Introduction
The most malignant tumour of the liver is
Hepatocellular carcinoma (HCC). The male/female rate in HCC is
reported to be constantly unbalanced towards the male sex, with
more or less marked differences, depending on the aetiology, with
Hepatitis B being characterised by more marked imbalance than
Hepatitis C (1,2). A consequence of this constant
imbalance is that HCC is the third or fourth most frequent solid
neoplasm in men for incidence and mortality while in women it is
not even among the top 10 (3).
Possible explanations for this finding, in addition to those
indicated above, have been put forward, most of them centred on the
different hormonal statuses: pro-inflammatory action of androgens
in males (4,5), oestrogen protection against excessive
inflammation in females offered by the prolonged fertile period
(6,7). In addition to the epidemiological and
clinical data, the relevance of hormonal status had been
demonstrated experimentally as well. In a seminal study, Naugler
et al (8) showed that the
oestrogen-mediated inhibition of Interleukin (IL)-6 production by
Kupffer cells reduced liver cancer risk in females. While the
suggestion put forward by the authors that these findings might be
used to prevent HCC in males cannot be easily applied to the
clinical practice since it would imply the feminization of males,
the importance of this demonstration remains unambitious. These
data clearly indicate that a different gender predisposition to HCC
development exists. However, although several studies have
investigated gene expression in patients with HCC, only very few,
if at all, have evaluated transcriptomic characteristics in
relation to sex and the possible relationship between the altered
genes with oestrogens in the attempt to identify features that
could be distinctive between males and females. In a prospective
study of a very well characterised cohort of patients with HCC at
first diagnosis, we performed an exhaustive transcriptomic
analysis, which allowed us to identify a neoangiogenic
transcriptomic signature able to accurately identify rapidly
growing and severe HCC cases (9).
In the current study, we have explored the annotated
HCC database of the prospective study to understand the differences
in gene expression in relation to sex. We found a low number of
genes differentially expressed between males and females. An even
lower number of genes was differentially expressed in the subgroup
of the HCV-positive subjects. Of these genes, sineoculis homeobox
homolog 1 (SIX1), ADH1C, and GPR19 were found to be possible
oestrogen targets (10). However,
only SIX1 was found in HCV-positive women that have a significant
correlation with both tumour growth speed and survival. Thus, we
have explored the relationship between SIX1 and the clinical,
pathologic, and transcriptomic features of HCV-positive patients
with HCC in relation to sex as this could yield pertinent
indications on various mechanisms of liver carcinogenesis between
males and females.
Materials and methods
Patients
We had identified 78 patients at first diagnosis of
HCC in a previously reported prospective study on patients with
liver cirrhosis on surveillance for HCC (9). After identification of a suspect
liver lesion, patients underwent two computed tomography (CT) scans
with ad hoc protocol at baseline and after six weeks (no therapy in
between) to define the doubling time of tumour size and imaging
traits. HCC doubling time (DT) was calculated as t ×
log2/logV1-logV0. At baseline, after acquiring informed consent,
they underwent paired HCC/surrounding-tissue biopsies for
microarray analysis (Agilent Whole Human Genome Oligo Microarrays),
histochemical (Ki67, CD34, e-cadherin), and histologic evaluation
[Edmondson-Steiner (E-S) grading, inflammatory intensity].
Survival, disease-free survival after down-staging and
transplant-free survival (Kaplan-Meier) were analysed in relation
to imaging and molecular data. A transcriptomic signature capable
of separating aggressive HCC from bland HCC was identified.
Detailed data are reported in (9).
For the present study, microarray data were
re-analysed by bivariate regression analysis according to sex; the
genes' differential levels of expression were recorded. The
relationship between gender-related expression and transcriptomic
signature was also evaluated. We also verified whether the
differentially expressed genes between females and males were known
oestrogen targets (10). As the
females were HCV-positive only, further analysis was restricted to
the HCV-positive group.
Determination of serum cytokines
According to the manufacturer's instructions,
Visfatin (Phoenix Pharmaceutical, Inc., Burlingame, CA, USA), IL-1
α, IL-1β, IL-10, tumour necrosis factor (TNF)-α (Aushon Biosystems,
Billerica, MA, USA), Adiponectin and Insulin (DRG International,
Springfield, NJ, USA), Hepatocyte growth factor (HGF) (Aushon
Biosystems, Inc., Billerica, MA, USA), and Insulin-like growth
factor (IGF)-2 (Boster Biological Technology, Pleasanton, CA, USA),
and were measured in duplicate We also tested serum CYFRA21-1 with
the human cytokeratin fragment antigen 21-1 (CYFRA21-1) ELISA kit
(Cusabio Biotech Co., Ltd., Wuhan, P.R. China) in accordance with
the instructions of the manufacturer. Serum IL-6, TNF-α, IL-8,
VEGF, Ang1, Ang2, and TGF-β1 levels were determined with the
quantikine/high-sensitivity enzyme-linked immunosorbent assay kit
(R&D Systems, Minneapolis, MN, USA). Absorbances were measured
at 450 and 490 nm using an automatic microplate reader (Multiskan
EX; Thermo Fisher Scientific, Inc, Waltham, MA, USA), with
background subtraction at 570 and 650 nm, respectively.
Immunohistochemistry
A portion of the formalin-fixed paraffin-embedded
liver tissue samples, obtained at enrolment in the study, was used
for SIX1′s the immunohistochemical evaluation. After
deparaffinization and rehydration, antigen unmasking was performed
with 1 mM EDTA buffer, pH 8, at 98°C for 15 mi. Then, these
sections were incubated in methanol 5% and
H2O2 1% for five minutes for blocking
endogenous peroxidases, whereas nonspecific sites were blocked
using a blocking solution reagent with bovine serum albumin 3% for
30 min at room temperature. Sections were incubated with mouse
anti-SIX1 (AB252224; Abcam) primary antibody at a working dilution
of 1:60. Thereafter, sections were incubated with prediluted
OmniMap anti-mouse horseradish peroxidase-conjugated secondary
antibody (Ventana Medical Systems from Tucson, AZ), for 20 min in a
humidity chamber as well as with detection kit reagents (ultra-view
universal horseradish peroxidase multimer and diaminobenzidine
[DAB] chromogen, Ventana Medical Systems) in compliance with the
manufacturer's instructions. Subsequently, these sections were
counterstained with haematoxylin, dehydrated, and permanently
mounted for microscopic examination). To obtain the intensity value
of the DAB signal, images of stained liver tissue were processed
with ImageJ software (http://rsbweb.nih.gov/.
