-SCs or HDFs, 3×106, 100 µl) were injected into
subcutaneous tissue on the top of the head of 20 healthy female
BALB-nu-nu mice (age, 4 weeks; mean weight, 15 g; Shimizu
Laboratory Supplies Co., Ltd.), as previously described (27). All mice were kept in the animal
center of Okayama University, Okayama, Japan (25°C; 50–60%
humidity; 12/12-h light/dark cycle) and had free access to food and
water. The health and behavior of mice were assessed by daily. The
animals were kept for 4 weeks after cancer cell transplantation. If
the transplanted tumor reached ≥10 mm in diameter or animals
exhibited weight loss ≥20% over a 3-day period or decreased food
and water intake or motility, the experiment was terminated
immediately and the animal was euthanized. The mice were divided
into four groups (n=5) as follows: i) HSC-3; ii) HSC-3 + G-SCs;
iii) HSC-3 + P-SCs and iv) HSC-3 + HDFs. The data from one mouse in
each group with poor tumor formation were removed based on the size
of tumor and the pathological diagnosis and the data from the
remaining four mice in each group were retained for analysis.
Hematoxylin and eosin (H&E)
staining
Mice were euthanized by excess inhalation of
isoflurane (concentration >5%) after 4 weeks. Cardiac arrest was
confirmed by pulse palpation prior to cervical dislocation. The
complete heads of the mice were collected, fixed with 4% PFA for 12
h in room temperature and soaked in 10% EDTA for 4 weeks at 4°C.
The samples were processed using paraffin wax and cut into 5 µm
sections, which were used for H&E staining (Carrazi'
hematoxylin, Muto Pure Chemicals Co., Ltd, 5 min and room
temperature; eosin, Muto Pure Chemicals Co., Ltd, 7 min and room
temperature) and imaged using a BX51 bright-field microscope
(magnification, ×40; Olympus Corporation).
TRAP staining of tissue
The 5-µm sections from each group were used for TRAP
staining using a TRAP staining kit (cat. no. AK04F; Cosmo Bio Co.,
Ltd.) according to the aforementioned method. A total of five
images of bone invasion regions were acquired using the
bright-field microscope (magnification, ×40) for TRAP-positive cell
counts using ImageJ software.
Immunohistochemistry (IHC)
The 5 µm thick sections from each group were
subjected to antigen retrieval [microwave 1 or 8 min (350 W); 0.01
M tri-sodium citrate buffer (pH 6) and 0.01 M Dako Target Retrieval
Solution (pH 9; cat. no. S2367; Agilent Technologies, Inc.)],
blocked with 10% normal serum (Vector Laboratories) for 20 min at
room temperature. Sections were incubated with primary antibodies
as follows: Mouse anti-MMP-9 (1:20; cat. no. F-69; Kyowa Pharma
Chemical Co., Ltd.), anti-membrane-type 1 MMP (MT1-MMP; 1:20; cat.
no. F-86; Kyowa Pharma Chemical Co., Ltd.), anti-Snail + SLUG
(1:200; cat. no. ab180714; Abcam), anti-RANKL (1:100; cat. no.
bs-0747R; BIOSS) and anti-PTHrP (1:100, cat. no. 10817-1-AP;
ProteinTech Group, Inc.) overnight at 4°C. The secondary antibodies
were avidin-biotin complexes from mouse (cat. no. PK-6102) and
rabbit (cat. no. PK-6101) ABC kit; Blocking serum (normal serum,
diluted with TBS, at 1:75); biotinylated secondary antibody
(diluted with normal serum, at 1:200); reagent A (avidin, ABC
Elite, vector Laboratories, Inc.) and reagent B (biotinylated HRP,
ABC Elite, Vector Laboratories, Inc., diluted with TBS, at 1:55)
were added and incubated for 1 h at room temperature after washing
with TBS three times and visualized using Histofine DAB substrate
(Nichirei Biosciences, Inc.) at room temperature. A total of ten
images (magnification, ×40) of each mouse was acquired using a
bright-field microscope to assess MMP-9, MT1-MMP, Snail, PTHrP and
RANKL protein expression levels using IHC score by ImageJ software
(version 1.53; National Institutes of Health). IHC scores were
calculated as follows: IHC score=positive cell percentage score ×
intensity score. The positive cell percentage score was defined as
follows: 0, <1; 1, 1–24; 2, 25–49; 3, 50–74 and 4, 75–100%. The
intensity score was defined as follows: 0, negative; 1, weak (light
yellow); 2, moderate (brown) and 3, strong staining (dark brown)
(28). The staining results were
assayed by two independent pathologists and and calculated as the
mean value if the results were different between the two
pathologists.
Bioinformatics analysis of microarray
data
RNA was extracted from the cultured G-SCs and P-SCs
by miRNeasy micro kit (Qiagen, Inc.) and quantified using NanoDrop
One (NanoDrop; Thermo Fisher Scientific, Inc) and
BioAnalyzerRNA6000 Nano (Agilent Technologies, In). cDNA synthesis,
cRNA labeling and amplification were conducted using the Low Input
Quick Amp Labeling Kit (Agilent Technologies, Inc.), and the
purification of labeled cRNAs was conducted using RNeasy mini spin
column (Qiagen, Inc.). Finally, the microarray (SurePrint G3 Human
8×60k ver.3.0; Agilent Technologies, Inc.) was scanned using a
G4900DA Microarray Scanner (Agilent Technologies, Inc.). The
differentially expressed genes (DEGs) in P-SCs compared with those
in G-SCs were analyzed using GeneSpring GX14.9.1 (Agilent) with a
cut-off value of LogFC >1
(ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE174595). Gene Ontology
(GO) enrichment analysis was performed to define the biological
process of upregulated (up)-DEGs in P-SCs using Cytoscape 3.7.2
(cytoscape.org/) with a cut-off value of adjusted P<0.05. The
protein-protein interaction (PPI) network was produced to identify
the hub genes using STRING (string-db.org/) and cytoHubba (version
0.1, cytoscape.org/apps/cytohubba) plugin for Cytoscape 3.7.2 with
the cut-off value of combined score >0.4.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 9 (GraphPad Software, Inc.). The cell experiments were
repeated three times and the animal experiments were repeated in
five independent mice. One-way ANOVA followed by Tukey's post hoc
test was used for parametric data analysis (mean ± SD).
Kruskal-Wallis followed by Dunn's test was used to analyze
non-parametric data (median and interquartile range). P<0.05 was
considered to indicate a statistically significant difference.
Results
G-SCs promote and P-SCs inhibit bone
resorption
As both GOSCC and other origins of OSCC induce bone
invasion or metastasis, the human OSCC HSC-3 cell line was selected
as a model to assess the association between G-SCs/P-SCs and
different origins of OSCC. The effects of G-SCs, P-SCs and HDFs on
bone resorption of OSCC cells in vivo were assessed using
H&E staining. The length of bone resorption in the HSC-3 +
G-SCs group was markedly higher than that in the HSC-3, HSC-3 +
HDFs groups and HSC-3+P-SCs groups. In addition, a notable
keratinized area was observed in the HSC-3 + P-SCs group (Fig. 1A and B). Clinically, there are
three types of bone invasion: Erosive, infiltrative and mixed. In
erosive type of bone invasion, there is stroma between cancer cells
and the bone. In infiltrative bone invasion, the cancer cells
directly touch bone tissue without any stroma between them. Mixed
bone invasion involves both erosive and infiltrative bone invasion
(7). The remaining bone tissue of
the HSC-3 and HSC-3 + G-SCs groups demonstrated erosive areas,
which were bone resorption areas caused by cancer stroma and
infiltrative areas that were directly resorbed by cancer cells. The
HSC-3 + HDFs group also demonstrated both erosive and infiltrative
areas; however, the bone resorption area was primarily within the
erosive area. Moreover, the HSC-3 + P-SCs group demonstrated only
erosive areas (Fig. 1C). Since
these cell line-derived xenograft (CDX) models contained erosive
and infiltrative areas, they demonstrated histological findings
similar to those of actual human OSCC (7); therefore, these CDX models were
appropriate to mimic bone resorption in OSCC. Based on bone
resorption length and invasion type, G-SCs exerted a promoting
effect on bone invasion of OSCC, whereas P-SCs exerted an
inhibitory effect and HDFs exerted a minimal effect.
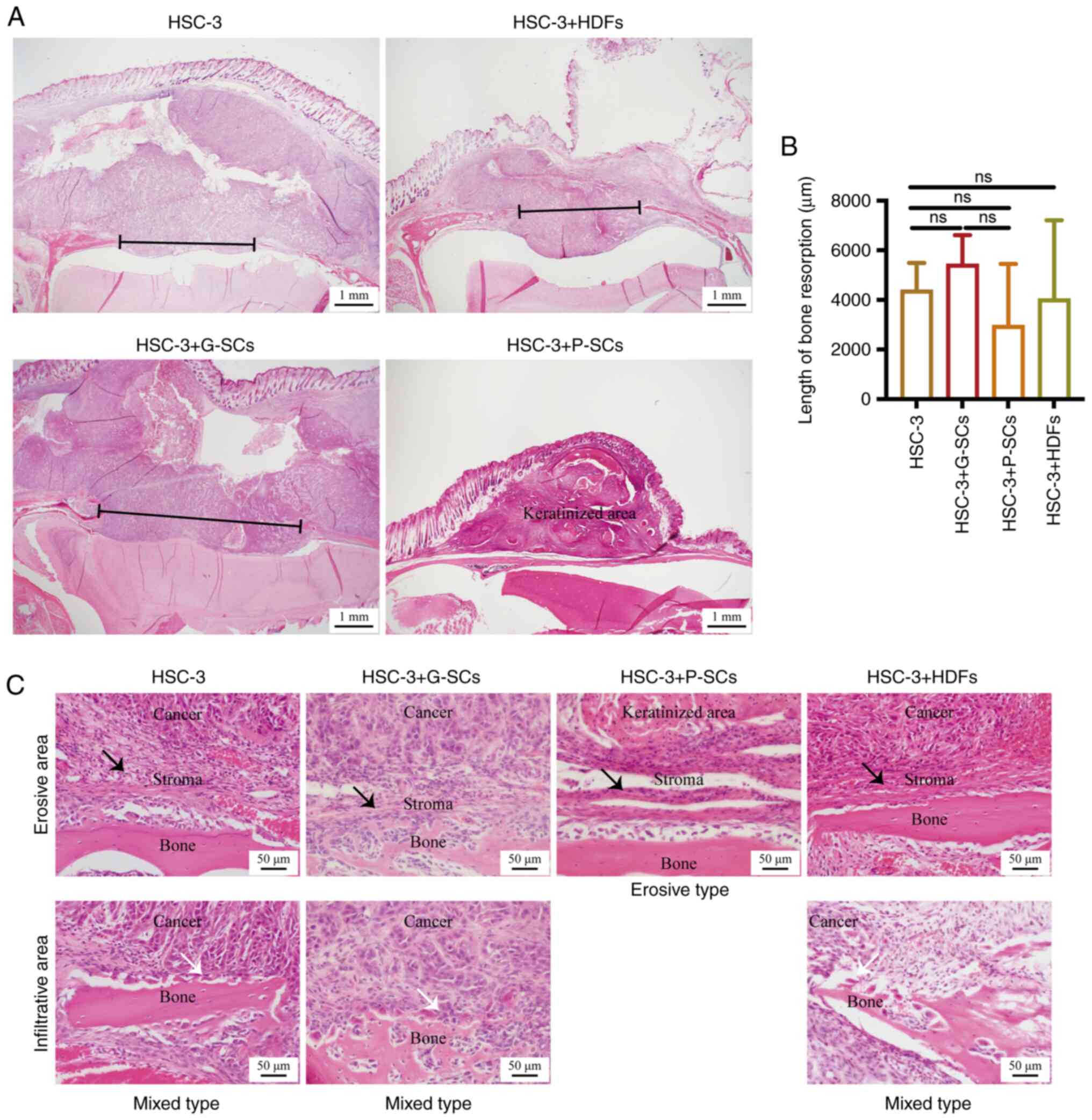 | Figure 1.H&E staining to assess the
effects of G-SCs, P-SCs and HDFs on bone invasion in OSCC in
vivo. (A) H&E staining was used to assess bone mass. Black
line, bone resorption. (B) Quantification of length of bone
resorption. Data are presented as mean ± SD (n=4). Statistical
analysis was performed using one-way ANOVA followed by Tukey's post
hoc test. (C) H&E staining was used to assess the type of OSCC
bone invasion. Black arrow, erosive area; white arrow, infiltrative
area. H&E, hematoxylin and eosin; G-SCs, gingival
tissue-derived stromal cells; P-SCs, periodontal ligament
tissue-derived stromal cells; HDFs, human dermal fibroblasts; OSCC,
oral squamous cell carcinoma; ns, not significant. |
G-SCs exert a more prominent promoting
effect on invasion and EMT of HSC-3 in the erosive area of OSCC
bone invasion region than P-SCs in vivo
IHC staining was used to assess MMP-9 and MT1-MMP
protein expression levels to evaluate the effects of G-SCs, P-SCs
and HDFs on invasion of HSC-3 cells in the OSCC bone invasion
region (Fig. 2A and C). IHC scores
of MMP-9 and MT1-MMP were significantly higher in the HSC-3 + G-SCs
group compared with the HSC-3 and HSC-3 + P-SCs groups and there
was little difference between the HSC-3, HSC-3 + P-SCs and HSC-3 +
HDFs groups (Fig. 2B and D).
Furthermore, osteoclasts on the bone surface were both MMP-9- and
MT1-MMP-positive and the intensity of MMP-9 and MT1-MMP in the
HSC-3 + G-SCs group was markedly higher than that in the other
groups. IHC staining was used to assess Snail protein expression
levels to evaluate the effects of G-SCs, P-SCs and HDFs on EMT of
HSC-3 cells in the erosive area of OSCC bone invasion region
(Fig. 2E). IHC score in the HSC-3
+ G-SCs group was significantly higher compared with HSC-3 and
HSC-3 + P-SCs groups but there was little difference between other
groups (Fig. 2F). These results
demonstrated that G-SCs promoted invasion and EMT of HSC-3 cells in
the erosive area of the OSCC bone invasion region, whereas P-SCs
and HDFs exerted a minimal effect.