Determination of oestradiol and
testosterone levels
Serum levels of oestradiol and testosterone were
determined by ELISA kits (Ella, Bio-Techne, Milan) following the
manufacturer's instructions.
MicroRNA analysis
RNAs from the aforementioned cohort of patients were
evaluated for miRNA expression and then assayed by quantitative PCR
using miRCURY LNA RT Kit (Hilden, Germany). Notably, 10 ng total
RNA served as a template for a 10 µl reverse transcription reaction
using miRCURY LNA miRNA SYBR Green PCR Kit and miRCURY LNA miRNA
PCR Assays (Qiagen). For each miRNA, reactions were done in
duplicate on a LightCycler 480 (Roche Applied Science) using the
manufacturer's recommendations for cycling parameters. The
expression levels of miRNA were measured using the quantification
cycle values (Cq values). U6 RNA and RNU5G were taken as controls,
and the assays were quantified by the 2−ΔΔCq method
(11). We used this method to
analyse the relative expression of the miRNAs of interest relative
to two endogenous controls and relative to the corresponding
cirrhotic non-tumour liver tissue.
Statistical analysis
A comparison between dichotomous and continuous
variables was drawn by employing Fisher's exact test, bivariate
(Pearson) correlation analysis, and t-test (paired or unpaired),
respectively. In case of SIX1 upregulation and survival, the
cumulative probability of overall survival was evaluated by the
Kaplan-Meier method. Patients were censored at the time of LT,
death, or the last available follow-up. Differences in observed
probability were assessed using the log-rank test or by two-stage
hazard rate comparison method in case of crossing survival curves
(12).
The Cox proportional method was utilised for
identifying risk factors for the overall survival and growth speed.
The variables that underwent testing included: sex, age, E-S
grading, presence of macrovascular invasion assessed by CT scan,
multifocality at baseline, platelets level, and α-fetoprotein
levels. For survival analysis, albumin, creatinine, and bilirubin,
were used as additional independent variables. The PASW Statistics
28 program (IBM Corp., Armonk, NY) was used for statistical
analysis.
Results
Cohort's description
Seventy-eight patients with liver cirrhosis
(Child-Pugh A: n=53; B: n=23; C: n=2) on surveillance for HCC were
enrolled in the study reported in (9). Sixty-one (78.2%) of these patients
were males. Of these, 10 (12.8%) were Hepatitis B positive, 11
(14.1%) had alcohol-related CLD, and 11 (14.1%) had dysmetabolic
CLD. Hepatitis C emerged as the most common aetiology, with 46
(58.9%) patients being HCV-positive [29 male (63.0%) and 17 females
(37.0%)]. In females, HCV was the only represented aetiology. We,
therefore, focused on the analysis of gender-related aspects of
HCV-positive patients.
The HCV-positive cohort's mean age was 68.9±9.2,
with males significantly younger than females (M vs. F: 64.0±14.8
vs. 71.0±8.3, P=0.001). Considering the whole cohort of
HCV-positive patients, survival was not significantly different
between males and females (males vs. females: 36.8±22.7 vs.
39.5±33.2, P=0.705).
Demographic and clinical data of the HCV-positive
cohort and the gender-based main HCC characteristics in
HCV-positive patients are summarised in Table I. No substantial differences were
found in HCC, although a trend towards a higher percentage of E-S
grade 3 in females was observed. The significantly higher AFP
levels in females at presentation are in line with this finding
(Table I).
 | Table I.Demographic and clinical
characteristics (including those related to HCC) of HCV-positive
patients at presentation according to sex. |
Table I.
Demographic and clinical
characteristics (including those related to HCC) of HCV-positive
patients at presentation according to sex.
| Variable | Whole cohort
(n=46) | Males (n=29) | Females (n=17) | P-value |
|---|
| Age, years | 68.9±9.2 | 64.0±14.8 | 71.0±8.3 | 0.001 |
| Child-Pugh
score |
|
|
| 0.549 |
| A | 38 | 23 | 15 |
|
| B | 8 | 6 | 2 |
|
| MELD | 11.4±3.2 | 11.6±3.2 | 10.6±3.2 | 0.264 |
| AFP, ng/ml | 122±596 | 109±387 | 1.949±6.931 | 0.049 |
| Bilirubin, mg% | 1.8±3.1 | 1.9±3.2 | 1.6±2.6 | 0.810 |
| Albumin, g/l | 3.6±0.56 | 3.5±0.59 | 3.6±0.44 | 0.533 |
| Creatinine,
mg/dl | 0.95±0.36 | 1.0±0.33 | 0.771±0.24 | 0.019 |
| INR | 1.30±0.18 | 1.31±0.18 | 1.29±0.19 | 0.639 |
| Platelets,
×103/mm3 | 114±61 | 112±61 | 120±58 | 0.655 |
| HCC doubling time,
days | 109±100 | 114±111 | 102±81 | 0.702 |
| Tumour volume, log
cm3 | 3.8±0.9 | 3.7±1.1 | 3.9±7.2 | 0.613 |
| Multifocality at
baseline (%) | 7 (15.2) | 6 (20.7) | 1 (5.9) | 0.391 |
| AFP >400 ng/ml
(%) | 3 (6.5) | 0 | 3 (17.6) | 0.525 |
| Macrovascular
invasion, n (%) | 3 (6.5) | 2 (6.9) | 1 (5.9) | 0.558 |
| BCLC class (%) |
|
|
| 0.653 |
| A | 32 (69.6) | 19 (65.5) | 13 (76.5) |
|
| B | 9 (19.6) | 6 (20.7) | 3 (17.6) |
|
| C | 5 (10.9) | 4 (13.8) | 1 (5.9) |
|
| Edmondson-Steiner
grade (%) |
|
|
| 0.086 |
| 1 | 16 (34.8) | 11 (37.9) | 5 (29.4) |
|
| 2 | 17 (37.0) | 13 (44.8) | 4 (23.5) |
|
| 3 | 13 (28.3) | 5 (17.2) | 8 (47.1) |
|
| Treatment (%) |
|
|
| 0.743 |
|
Supportive care | 13 (28.3) | 6 (20.7) | 7 (41.2) |
|
| Liver
transplant | 1 (2.2) | 1 (3.4) | 0 |
|
|
Resection | 2 (4.3) | 1 (3.4) | 1 (5.9) |
|
|
TACE | 17 (36.9) | 11 (37.9) | 6 (35.3) |
|
|
RFA | 10 (21.7) | 8 (27.6) | 2 (11.8) |
|
|
Sorafenib | 2 (4.3) | 1 (3.4) | 1 (5.9) |
|
|
Sequential treatments | 1 (2.2) | 1 (3.4) | 0 |
|
Global gene expression according to
gender
We analysed the original database comprising all
genes that had been found differentially expressed in tumour tissue
when compared to the non-tumoral cirrhotic tissue for differences
in the expression level according to sex. In the entire cohort, we
found 198 genes differentially expressed between males and females,
irrespective of aetiology. Table
SI lists the 133 genes differentially expressed in the
HCV-positive cohort. Seventy-six were up regulated in females and
59 in males. Three genes (SIX1, GPR19, ADH1C) were defined by
micro-array and high throughput sequencing technologies as possible
oestrogen targets (10). Since
SIX1 was the only one which was significantly related with both
higher HCC growth speed and lower survival in females (see below),
we focused our attention on it. SIX1 transcriptomic expression
levels, from non-tumour and tumour tissue, stratified based on sex,
are shown in Fig. 1A. Levels of
expression in non-tumour tissue were not significantly different
between males and females while a highly significant,
straightforward difference was present in tumour tissue, with
females exhibiting the highest levels of expression (P<0.001).