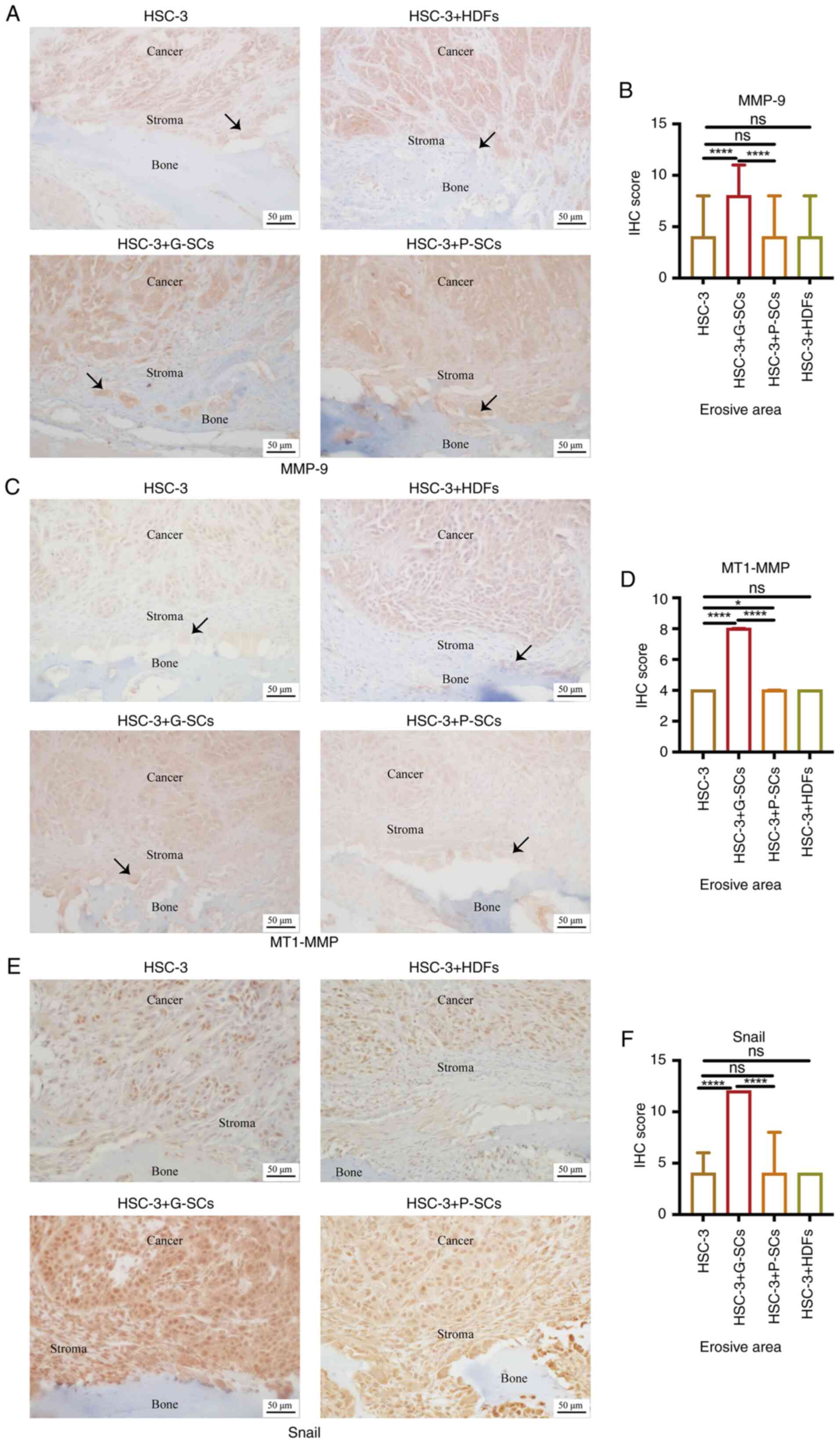 | Figure 2.Effect of G-SCs, P-SCs and HDFs on
invasion and EMT of HSC-3 cells in OSCC bone invasion region.
Immunohistochemical staining was used to (A) assess and (B)
quantify effect of G-SCs, P-SCs and HDFs on protein expression
levels of MMP9. Immunohistochemical staining was used to (C) assess
and (D) quantify effect of G-SCs, P-SCs and HDFs on protein
expression levels of MT1-MMP. Immunohistochemical staining was used
to (E) assess and (F) quantify effect of G-SCs, P-SCs and HDFs on
protein expression levels of Snail. Black arrow, osteoclast. Data
are presented as median and interquartile range (n=4). Statistical
analysis was performed using Kruskal-Wallis followed by Dunn's
test. *P<0.05 and ****P<0.0001. G-SCs, gingival
tissue-derived stromal cells; P-SCs, periodontal ligament
tissue-derived stromal cells; HDFs, human dermal fibroblasts; OSCC,
oral squamous cell carcinoma; ns, not significant; EMT,
epithelial-mesenchymal transition; MMP, matrix metalloproteinase;
MT1-MMP, membrane type 1 MMP. |
Crosstalk between G-SCs and HSC-3
cells induces osteoclastogenesis more effectively than crosstalk
between P-SCs and HSC-3 cells
TRAP staining was used to test the effects of G-SCs,
P-SCs and HDFs on activation, differentiation and proliferationn of
osteoclasts in vitro. There were only small round
osteoclasts with a single nucleus in the RAW264.7 group as well as
RAW264.7 cells with G-SCs and P-SCs (Fig. 3A). The percentage of TRAP-positive
osteoclasts in the RAW264.7 group was significantly higher than in
the RAW264.7 + HDFs, RAW264.7 + G-SCs and RAW264.7 + P-SCs groups.
There was no significant difference between the RAW264.7 + G-SCs
and RAW264.7 + P-SCs groups (Fig.
3C). The total number of osteoclasts was counted to assess the
effects of G-SCs, P-SCs and HDFs on osteoclast proliferation in
vitro. The total number of osteoclasts in the RAW264.7 + G-SCs
and RAW264.7 + P-SCs groups was markedly higher than that in the
RAW264.7 + HDFs group and significantly higher than that in the
HSC-3 group. There was no significant difference between RAW264.7 +
G-SCs and RAW264.7 + P-SCs groups (Fig. 3E). TRAP staining was used to assess
the effects of G-SCs, P-SCs and HDFs on activation and cell
proliferation of osteoclasts after crosstalk with HSC-3 cells in
vitro. A previous study reported that differently shaped
osteoclasts exert different effects on bone resorption; therefore,
the shapes of osteoclasts in different groups were evaluated
(29). The RAW264.7 + HSC-3,
RAW264.7 + HSC-3 + P-SCs and RAW264.7 + HSC-3 + HDFs groups
primarily presented round-shaped osteoclasts whereas the RAW264.7 +
HSC-3 + G-SCs group presented round and triangular-shaped
osteoclasts (Fig. 3B). The
percentage of TRAP-positive osteoclasts in the RAW264.7 + HSC-3 +
G-SCs group was significantly higher than that in the RAW264.7 +
HSC-3 and RAW264.7 + HSC-3 + P-SCs groups and markedly higher than
that in the RAW264.7 + HSC-3 + HDFs group. There was no significant
difference between the RAW264.7 + HSC-3 and RAW264.7 + HSC-3 +
P-SCs groups (Fig. 3D). The total
number of osteoclasts in the RAW264.7 + HSC-3 + P-SCs group was
significantly higher than that in the RAW264.7 + HSC-3 and RAW264.7
+ HSC-3 + G-SCs groups and was markedly higher than that in the
RAW264.7 + HSC-3 + HDFs group (Fig.
3F). TRAP-positive cells in the RAW264.7 + HSC-3, RAW264.7 +
HSC-3 + G-SCs, RAW264.7 + HSC-3 + P-SCs, and RAW264.7 + HSC-3 +
HDFs groups were markedly larger than those in RAW264.7, RAW264.7 +
G-SCs, RAW264.7 + P-SCs and RAW264.7 + HDFs groups. TRAP staining
was used to assess the proportion of activated osteoclasts on the
bone surface to evaluate the effects of G-SCs, P-SCs and HDFs on
osteoclast activation following crosstalk with HSC-3 in
vivo. The HSC-3 group presented mainly round and ellipse-shaped
osteoclasts on the bone surface. HSC-3 + P-SCs primarily presented
ellipse-shaped osteoclasts and HSC-3 + HDFs mainly presented round
and ellipse-shaped osteoclasts. The HSC-3 + G-SCs group presented
round, ellipse and triangular osteoclasts (Fig. 3G). There was little difference in
size of osteoclasts between groups (Fig. 3G). The number of TRAP-positive
osteoclasts on the bone surface in the HSC-3 + G-SCs group was the
highest, and there was no marked difference between the HSC-3,
HSC-3 + P-SCs and HSC-3 + HDFs groups (Fig. 3H). Therefore, these data suggested
that G-SCs, P-SCs and HDFs promoted cell proliferation and
inhibited activation of osteoclasts and there was little difference
between the effects of G-SCs and P-SCs. Following crosstalk with
HSC-3 cells in vitro, G-SCs promoted activation and
inhibited proliferation of osteoclasts, whereas P-SCs promoted
proliferation of osteoclasts and exerted minimal effect on the
activation of osteoclasts. HDFs exerted a minimal effect on
activation and proliferation of osteoclasts following crosstalk
with HSC-3 cells. These data suggested that crosstalk between HSC-3
cells and normal stroma may have increased size of activated
osteoclasts in vitro. Moreover, the crosstalk between G-SCs
and HSC-3 promoted activation of osteoclasts on the bone surface in
the erosive area of the OSCC bone invasion region by regulating the
shape and number of osteoclasts, whereas P-SCs and HDFs exerted a
minimal effect in vivo.
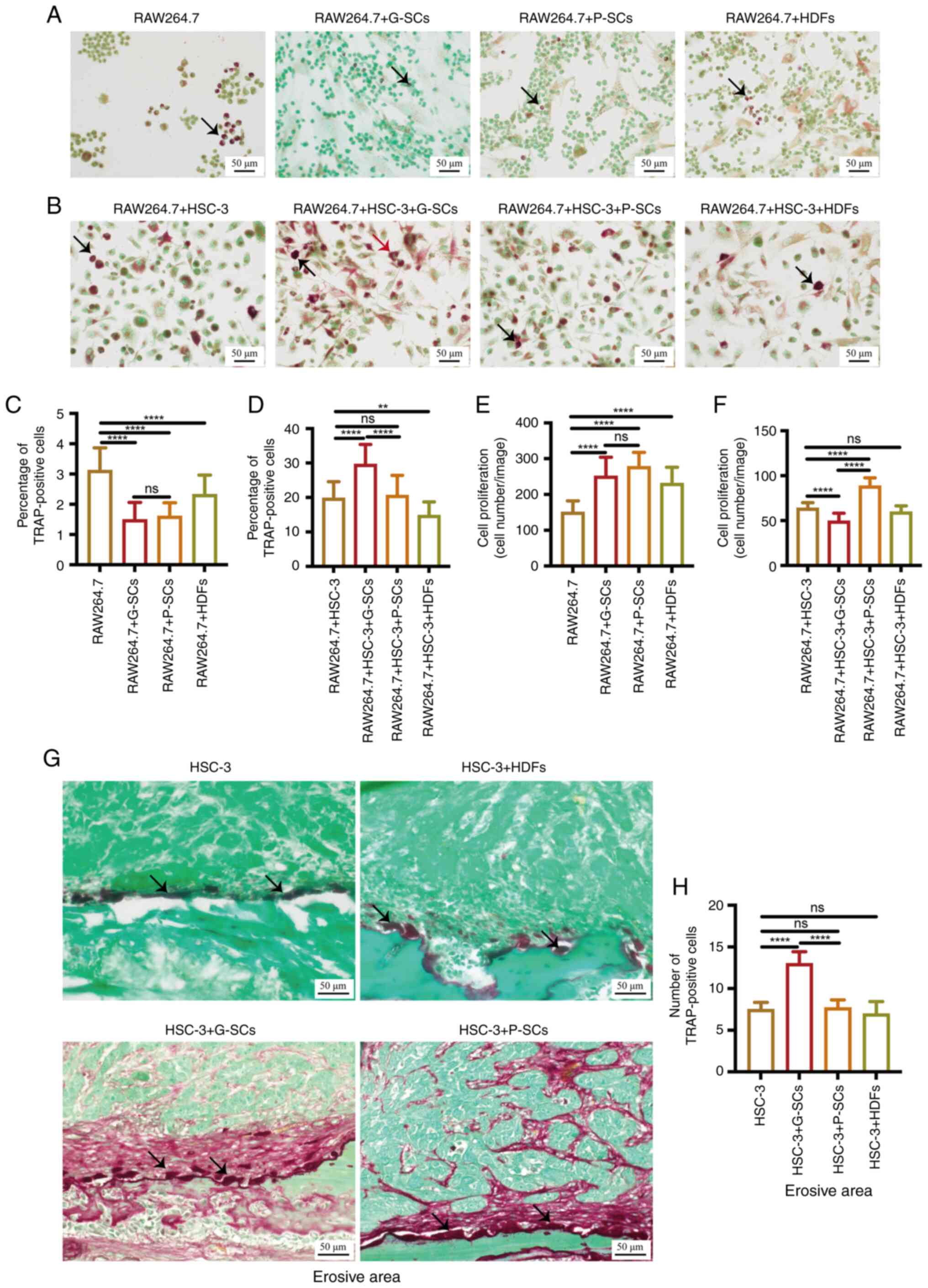 | Figure 3.Effects of G-SCs, P-SCs and HDFs on
activation of osteoclasts following crosstalk with HSC-3 cells both
in vitro and in vivo. (A) TRAP staining was used to
assess the effects of G-SCs, P-SCs and HDFs independently on
activation and cell proliferation of osteoclasts in vitro.
(B) TRAP staining was used to assess the effects of G-SCs, P-SCs
and HDFs on activation and cell proliferation of osteoclasts
following crosstalk with HSC-3 in vitro. Black arrow, round
osteoclast; red arrow, triangular osteoclast. (C) Quantification of
the percentage of TRAP-positive osteoclasts in RAW264.7, RAW264.7 +
G-SCs, RAW264.7 + P-SCs and RAW264.7 + HDFs groups. (D)
Quantification of the percentage of TRAP-positive osteoclasts in
RAW264.7 + HSC-3, RAW264.7 + HSC-3 + G-SCs, RAW264.7 + HSC-3 +
P-SCs and RAW264.7 + HSC-3 + HDFs groups. (E) Quantification of
osteoclasts proliferation in RAW264.7, RAW264.7 + G-SCs, RAW264.7 +
P-SCs, and RAW264.7 + HDFs groups. (F) Quantification of
osteoclasts proliferation in RAW264.7 + HSC-3, RAW264.7 + HSC-3 +
G-SCs, RAW264.7 + HSC-3 + P-SCs, and RAW264.7 + HSC-3 + HDFs
groups. Data are presented as mean ± SD (n=3). (G) TRAP staining
was used to assess the effects of G-SCs, P-SCs and HDFs on
activation of osteoclasts on the bone surface following crosstalk
with HSC-3 in vivo. Black arrow, activated osteoclast. (H)
Quantification of TRAP-positive osteoclasts. Data are presented as
mean ± SD (n=4). Statistical analysis was performed using one-way
ANOVA followed by Tukey's post hoc test. **P<0.01 and
****P<0.0001. G-SCs, gingival tissue-derived stromal cells;
P-SCs, periodontal ligament tissue-derived stromal cells; HDFs,
human dermal fibroblasts; TRAP, tartrate-resistant acid
phosphatase; ns, not significant. |
G-SCs exert a more prominent promoting
effect on RANKL and PTHrP protein expression levels in HSC-3 cells
in the erosive area of OSCC bone invasion region than P-SCs
As RANKL regulates osteoclast activation (13), IHC staining was used to assess the
effects of G-SCs, P-SCs and HDFs on RANKL protein expression levels
in HSC-3 cells in the OSCC bone invasion region (Fig. 4A). IHC score in the HSC-3 + G-SCs
group was markedly higher than that in the HSC-3 + HDFs group and
significantly higher than in the HSC-3 and HSC-3 + P-SCs groups.
There was no significant difference between the HSC-3 and HSC-3 +
HDFs groups (Fig. 4B). As PTHrP
not only recruits osteoclasts from peripheral blood but also
regulates RANKL expression (30),
IHC staining was used to assess the effects of G-SCs, P-SCs and
HDFs on PTHrP protein expression levels in HSC-3 cells in the
erosive area of OSCC bone invasion region (Fig. 4C). IHC score in the HSC-3 + G-SCs
group was higher than that in the HSC-3 + P-SCs group and markedly
higher than that in the HSC-3 and HSC-3 + HDFs groups. There was no
significant difference between HSC-3 and HSC-3 + HDFs groups
(Fig. 4D). These data demonstrated
that both G-SCs and P-SCs promoted RANKL and PTHrP expression in
HSC-3 cells in the OSCC bone invasion region. G-SCs exerted a
significant promoting effect compared with P-SCs, whereas HDFs
exerted no significant effect.