The expression level between non-tumour and tumour tissue was
significantly different in both males and females (P<0.001). We
also evaluated DACH1 expression since many studies suggested that
it had a correlation with SIX1 expression (13,14).
Although DACH1 expression was not significantly different between
males and females, a significant inverse relationship between SIX1
and DACH1 was observed in males, both in the HCV-positive males
(males: r=−0.532, P=0.003; females: r=−0.055, P=0.780, Pearson
bivariate correlation) and in the entire cohort (r=−0.395, P=0.002,
Pearson correlation). The gene TGF-β (14) is also known to interact with SIX1.
Upon being upregulated, SIX1 can switch TGF-β signalling to the
prometastatic phenotype. In females, we found a notable positive
relationship between upregulated SIX1 and TGF-β (r=0.505, P=0.006,
Pearson correlation) that was absent in males (r=-.375; P=0.060,
Pearson correlation).
Immunohistochemical evaluation of
SIX1
Immunohistochemical evaluation of SIX1 in the paired
HCC/non-tumour tissue biopsies showed a significantly higher
expression in tumoral tissue of female patients when compared to
that of males (Figs. 1B and
S1). The expression level was not
different in nontumoral tissue between males and females while a
significant difference was present between SIX1 expression between
tumoral and nontumoral liver tissue in females (Fig. 1B).
Correlations between SIX1 expression
and pathologic features
A distinctive pattern of relationships was found
between sex, pathologic features, and SIX1 expression. E-S grading
only was positively correlated with increased SIX1 expression in
HCV-positive males while neither inflammatory intensity, maximal
nodule size, Ki67, e-cadherin gene nor protein expression in HCC
had such a correlation. On the contrary, inflammatory intensity,
e-cadherin gene and protein expression were significantly
correlated in HCV-positive females, with e-cadherin having an
inverse relationship with SIX1.
On the contrary, inflammatory intensity, e-cadherin
gene, and protein expression were significantly correlated in
HCV-positive females, with e-cadherin having an inverse
relationship with SIX1.
SIX1 transcriptomic upregulation
relates with higher growth speed in females
A higher SIX1 expression had a strong relationship
with the probability of having a higher growth speed both in the
group of HCV-positive patients (P=0.007, log-rank test) and the
entire cohort of patients (P=0.014, log-rank test). However, in the
former group, stratification by gender showed that the probability
of having a higher growth speed was highly significant in females
(P=0.009, log-rank test), but was absent in males (P=0.228,
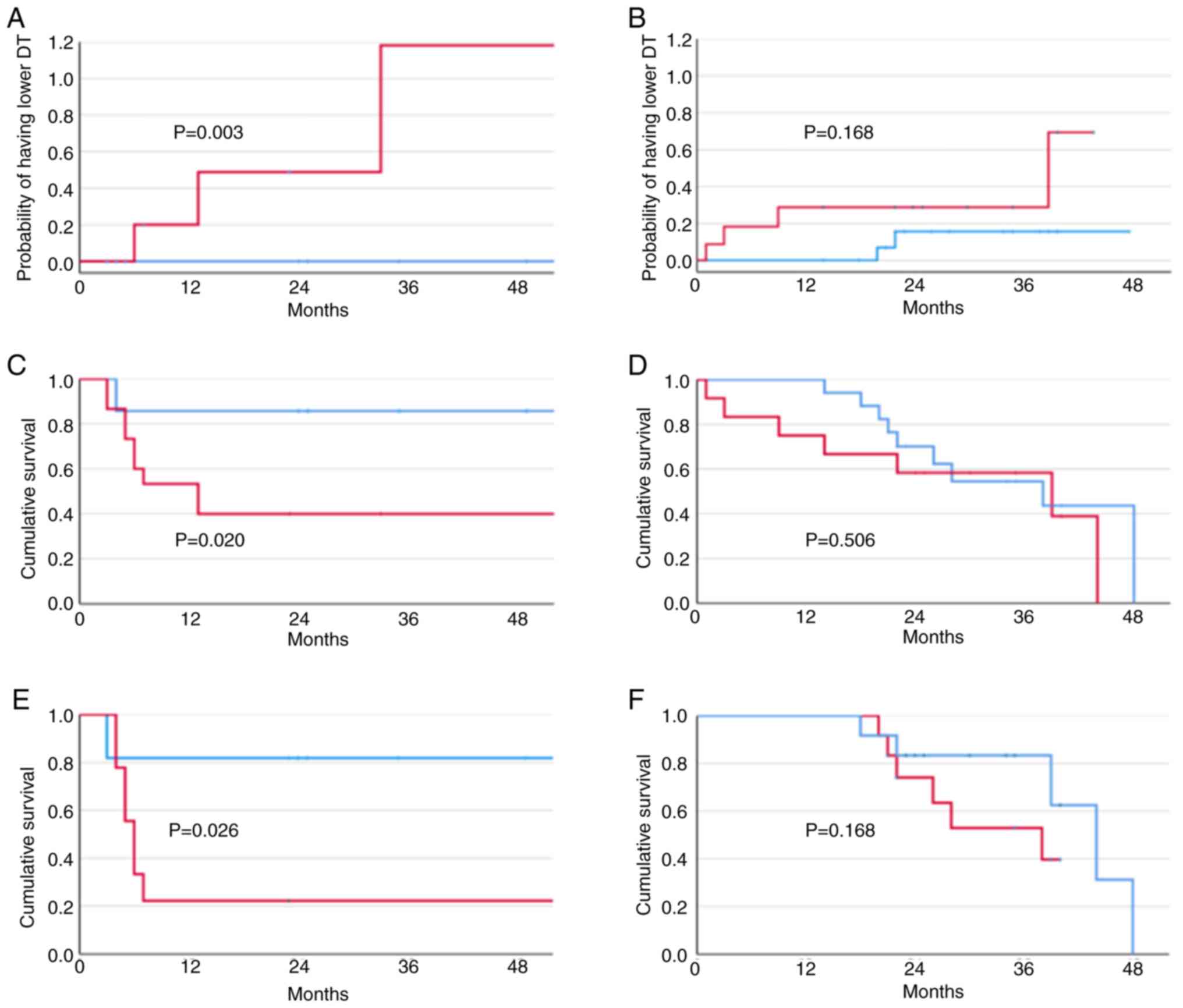
log-rank test) (Fig. 2A and
B).
HCC doubling time in the six weeks after diagnosis
was significantly inversely related to SIX1 expression levels in
females (r=−0.575, P=0.001, Pearson correlation). No significant
relationship was found in males (r=−0.309, P=0.103, Pearson
correlation). The transcriptomic signature was related in the
initial report to survival as well as growth speed (9). Three of the individual components of
the transcriptomic signature (ANGPT2, DLL4, ESM1, NETO2, NR4A1)
were differentially related with SIX1 expression in the
HCV-positive cohort as a whole: ANGPT2 (r=0.292, P=0.026, Pearson
correlation); DLL4 (r=0.344, P=0.008, Pearson correlation); ESM1
(r=0.459; P<0.001, Pearson correlation). Stratifications by sex
displayed that in females, SIX1 had a significant relationship with