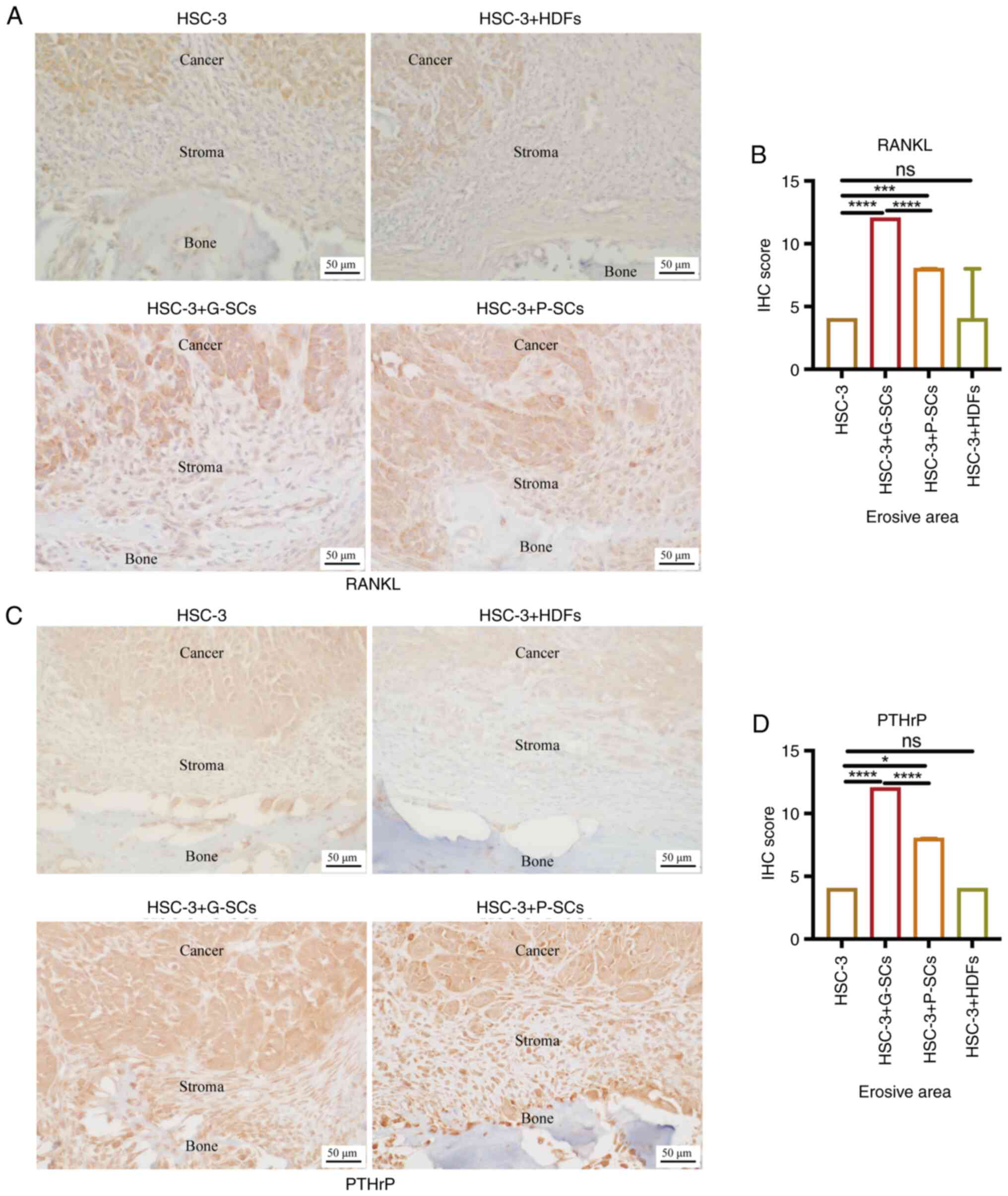 | Figure 4.Effects of G-SCs, P-SCs and HDFs on
the protein expression levels of RANKL and PTHrP in HSC-3 cells in
the erosive area of OSCC bone invasion region. Immunohistochemical
staining was used to (A) assess and (B) quantify effect of G-SCs,
P-SCs and HDFs on protein expression levels of RANKL.
Immunohistochemical staining was used to (C) assess and (D)
quantify effect of G-SCs, P-SCs and HDFs on protein expression
levels PTHrP in HSC-3 cells in the erosive area of OSCC bone
invasion region. Data are presented as median and interquartile
range (n=4). Statistical analysis was performed by Kruskal-Wallis
followed by Dunn's test. *P<0.05, ***P<0.001 and
****P<0.0001. IHC, immunohistochemistry; G-SCs, gingival
tissue-derived stromal cells; P-SC, periodontal ligament
tissue-derived stromal cell; HDFs, human dermal fibroblasts; OSCC,
oral squamous cell carcinoma; ns, not significant. |
Cylin-dependent kinase 1 (CDK1),
insulin (INS), aurora kinase A (AURKA), cyclin B1 (CCNB1) and DNA
topoisomerase IIα (TOP2A) are potential genes underlying the
differential effects of G-SCs and P-SCs on OSCC bone invasion
following crosstalk with HSC-3 cells in vivo
The differential effects between G-SCs and P-SCs
were evaluated using microarray analysis of DEGs. The biological
processes of up-DEGs in P-SCs were analyzed by GO enrichment
analysis, which indicated that these up-DEGs were primarily
associated with biological processes such as ‘cell
differentiation’, ‘cell migration’, ‘blood vessel development’ and
‘skeletal system development’, which are associated with bone
invasion (Fig. 5A). The degree of
differentiation leads to differential effects of G-SCs and P-SCs on
OSCC bone invasion following crosstalk with HSC-3 cells in
vivo and ‘cell differentiation’ was the most relevant
biological process. Furthermore, hub genes in ‘cell
differentiation’ were identified using the PPI network;
CDK1, nucleolar and spindle associated protein 1
(NUSAP1), centromere protein F (CENPF), assembly
factor for spindle microtubules (ASPM), AURKA, INS,
CCNB1, TOP2A, anillin actin binding protein (ANLN) and
Rac GTPase activating protein 1 (RACGAP1) were the top 10
hub genes involved in cell differentiation (Fig. 5B). As demonstrated by coloration of
the hub genes, CDK1, INS, AURKA, CCNB1 and TOP2A were
more relevant to ‘cell differentiation’, which suggested that they
may underlie the different effects of G-SCs and P-SCs on OSCC bone
invasion following crosstalk with HSC-3 cells.
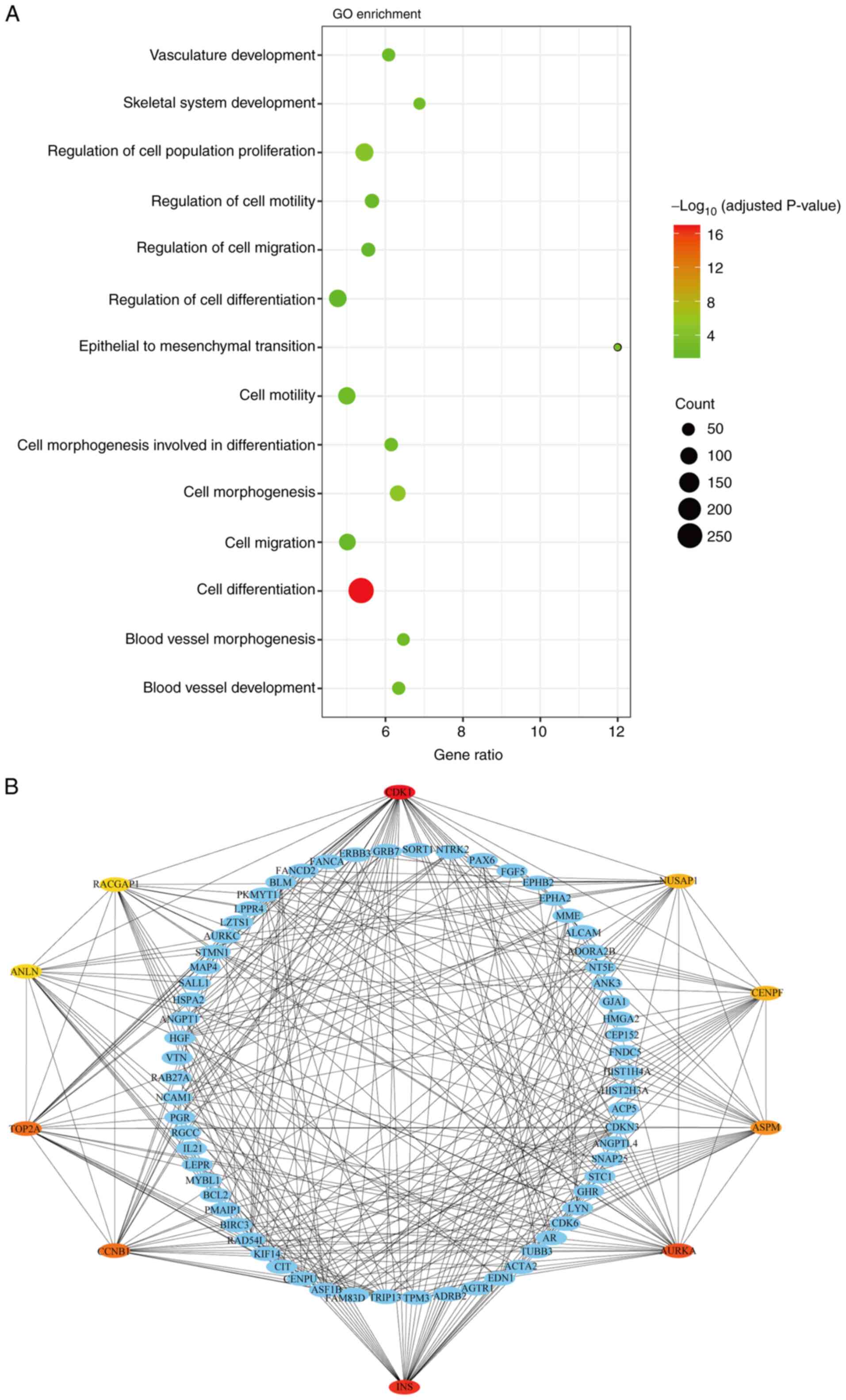 | Figure 5.Identification of potential genes
underlying the differential effects of G-SCs and P-SCs on OSCC bone
invasion following crosstalk with HSC-3 cells in vivo. (A)
Biological processes associated with upregulated differentially
expressed genes in P-SCs was assess using Gene Ontology enrichment
analysis (only presenting the potential biological process
underlying the differential effects of G-SCs and P-SCs on OSCC bone
invasion following crosstalk with HSC-3 in vivo). (B)
Protein-protein interaction network was used to identify the hub
genes in biological process of ‘cell differentiation’. G-SC,
gingival tissue-derived stromal cells; P-SCs, periodontal ligament
tissue-derived stromal cells; OSCC, oral squamous cell carcinoma;
CDK1, cylin-dependent kinase 1; INS, insulin; AURKA, aurora kinase
A; CCNB1, cyclin B1; TOP2A, DNA topoisomerase IIα; NUSAP1,
nucleolar and spindle-associated protein 1; CENPF, centromere
protein F; ASPM, assembly factor for spindle microtubules; ANLN,
anillin actin binding protein; RACGAP1, Rac GTPase activating
protein 1. |
Discussion
The results of the present study suggested that both
G-SCs and P-SCs exerted differential effects on the bone invasion
of OSCC, which may cause by their differential effects on the
differentiation degree of OSCC. CDK1, INS, AURKA, CCNB1 and
TOP2A have much potential to underlie this differential
effect on the differentiation degree of OSCC. However, the effect
of G-SCs and P-SCs on the bone invasion of OSCC need to be examined
on the other types of oral cancer cell lines and the detail
function of different size and shape of osteoclasts in the OSCC
bone invasion need to be further studied. Finally, the detail role
and function of CDK1, INS, AURKA, CCNB1 and TOP2A in
G-SCs and P-SCs on bone invasion of OSCC need to be further
investigated.
Epithelial tumors influence neighboring normal
stroma and utilize it for tumor tissue extension (5). However, the effects of different
properties of the normal stroma on bone resorption have not yet
been fully elucidated. Our previous studies assessed the effect of
different subtypes of cancer stroma on bone invasion of both HSC-2
and HSC-3 cells (20–22). Furthermore, our previous study also
assayed the effects of normal stroma G-SCs and P-SCs on bone
invasion of OSCC but did not provide a detailed description of the
bone invasion type and potential regulatory mechanism (5). In the present study, G-SCs and P-SCs
were mixed with HSC-3 in a 3:1 ratio and were used to generate an
animal model. Bone invasion of OSCC is classified into three types:
Erosive, infiltrative and mixed (31,32).
A previous study reported that the interaction between
cancer-associated fibroblasts and cancer cells promoted bone
invasion and prognosis of OSCC (27). The present study demonstrated that
bone invasion of the HSC-3 + G-SCs group was of the mixed type,
whereas the bone invasion of the HSC-3 + P-SCs group was erosive.
This suggested that P-SCs induced change of bone invasion of HSC-3
cells from a mixed to erosive type. Based on degree of bone
resorption, G-SCs promoted bone invasion, whereas P-SCs exerted an
inhibitory effect. Furthermore, keratinized cancer cells were
observed in the bone invasion region of the HSC-3 + P-SCs group.
This type of cancer cell has low invasion and migration ability,
which makes it difficult for them to approach the bone surface by
EMT and results in bone invasion type changing from the mixed to
erosive type (20). Moreover, HDFs
in the present study were selected as controls; there was no
significant difference between the HSC-3 and HSC-3 + HDFs groups.
These results demonstrated that HDFs exerted a minimal effect on
bone invasion of HSC-3 cells in vivo.
Bone invasion in OSCC is mediated by osteoclasts
(33). A previous study generated
animal models using the human tongue SCC SCC-25 cell line and
reported different osteoclast shapes on the bone resorption surface
(29). Our previous study
demonstrated that the HSC-3 + squamous cell carcinoma-associated
stromal cells (SCC-SCs) group contained triangular and round
osteoclasts, whereas the HSC-3 + verrucous SCC-SCs group presented
round, ellipse and triangular osteoclasts. In the present study,
there were only round and ellipse-shaped osteoclasts in HSC-3 and
HSC-3 + HDFs groups. Therefore, triangular osteoclasts demonstrated
the best bone invasion ability, which indirectly confirmed the
results of the aforementioned study (29). In the present study, RAW264.7 +
G-SCs and RAW264.7 + P-SCs groups primarily presented round
osteoclasts. Following crosstalk with HSC-3 cells in vitro,
only the RAW264.7 + HSC-3 + G-SCs group demonstrated triangular
osteoclasts and only round osteoclasts were observed in the other
groups. Triangular osteoclasts were also demonstrated in the HSC-3
+ G-SCs group in vivo and only round or ellipse-shaped
osteoclasts were observed in the other groups. The crosstalk
between G-SCs and HSC-3 cells promoted activation of osteoclasts
and inhibited cell proliferation, whereas the crosstalk between
P-SCs and HSC-3 cells promoted proliferation of osteoclasts and
exerted a minimal effect on activation of osteoclasts. Crosstalk
between HSC-3 cells and normal stroma exerted a more prominent
promoting effect on the size of activated osteoclasts than the
RAW264.7, RAW264.7 + G-SCs, RAW264.7 + P-SCs and RAW264.7 + HDFs
groups in vitro. To the best of our knowledge, the present
study is the first to demonstrate that normal stroma surrounding
OSCC and experimental cancer stroma derived from OSCC induce
similar responses in osteoblasts, which suggested that normal
stroma may also serve a functional role in bone remodeling in OSCC.
In the animal experiments, the number of activated osteoclast cells
was significantly higher in the HSC-3 + G-SCs group and there was
little difference between other groups. These data suggested that
G-SCs promoted bone invasion of OSCC by regulating the shape,
number and size of osteoclasts on the bone surface, whereas P-SCs
exerted a minimal effect on the shape and number of osteoclasts on
the bone surface. It has been previously reported that exposure to
RANKL facilitates osteoclastogenesis in the murine macrophage cell
line RAW264.7 (34). However, the
association between OSCC bone invasion and osteoclasts from
peripheral blood requires further investigation.