ANGPT2 (r=0.378, P=0.043, Pearson correlation); NETO2 (r=0.612,
P<0.001, Pearson correlation); and ESM1 (r=0.376, P=0.044,
Pearson correlation) while a significant relationship was present
in males with DLL4 (r=0.499, P=0.015, Pearson correlation), and
ESM1 (r=0.496; P=0.006, Pearson correlation). Quantitative analysis
of the relationship between median SIX1 values and dichotomic value
of the transcriptomic score (bland vs. aggressive) showed that all
seven women with lower median SIX1 levels had a bland HCC while
four of them with higher median values had an aggressive HCC
(P=0.029). No difference was found in males between those with
higher and lower SIX1 median levels (P=0.158).
Factors predictive of growth speed at
Cox regression analysis
Of the variables tested (sex, age, E-S grading,
presence of macrovascular invasion at CT scan, multifocality at
baseline, platelet level, α-fetoprotein levels, SIX1 levels), only
the presence of macrovascular invasion at CT scan (HR: 6.670, 95%
CI 1.689 to 4.505, P=0.007), E-S grading (HR: 3–909, 95% CI 1.371
to 11.143, P=0.011), multifocality at baseline (HR: 2.761, 95% CI
1.692 to 6.382, P<0.001), and SIX1 levels (HR: 6.024, 95% CI
1.314 to 27.622, P=0.021) were predictive of growth speed at
univariate Cox regression analysis. As all the significant factors
were collinear, multivariable analysis was not performed. Analysis
of the Cox results for the different variables based on sex
revealed that only multifocality at baseline was significant in
both males and females while there was a highly significant
relationship in females only for the presence of macrovascular
invasion at CT scan and SIX1 (females: macrovascular invasion: HR:
9.721, 95% CI 1.367 to 69.111, P=0.023; SIX1: HR: 2.803, 95% CI
1.132 to 6.382, P=0.026; males: macrovascular invasion: HR: 3.097,
95% CI 0.343 to 28.000, P=0.314; SIX1: HR: 1.215, 95% CI 0.098 to
1.645, P=0.207).
SIX1 upregulation relates with lower
survival in females
No significant difference was observed in the entire
HCV-positive population regarding survival in relation to SIX1
upregulation (P=0.119, log-rank test). However, stratifying the
HCV-positive cohort by sex, a significantly lower survival was
present in HCV-positive females (P=0.020, log-rank test). On the
other hand, no difference was found in males (P=0.772, log-rank
test) (Fig. 2C and D). The same
finding was obtained after stratifying survival by median
histochemical SIX1 values (Fig. 2E and
F).
Factors predictive of survival at Cox
regression analysis
Of the variables examined at univariate analysis
(sex, age, E-S grading, presence of macrovascular invasion assessed
by CT scan, multifocality at baseline, the presence of ascites,
encephalopathy, platelet level, α-fetoprotein levels, albumin,
creatinine, and bilirubin) in the whole HCV-positive cohort, only
the number of nodules at entry into the study HR: 1.840, 95% CI
1.363 to 2.484, P=0.001), albumin levels (HR: 0.335, 95% CI 0.158
to 0.712, P=0.004), and SIX1 upregulation (HR: 2.288, 95% CI 1.058
to 4.947, P=0.035) were found to have a significant relationship
with survival. Multivariable analysis was prevented by the
collinearity between the three significant factors. Analysis after
stratification by sex revealed that for both males and females, the
number of nodules at entry was significantly related with survival
(women: HR 1.791, 95% confidence interval, 1.160 to 2.765, P=0.009,
men: HR 1.715, 95% confidence interval, 1.127 to 2.609, P=0.012).
Albumin and SIX1 upregulation were significant for females only
(SIX1: women HR 5.034, 95% CI 1.083 to 23.387, P=0.039, men: HR
1.407, 95% confidence interval, 0.508 to 3.895, P=0.511; albumin:
women HR 0.086, 95% confidence interval, 0.019 to 0.398, P=0.002,
men: HR 0.610, 95% confidence interval, 0.196 to 1.899,
P=0.394).
Relationship between SIX1 upregulation
and miRNA
In the entire HCV-positive cohort, only miR-421,
miR-9-5p, and miR-19b-1-5p were found to have a significant
relationship with SIX1 upregulation. However, when the HCV-positive
cohort was stratified by sex, a very distinctive and different
miRNA pattern was present in relation to SIX1. In males, only
miR-421 and miR-9-5p were related with SIX1. In females, several
more miRNAs were found to be related with SIX1: miR-181b,
miR-503-5p, and miR-125b had a direct relationship, while
miR139-5p, miR-26b, let7c-3p, and let7c-5p had an inverse
relationship (Tables II and
III).
 | Table II.Correlation between SIX1 and miRNA
expression in HCV-positive males and females. |
Table II.
Correlation between SIX1 and miRNA
expression in HCV-positive males and females.
| MicroRNA | Males | P-value | Females | P-value |
|---|
| miR-421 | 0.431 | 0.036 | 0.162 | 0.461 |
| miR-9-5p | 0.429 | 0.041 | 0.478 | 0.033 |
| miR-181b | −0.136 | 0.528 | 0.578 | 0.006 |
| miR-503-5p | −0.021 | 0.924 | 0.626 | 0.002 |
| miR-125b | −0.251 | 0.248 | −0.437 | 0.037 |
| miR-19b-1-5p | 0.258 | 0.235 | 0.509 | 0.018 |
| miR-139-5p | −0.084 | 0.703 | −0.438 | 0.037 |
| let-7c-3p | −0.011 | 0.963 | −0.536 | 0.027 |
| let-7c-5p | −0.065 | 0.762 | −0.440 | 0.035 |
| miR-26b | −0.131 | 0.543 | −0.465 | 0.025 |
| miR-1303 | 0.257 | 0.226 | −0.409 | 0.053 |
 | Table III.Relationship between median SIX1
expression values and level of expression of the significantly
associated miRNA at Pearson bivariate correlation. |
Table III.
Relationship between median SIX1
expression values and level of expression of the significantly
associated miRNA at Pearson bivariate correlation.
|
|
| Six1 |
|
|---|
|
|
|
|
|
|---|
| miRNA | Median miRNA
value | Lower median
value | Higher median
value | P-value |
|---|
| Males |
|
|
|
|
|
miR-421 | 0.380 | Downregulated | Normally
regulated | 0.009 |
|
miR-9-5p | 0.135 | Downregulated | Normally
regulated | 0.049 |
| Females |
|
|
|
|
|
miR-9-5p | 0.135 | Downregulated | Normally
regulated | 0.025 |
|
miR-181b | −0.340 | Downregulated | Upregulated | 0.003 |
|
miR-503-5p | −0.780 | Normally
regulated | Highly
downregulated | 0.004 |
|
miR-125b | −0.880 | Downregulated | Extremely
downregulated | 0.037 |
|
miR-19b-1-5p | 0.654 | Downregulated | Upregulated | 0.001 |
|
miR-26b | −0.980 | Normally
regulated | Normally
regulated | 0.025 |
|
let7c-3p | −0.650 | Upregulated | Downregulated | 0.017 |
|
let7c-5p | −0.310 | Upregulated | Downregulated | 0.017 |
|
miR-139-5p | −0.910 | Normally
regulated | Extremely
downregulated | 0.037 |
Oestradiol and testosterone
levels
No significant difference was present between males
and females in the level of circulating oestradiol or testosterone
(Fig. S2). However, a
significantly lower concentration of circulating oestradiol was
found in those overexpressing SIX1 (P<0.001 when stratifying by
SIX1 median levels, within the female group), while testosterone
had higher, although only borderline significant, levels (P=0.069).
Females with HCCs overexpressing SIX1 were found to have
significantly higher TGF-β1 and HGF levels and lower Visfatin
levels.
Of the large panel of 17 cytokines tested, very
different results were found in males and females in relation to
SIX1 expression. In females TGF-β1 (r=0.428, P=0.029, Pearson
correlation) and HGF (r=0.639, P<0.001, Pearson correlation)
were positively correlated with SIX1 levels while Visfatin
(r=−0.599, P=0.002 Pearson correlation) was inversely related. A
positive relationship between both TGF-β1 and SIX1 was found within
AFP (r=0.595, P=0.001, and r=0.378, P=0.048, respectively, Pearson
correlation).
In males, none of the cytokines tested but HGF was
found to have a positive correlation with SIX1 levels (r=0.462,
P=0.017 Pearson correlation). Visfatin levels inversely related
with BMI (r=−0.573, P<0.001 Pearson correlation) in females but
not in males were.
Discussion
In this study, we have shown that addressing the
onset and the course of HCC without stratifying by sex can be
grossly misleading. Evaluation of the HCC data keeping female and
male patients together would have caused us to miss very
distinctive features that became obvious during a separate sex
analysis. We performed a detailed evaluation of gene expression in
males and females from a very well-characterised cohort of HCC
patients at first HCC diagnosis, restricting the analysis to the
HCV-positive subgroup, as no females with other aetiologies were
represented in the whole cohort (9). After evaluating which genes were
differentially expressed between males and females in the entire
cohort, we then assessed those that were differentially expressed
in the HCV-positive cohort and which, of these, were possible
targets of oestrogen action. Accordingly, we identified three
differentially expressed genes (SIX1, GPR19, ADH1C), which are
reported as possible oestrogen targets (10). We focused our attention on SIX1 as
it was the only one, of the three indicated above, that was
significantly associated with growth speed and survival in
females.