The bone invasion of OSCC is divided into initial,
bone resorption and final phase (30). MMPs and EMT serve a key role in the
bone invasion of OSCC. Cancer cells disrupt extracellular matrix by
secreting MMPs and approach the bone surface via EMT (35–37).
Our previous study suggested that cancer stroma promotes expression
of MMP-9, MT1-MMP and Snail in HSC-3 cells in the OSCC bone
invasion region (22). In the
present study, G-SCs promoted MMP-9, MT1-MMP and Snail protein
expression levels in HSC-3 cells in OSCC bone invasion regions,
whereas P-SCs exerted a minimal effect. These data suggested that
G-SCs caused promote cancer cells to approach the bone surface by
secreting MMP-9 and MT1-MMP and, promoting EMT, whereas P-SCs
exerted a minimal effect.
The bone resorption phase is mediated by activated
osteoclasts (30). Osteoclast
activation is regulated by colony-stimulating factor 1 and RANKL
(38). A previous study reported
that both oral cancer cells and cancer-associated fibroblasts
promote osteoclastogenesis by enhancing RANKL expression (39). Our previous study demonstrated that
cancer stroma promotes RANKL expression in HSC-3 cells in OSCC bone
invasion regions (22). In the
present study, G-SCs and P-SCs promoted RANKL expression in HSC-3
cells in the OSCC bone invasion region, with G-SCs exerting a
significantly greater effect than that exerted by P-SCs. PTHrP
mediates RANKL expression (38)
and recruitment of osteoclasts from circulating blood (30). In the present study, both G-SCs and
P-SCs significantly promoted PTHrP expression in HSC-3 cells in
OSCC bone invasion regions compared with HSC-3 cells, with G-SCs
exerting a significantly greater effect than P-SCs. P-SCs exerted a
minimal effect on the initial phase of the bone invasion based on
the MMP9, MT1-MMP, and Snail expression; however, they had a
promoting effect on bone resorption phase of bone invasion of OSCC
based on the RANKL and PTHrP expression, which suggested that
little bone invasion occurred in the HSC-3 + P-SCs group. These
data suggested that P-SCs exerted an inhibitory effect on bone
invasion of OSCC cells. Previous studies have also reported that
PDL cells regulate bone resorption and formation by secreting
cytokines (40,41). Here, however, P-SCs exerted an
inhibitory effect on bone resorption in oral cancer, rather than a
promoting effect, as observed in conditions such as inflammation
and diabetes (42,43). Therefore, it can be hypothesized
that bone invasion in OSCC was primarily caused by cancer rather
than PDL cells and P-SCs indirectly affected OSCC bone invasion by
interacting with cancer cells instead of directly regulating bone
resorption.
The effects of G-SCs and P-SCs on bone invasion in
OSCC were caused by effects on the degree of differentiation of
HSC-3 cells in the OSCC bone invasion regions. P-SCs promoted
differentiation of HSC-3 cells in the bone invasion regions,
whereas G-SCs exerted an inhibitory effect. Therefore, G-SCs were
used as the control group to assess DEGs in P-SCs using microarray
analysis. The biological processes of up-DEGs in P-SCs were
evaluated using GO enrichment analysis. The biological processes
associated with differentiation, migration, vessel formation and
bone development are considered to affect bone invasion in OSCC
(22,23,30).
Therefore, the biological processes were assessed and the most
up-DEGs were enriched in ‘cell differentiation’. PPI was used to
identify hub genes involved in ‘cell differentiation’, which
indicated that CDK1, NUSAP1, CENPF, ASPM, AURKA, INS, CCNB1,
TOP2A, ANLN and RACGAP1 were the top 10 hub genes.
CDK1, INS, AURKA, CCNB1 and TOP2A were most relevant
to cell differentiation.
In conclusion, G-SCs promoted bone invasion in OSCC,
whereas P-SCs exerted an inhibitory effect. The differential
effects of G-SCs and P-SCs on bone invasion in OSCC were assessed
using differentiation of HSC-3 cells in OSCC bone invasion regions.
CDK1, INS, AURKA, CCNB1 and TOP2A were potential
genes that underlie the differential effects on differentiation of
HSC-3 cells in OSCC bone invasion regions. To the best of our
knowledge, the present study is the first to provide in vivo
and in vitro evidence that normal stromal characteristics
serve a key role in bone invasion in OSCC. The present data
suggested that the normal stroma may be promising as a novel
therapeutic target for bone invasion. These findings may provide a
potential regulatory mechanism for normal stroma regulation of bone
invasion of OSCC.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Japan Society for Promotion
of Science KAKENHI Grants-in-Aid for Scientific Research (grant
nos. JP20K10094, JP21K10043, JP21K17089, JP19K19159, JP20H03888 and
JP22K10170).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The microarray data generated in the present study can be
found in the Gene Expression Omnibus under accession number
GSE174595 or at
ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE174595.
Authors' contributions
QS designed the study. QS, KT, HO, HK, MWO, YI, SSu,
MF, SSa, KN and HN performed experiments and data analysis. QS and
KT constructed figures. QS wrote the manuscript. KN and HN
supervised the study and contributed to data interpretation and
manuscript revision. KT and HN confirm the authenticity of all the
raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Okayama University (project identification code
1703-042-001). All animal experiments were performed in accordance
with the relevant guidelines and regulations approved by the
institutional committee of Okayama University (approval no.
OKU-2022354).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar
|
|
2
|
Lee KC, Chuang SK, Philipone EM and Peters
SM: Which clinicopathologic factors affect the prognosis of
gingival squamous cell carcinoma: A population analysis of 4,345
cases. J Oral Maxillofac Surg. 77:986–993. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kuk SK, Yoon HJ, Hong SD, Hong SP and Lee
JI: Staging significance of bone invasion in small-sized (4cm or
less) oral squamous cell carcinoma as defined by the American joint
committee on cancer. Oral Oncol. 55:31–36. 2016. View Article : Google Scholar
|
|
4
|
Russmueller G, Moser D, Würger T, Wrba F,
Christopoulos P, Kostakis G, Seemann R, Stadler V, Wimmer G, Kornek
G, et al: Upregulation of osteoprotegerin expression correlates
with bone invasion and predicts poor clinical outcome in oral
cancer. Oral Oncol. 51:247–253. 2015. View Article : Google Scholar
|
|
5
|
Omori H, Shan Q, Takabatake K, Nakano K,
Kawai H, Sukegawa S, Tsujigiwa H and Nagatsuka H: The origin of
stroma influences the biological characteristics of oral squamous
cell carcinoma. Cancers (Basel). 13:34912021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martin CK, Dirksen WP, Shu ST, Werbeck JL,
Thudi NK, Yamaguchi M, Wolfe TD, Heller KN and Rosol TJ:
Characterization of bone resorption in novel in vitro and in vivo
models of oral squamous cell carcinoma. Oral Oncol. 48:491–499.
2012. View Article : Google Scholar
|
|
7
|
Wong RJ, Keel SB, Glynn RJ and Varvares
MA: Histological pattern of mandibular invasion by oral squamous
cell carcinoma. Laryngoscope. 110:65–72. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ishikuro M, Sakamoto K, Kayamori K, Akashi
T, Kanda H, Izumo T and Yamaguchi A: Significance of the fibrous
stroma in bone invasion by human gingival squamous cell carcinomas.
Bone. 43:621–627. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Teitelbaum SL and Ross FP: Genetic
regulation of osteoclast development and function. Nat Rev Genet.
4:638–649. 2003. View
Article : Google Scholar
|
|
10
|
Takayanagi H: Osteoimmunology: Shared
mechanisms and crosstalk between the immune and bone systems. Nat
Rev Immunol. 7:292–304. 2007. View
Article : Google Scholar
|
|
11
|
Teitelbaum SL: Osteoclasts: What do they
do and how do they do it? Am J Pathol. 170:427–435. 2007.