According to SIX1 expression (which was also
confirmed at the proteomic level), analysis of the relationship of
males and females with pathologic features showed that the only
significant association in males was with E-S grading. None of the
other pathologic features considered (inflammatory activity,
e-cadherin expression) was significantly related. In females, no
relationship was found between upregulated SIX1 and E-S grading
while other features such as increased inflammatory activity and
decreased e-cadherin expression (both at RNA and at protein level)
were significantly associated. This last feature is particularly
interesting given the relationship we found, in females only,
between SIX1 upregulation and TGF-β up regulation in tumour tissue
and increased levels in serum. In this regard, Micalizzi et
al (15,16) demonstrated in breast cancer that
while TGF-β upregulation is sufficient to induce
epithelial-mesenchymal transition (EMT), SIX1 upregulation is
required to determine the switch of TGF-β signalling to the
prometastatic phenotype. Meanwhile, Min and Wei (17) demonstrated that silencing SIX1 was
able to inhibit TGF-β/Smad2/3 pathway, suppressing EMT. Liu et
al (18) hand showed that SIX1
enhances the TGF-β signalling pathway by upregulating TGFβ-R2
expression and that deletion of Six1 in cancer cells significantly
reduced tumour growth in an immune-dependent manner with enhanced
antitumor immunity in the TME. The addition of SIX1 upregulation is
a further piece of knowledge to the complex microenvironment that
we already described for TS-positive aggressive HCC, i.e. marked
PD-1 and PD-L1 upregulation, prominent EMT, and clear-cut
activation of TGFβ1 signalling (19). In this subgroup of HCV-patients
females with aggressive HCC, we found a positive relationship
between higher circulating TGFβ levels and upregulated SIX1 as well
as a positive relationship between them and significantly higher
AFP levels. In addition, several genes composing the transcriptomic
signature (ANGPT2, NETO2, ESM1) were also upregulated and had a
positive relationship with SIX1 upregulation in females HCC, which
were also characterised by higher growth speed and lower survival.
All these features point toward an increased biologic
aggressiveness for HCC overexpressing SIX1 in females.
The SIX1 gene encodes a homeodomain-containing
transcription belonging to the 6th family of homeoproteins. SIX1
was found to be linked to the development of tissues and organs,
thus potentially promoting the proliferation and survival of
precursor cells before cell differentiation (20). An important role in cell apoptosis
has also been reported (21,22).
In human cancer, elevated levels of SIX1 mRNA were found in early
and late-stage ovarian cancer (21). SIX1 upregulation has been also
associated with poor prognosis in breast cancer (23), in gastric cancer (24), and in colorectal cancer (25). More recently, a few studies have
reported that SIX1 has a relevant role in HCC as well: SIX1
upregulation was identified as an independent poor prognostic
factor of HCC (26,27). Cheng et al (22) demonstrated that SIX1 upregulation
was linked to tumorigenesis and that its suppression, coupled with
induction of DACH1 upregulation, inhibited the progression of HCC
both in vitro and in vivo. In our series, a
significant relationship between SIX1 and DACH1 was present in the
whole HCV-positive cohort and in the HCV-positive males but not in
females. This could be indicative of the fact that the mechanisms
proposed by the authors, i.e. suppression of tumorigenesis via p53
up-regulation, via SIX1 inhibition, and DACH1 upregulation, might
be valid for males only while in females SIX1 could act via other
mechanisms. Accordingly, a contrasting gene expression pattern
comparable to our results was shown in male and female breast
cancer (28). In this study, DACH1
and SIX1 had contrasting expression pattern in males and females
and comparable opposite prognostic implication. Similar to other
findings, females with breast cancer overexpressing SIX1 had more
aggressive disease and severe prognosis.
SIX1 is known to directly interact with oestrogens
(10). Its role in hormonal
carcinogenesis has been demonstrated in experimental as well as
human endometrial carcinogenesis (29). Neonatal exposure to phytoestrogens
or diethylstilbestrol could be followed in later life by aberrant
endometrial expression of SIX1 and eventually by endometrial
carcinoma (30,31). It is certainly difficult to
ascertain whether this group of HCV-positive women had had any
early hormonal exposure; it is more likely that if present, had
occurred later in life. On the other hand, no comparable data are
available for an organ, like the liver, which is a non-classical
target for oestrogen. The presence of α-oestrogen receptors in the
liver functionally identical to those of the classical target
organs (32) offers the potential
physiologic basis for similar mechanisms to occur. Interestingly,
women overexpressing SIX1 were found to have significantly lower
circulating concentrations of oestradiol and higher levels of
testosterone (although the latter did not reach full significance),
despite all having an age indicating advanced menopause. This
hormonal framework has already been elucidated in menopausal
HCV-positive women with advanced fibrosis and resistance to
antiviral therapy, thereby suggesting that this modification during
the course of chronic liver disease can contribute to the loss of
anti-inflammatory action linked with oestrogens, aggravated by the
contemporary increase of androgens hormones (33).
The pattern of the activated miRNA in association
with SIX1 was also very distinctive between males and females.
While we found only miR-421 and miR-9-5p positively upregulated in
males, a much larger number was found in females. Some upregulated
(miR-181b, miR-19b_1_5p) whereas most of them were down-regulated
(miR-139-5p, miR-503-5p, miR-125b miR-26b, let7c-3p, let7c-5p), all
of them in association with SIX1 upregulation. The presence of this
relevant number of upregulated or downregulated miRNA was not
evident when the cohort was examined as a whole, probably because
the lower number of females in respect to males did not allow the
revelation of their specific patterns. These different miRNAs
combinations found exclusively in females are quite informative.
Meng et al (34) have
already described the combination of upregulated miR-181b and
downregulated let7 and suggested that it could represent a
molecular target in HCC and a possible therapeutic tool for
eradication of hepatocellular malignancies. A possible functional
role as an oncogene has been suggested for miR-181b (35). Zhou et al (36) showed that miRNA-181b was
significantly upregulated in response to TGF-β treatment in gastric
cancer cell lines via induction of Smad2/3 signalling.
Interestingly, SIX1 is known to activate the TGF-β/Smad2/3 pathway,
and silencing SIX1 blocks EMT via inhibition of TGF-β/Smad2/3
signals (17). Similarly,
downregulated miR-503-5p, as we have found in HCV-positive females
in association with upregulated SIX1, has been linked to increased
EMT (37) and increased HCC
progression (38). By contrast,
upregulated mir503 inhibits cellular proliferation and induces
apoptosis in HCC cells (39) and
can sensitize HCC cells to 5-fluorouracil (40). Concordantly, the downregulation of
miR-139-5p (41) and of miR-125b
(42) was associated with
increased EMT and increased metastatic capacity. A recent study in
patients with HCC revealed that the downregulation of miR-139-5p
resulted in poor survival (43).
In its entirety, the peculiar miRNA pattern evidenced in females
HCC overexpressing SIX1 points towards a specific activation of
pathways associated with relevant biologic aggressiveness of the
tumour, a feature that is concordant with the clinical course of
these patients.
The novelty of our findings is the association with
SIX1 upregulation in females, a relationship that was yet to be
explored previously. However, there are some limitations in his
study: one resides in the lack of an experimental demonstration of
the ability of SIX1 upregulation to modify hepatic cellular
reactivity toward a higher oncogenic ability. Nevertheless, our
findings seem strong enough to suggest the opportunity to conduct
an experimental exploration of the effect of SIX1 upregulation in
liver carcinogenesis in females. Secondly, an explanation for the
specific activation of SIX1 in females should be sought. A starting
point to explore could be represented by a careful epidemiologic
and anamnestic study in women with HCC to discover possible
hormonal exposure that could, as in the case of endometrial cancer,
offer a key to the interpretation of this selective
upregulation.
Overall, on the one hand, these data suggest a very
distinctive model for carcinogenesis, unique to HCV-positive women,
characterised by a marked downregulation of potentially protective
mechanisms against excess proliferation, EMT, and metastatic
capacity, and by a marked activation of potential oncogenes on the
other hand. All these mechanisms are in relation to a gene, SIX1,
which has a close relationship with estrogenic control. However, it
is not completely clear and deserves a further evaluation of how
this gene can influence liver carcinogenesis specifically in
females.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by Associazione Italiana sulla
Ricerca sul Cancro (AIRC; grant no. IG 2020-ID. 24858 project-P.I.
Villa Erica).
Availability of data and materials
Gene expression data are available at the Gene
Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo) under the accession
number: GSE54236. The other datasets used and/or analyzed during
the current study are available from the corresponding author on
reasonable request.
Authors' contributions
RMC, FM, SL, AR, SM and FD and GM performed
experiments, and analysed and interpreted experimental data. FS,
DR, AP and LDM identified and followed up with suitable patients
and compiled and analysed the clinical database. RMC, FM, MLMC, GG
and EV interpreted the results. RMC, FM, FS, MLMC and GG reviewed
the manuscript. RMC, FM, FS, DR, MLMC and GG approved the final
version of the manuscript. GG and EV confirm the authenticity of
all the raw data. EV conceived and designed the study and wrote the
paper. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was conducted according to the Declaration
of Helsinki and approved by or Ethics Committee of University
Hospital of Modena (IRB10/08_CE_UniRer; ClinicalTrials ID:
NCT01657695). Written informed consent was obtained from all
subjects involved in the study.
Patient consent for publication
Written informed consent included consent to
publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Villa E, Baldini GM, Pasquinelli C,
Melegari M, Cariani E, Di Chirico G and Manenti F: Risk factors for
hepatocellular carcinoma in Italy. Male sex, hepatitis B virus,
non-A non-B infection, and alcohol. Cancer. 62:611–615. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang X, El-Serag HB and Thrift AP: Sex
and race disparities in the incidence of hepatocellular carcinoma
in the United States examined through age-period-cohort analysis.
Cancer Epidemiol Biomarkers Prev. 29:88–94. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McGlynn KA, Petrick JL and El-Serag HB:
Epidemiology of hepatocellular carcinoma. Hepatology. 73 (Suppl
1):S4–S13. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ma WL, Lai HC, Yeh S, Cai X and Chang C:
Androgen receptor roles in hepatocellular carcinoma, cirrhosis, and
hepatitis. Endocr Relat Cancer. 21:R165–R182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Y, Xu A, Jia S and Huang J: Recent
advances in the molecular mechanism of sex disparity in
hepatocellular carcinoma. Oncol Lett. 17:4222–4228. 2019.PubMed/NCBI
|
|
6
|
Villa E: Role of estrogen in liver cancer.
Womens Health (Lond). 4:41–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi L, Feng Y, Lin H, Ma R and Cai X: Role
of estrogen in hepatocellular carcinoma: Is inflammation the key? J
Transl Med. 12:932014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Naugler WE, Sakurai T, Kim S, Maeda S, Kim
K, Elsharkawy AM and Karin M: Gender disparity in liver cancer due
to sex differences in MyD88-dependent IL-6 production. Science.
317:121–124. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Villa E, Critelli R, Lei B, Marzocchi G,
Cammà C, Giannelli G, Pontisso P, Cabibbo G, Enea M, Colopi S, et
al: Neoangiogenesis-related genes are hallmarks of fast-growing
hepatocellular carcinomas and worst survival. Results from a
prospective study. Gut. 65:861–869. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Berglund JA, Voelker R, Barber P, Diegel
J, Mahady A and Bodner M: RNA regulation by estrogen. Oregon Univ
Eugene; 2011, View Article : Google Scholar
|
|
11
|
Li H, Han D, Hou Y, Chen H and Chen Z:
Statistical inference methods for two crossing survival curves: A
comparison of methods. PLoS One. 10:e01167742015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martik ML and McClay DR: Deployment of a
retinal determination gene network drives directed cell migration
in the sea urchin embryo. Elife. 4:e088272015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Y, Han N, Zhou S, Zhou R, Yuan X, Xu
H, Zhang C, Yin T and Wu K: The DACH/EYA/SIX gene network and its
role in tumor initiation and progression. Int J Cancer.