View Article : Google Scholar
|
|
12
|
Suzuki T, Hayakawa T and Gomi K: GM-CSF
stimulates mouse macrophages and causes inflammatory effects in
vitro. J Hard Tissue Biol. 28:37–42. 2019. View Article : Google Scholar
|
|
13
|
Shibuya I, Takami M, Kawamoto M, Karakawa
A, Nakamura S and Kamijo R: Immunohistochemical analysis of the
distribution of RANKL-expressing cells and the expression of
osteoclast-related markers in giant cell tumor of bone. J Hard
Tissue Biol. 29:137–146. 2020. View Article : Google Scholar
|
|
14
|
Okamoto M, Hiura K, Ohe G, Ohba Y, Terai
K, Oshikawa T, Furuichi S, Nishikawa H, Moriyama K, Yoshida H and
Sato M: Mechanism for bone invasion of oral cancer cells mediated
by interleukin-6 in vitro and in vivo. Cancer. 89:1966–1975. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jimi E, Furuta H, Matsuo K, Tominaga K,
Takahashi T and Nakanishi O: The cellular and molecular mechanisms
of bone invasion by oral squamous cell carcinoma. Oral Dis.
17:462–468. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ono K, Akatsu T, Kugai N, Pilbeam CC and
Raisz LG: The effect of deletion of cyclooxygenase-2, prostaglandin
receptor EP2, or EP4 in bone marrow cells on osteoclasts induced by
mouse mammary cancer cell lines. Bone. 33:798–804. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kayamori K, Sakamoto K, Nakashima T,
Takayanagi H, Morita K, Omura K, Nguyen ST, Miki Y, Iimura T,
Himeno A, et al: Roles of interleukin-6 and parathyroid
hormone-related peptide in osteoclast formation associated with
oral cancers: Significance of interleukin-6 synthesized by stromal
cells in response to cancer cells. Am J Pathol. 176:968–980. 2010.
View Article : Google Scholar
|
|
18
|
Valkenburg KC, de Groot AE and Pienta KJ:
Targeting the tumour stroma to improve cancer therapy. Nat Rev Clin
Oncol. 15:366–381. 2018. View Article : Google Scholar
|
|
19
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takabatake K, Kawai H, Omori H, Qiusheng
S, Oo MW, Sukegawa S, Nakano K, Tsujigiwa H and Nagatsuka H: Impact
of the stroma on the biological characteristics of the parenchyma
in oral squamous cell carcinoma. Int J Mol Sci. 21:77142020.
View Article : Google Scholar
|
|
21
|
Shan Q, Takabatake K, Omori H, Kawai H, Oo
MW, Nakano K, Ibaragi S, Sasaki A and Nagatsuka H: Stromal cells in
the tumor microenvironment promote the progression of oral squamous
cell carcinoma. Int J Oncol. 59:722021. View Article : Google Scholar
|
|
22
|
Shan Q, Takabatake K, Kawai H, Oo MW,
Inada Y, Sukegawa S, Fushimi S, Nakano K and Nagatsuka H:
Significance of cancer stroma for bone destruction in oral squamous
cell carcinoma using different cancer stroma subtypes. Oncol Rep.
47:812022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shan Q, Takabatake K, Kawai H, Oo MW,
Sukegawa S, Fujii M, Nakano K and Nagatsuka H: Crosstalk between
cancer and different cancer stroma subtypes promotes the
infiltration of tumor-associated macrophages into the tumor
microenvironment of oral squamous cell carcinoma. Int J Oncol.
60:782022. View Article : Google Scholar
|
|
24
|
Quan J, Elhousiny M, Johnson NW and Gao J:
Transforming growth factor-β1 treatment of oral cancer induces
epithelial-mesenchymal transition and promotes bone invasion via
enhanced activity of osteoclasts. Clin Exp Metastasis. 30:659–670.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Flecknell PA: Laboratory animal
anesthesia. 3rd edition. Academic Press; San Diego, CA: pp.
191–192. 2009
|
|
26
|
Adams S and Pacharinsak C: Mouse
anesthesia and analgesia. Curr Protoc Mouse Biol. 5:51–63. 2015.
View Article : Google Scholar
|
|
27
|
An YZ, Cho E, Ling J and Zhang X: The
axin2-snail axis promotes bone invasion by activating
cancer-associated fibroblasts in oral squamous cell carcinoma. BMC
Cancer. 20:9872020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chuang FH, Hsue SS, Wu CW and Chen YK:
Immunohistochemical expression of RANKL, RANK, and OPG in human
oral squamous cell carcinoma. J Oral Pathol Med. 38:753–758. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Quan J, Hou Y, Long W, Ye S and Wang Z:
Characterization of different osteoclast phenotypes in the
progression of bone invasion by oral squamous cell carcinoma. Oncol
Rep. 39:1043–1051. 2018.PubMed/NCBI
|
|
30
|
Quan J, Johnson NW, Zhou G, Parsons PG,
Boyle GM and Gao J: Potential molecular targets for inhibiting bone
invasion by oral squamous cell carcinoma: A review of mechanisms.
Cancer Metastasis Rev. 31:209–219. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brown JS, Lowe D, Kalavrezos N, D'Souza J,
Magennis P and Woolgar J: Patterns of invasion and routes of tumor
entry into the mandible by oral squamous cell carcinoma. Head Neck.
24:370–383. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shaw RJ, Brown JS, Woolgar JA, Lowe D,
Rogers SN and Vaughan ED: The influence of the pattern of
mandibular invasion on recurrence and survival in oral squamous
cell carcinoma. Head Neck. 26:861–869. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ito M, Izumi N, Cheng J, Sakai H, Shingaki
S, Nakajima T, Oda K and Saku T: Jaw bone remodeling at the
invasion front of gingival squamous cell carcinomas. J Oral Pathol
Med. 32:10–17. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vincent C, Kogawa M, Findlay DM and Atkins
GJ: The generation of osteoclasts from RAW 264.7 precursors in
defined, serum-free conditions. J Bone Miner Metab. 27:114–119.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim H, Lee JH, Lee SK, Song NY, Son SH,
Kim KR and Chung WY: Chemerin treatment inhibits the growth and
bone invasion of breast cancer cells. Int J Mol Sci. 21:28712020.
View Article : Google Scholar
|
|
36
|
Woodward JK, Holen I, Coleman RE and
Buttle DJ: The roles of proteolytic enzymes in the development of
tumour-induced bone disease in breast and prostate cancer. Bone.
41:912–927. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
van der Pluijm G: Epithelial plasticity,
cancer stem cells and bone metastasis formation. Bone. 48:37–43.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Raggatt LJ and Partridge NC: Cellular and
molecular mechanisms of bone remodeling. J Biol Chem.
285:25103–25108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Elmusrati AA, Pilborough AE, Khurram SA
and Lambert DW: Cancer-associated fibroblasts promote bone invasion
in oral squamous cell carcinoma. Br J Cancer. 117:867–875. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ueda M, Goto T, Kuroishi KN, Gunjigake KK,
Ikeda E, Kataoka S, Nakatomi M, Toyono T, Seta Y and Kawamoto T:
Asporin in compressed periodontal ligament cells inhibits bone
formation. Arch Oral Biol. 62:86–92. 2016. View Article : Google Scholar
|
|
41
|
Nishigaki M, Yamamoto T, Ichioka H, Honjo
KI, Yamamoto K, Oseko F, Kita M, Mazda O and Kanamura N:
β-cryptoxanthin regulates bone resorption related-cytokine
production in human periodontal ligament cells. Arch Oral Biol.
58:880–886. 2013. View Article : Google Scholar
|
|
42
|
Kats A, Gerasimcik N, Näreoja T, Nederberg
J, Grenlöv S, Lagnöhed E, Desai S, Andersson G and Yucel-Lindberg
T: Aminothiazoles inhibit osteoclastogenesis and PGE2 production in
LPS-stimulated co-cultures of periodontal ligament and RAW 264.7
cells, and RANKL-mediated osteoclastogenesis and bone resorption in
PBMCs. J Cell Mol Med. 23:1152–1163. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu R, Bal HS, Desta T, Krothapalli N,
Alyassi M, Luan Q and Graves DT: Diabetes enhances periodontal bone
loss through enhanced resorption and diminished bone formation. J
Dent Res. 85:510–514. 2006. View Article : Google Scholar : PubMed/NCBI
|



