138:1067–1075. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Micalizzi DS, Christensen KL, Jedlicka P,
Coletta CD, Barón AE, Harrel JC, Horwitz KB, Billheimer D, Heichman
KA, Welm AL, et al: The Six1 homeoprotein induces human mammary
carcinoma cells to undergo epithelial-mesenchymal transition and
metastasis in mice through increasing TGF-beta signaling. J Clin
Invest. 119:2678–2690. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Micalizzi DS, Wang CA, Farabaugh SM,
Schiemann WP and Ford HL: Homeoprotein Six1 increases TGF-beta type
I receptor and converts TGF-beta signaling from suppressive to
supportive for tumor growth. Cancer Res. 70:10371–10380. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Min WP and Wei XF: Silencing SIX1 inhibits
epithelial mesenchymal transition through regulating TGF-β/Smad2/3
signaling pathway in papillary thyroid carcinoma. Auris Nasus
Larynx. 48:487–495. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu W, Gao M, Li L, Chen Y, Fan H, Cai Q,
Shi Y, Pan C, Liu J, Cheng L, et al: Homeoprotein SIX1 compromises
antitumor immunity through TGF-β-mediated regulation of collagens.
Cell Mol Immunol. 18:2660–2672. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Critelli R, Milosa F, Faillaci F, Condello
R, Turola E, Marzi L, Lei B, Dituri F, Andreani S, Sighinolfi P, et
al: Microenvironment inflammatory infiltrate drives growth speed
and outcome of hepatocellular carcinoma: A prospective clinical
study. Cell Death Dis. 8:e30172017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Christensen KL, Patrick AN, McCoy EL and
Ford HL: The six family of homeobox genes in development and
cancer. Adv Cancer Res. 101:93–126. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Behbakht K, Qamar L, Aldridge CS, Coletta
RD, Davidson SA, Thorburn A and Ford HL: Six1 overexpression in
ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and
is associated with poor survival. Cancer Res. 67:3036–3042. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng Q, Ning D, Chen J, Li X, Chen XP and
Jiang L: SIX1 and DACH1 influence the proliferation and apoptosis
of hepatocellular carcinoma through regulating p53. Cancer Biol
Ther. 19:381–390. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ford HL, Kabingu EN, Bump EA, Mutter GL
and Pardee AB: Abrogation of the G2 cell cycle checkpoint
associated with overexpression of HSIX1: A possible mechanism of
breast carcinogenesis. Proc Natl Acad Sci USA. 95:12608–12613.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jin J, Jin T, Quan M, Piao Y and Lin Z:
Ezrin overexpression predicts the poor prognosis of gastric
adenocarcinoma. Diagn Pathol. 7:1352012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kahlert C, Lerbs T, Pecqueux M, Herpel E,
Hoffmeister M, Jansen L, Brenner H, Chang-Claude J, Bläker H, Kloor
M, et al: Overexpression of SIX1 is an independent prognostic
marker in stage I–III colorectal cancer. Int J Cancer.
137:2104–2113. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kong J, Zhou X, Liu S, Jin T, Piao Y, Liu
C and Lin Z: Overexpression of sineoculis homeobox homolog 1
predicts poor prognosis of hepatocellular carcinoma. Int J Clin Exp
Pathol. 7:3018–3027. 2014.PubMed/NCBI
|
|
27
|
Chen K, Wei H, Pan J, Chen Z, Pan D, Gao
T, Huang J, Huang M, Ou M and Zhong W: Six1 is negatively
correlated with poor prognosis and reduces 5-fluorouracil
sensitivity via attenuating the stemness of hepatocellular
carcinoma cells. Eur J Pharmacol. 861:1725992019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cui Q, Kong D, Li Z, Ahiable P, Wang K, Wu
K and Wu G: Dachshund 1 is differentially expressed between male
and female breast cancer: A matched case-control study of clinical
characteristics and prognosis. Clinical Breast Cancer.
18:e875–e882. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Suen AA, Jefferson WN, Wood CE,
Padilla-Banks E, Bae-Jump VL and Williams CJ: SIX1 oncoprotein as a
biomarker in a model of hormonal carcinogenesis and in human
endometrial cancer. Mol Cancer Res. 14:849–858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jefferson WN, Padilla-Banks E, Phelps JY,
Gerrish KE and Williams CJ: Permanent oviduct posteriorization
after neonatal exposure to the phytoestrogen genistein. Environ
Health Perspect. 119:1575–1582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jefferson WN, Chevalier DM, Phelps JY,
Cantor AM, Padilla-Banks E, Newbold RR, Archer TK, Kinyamu HK and
Williams CJ: Persistently altered epigenetic marks in the mouse
uterus after neonatal estrogen exposure. Mol Endocrinol.
27:1666–1677. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rossini GP, Baldini GM, Villa E and
Manenti F: Characterization of estrogen receptor from human liver.
Gastroenterology. 96:1102–1109. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Villa E, Vukotic R, Cammà C, Petta S, Di
Leo A, Gitto S, Turola E, Karampatou A, Losi L, Bernabucci V, et
al: Reproductive status is associated with the severity of fibrosis
in women with hepatitis C. PLoS One. 7:e446242012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Meng F, Glaser SS, Francis H, DeMorrow S,
Han Y, Passarini JD, Stokes A, Cleary JP, Liu X, Venter J, et al:
Functional analysis of microRNAs in human hepatocellular cancer
stem cells. J Cell Mol Med. 16:160–173. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang B, Hsu SH, Majumder S, Kutay H, Huang
W, Jacob ST and Ghoshal K: TGFbeta-mediated upregulation of hepatic
miR-181b promotes hepatocarcinogenesis by targeting TIMP3.
Oncogene. 29:1787–1797. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou Q, Zheng X, Chen L, Xu B, Yang X,
Jiang J and Wu C: Smad2/3/4 pathway contributes to TGF-β-induced
MiRNA-181b expression to promote gastric cancer metastasis by
targeting Timp3. Cell Physiol Biochem. 39:453–466. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li B, Liu L, Li X and Wu L: miR-503
suppresses metastasis of hepatocellular carcinoma cell by targeting
PRMT1. Biochem Biophys Res Commun. 464:982–987. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xiao Z, Shen J, Zhang L, Li M, Hu W and
Cho C: Therapeutic targeting of noncoding RNAs in hepatocellular
carcinoma: Recent progress and future prospects. Oncol Lett.
15:3395–3402. 2018.PubMed/NCBI
|
|
39
|
Xiao Y, Tian Q, He J, Huang M, Yang C and
Gong L: MiR-503 inhibits hepatocellular carcinoma cell growth via
inhibition of insulin-like growth factor 1 receptor. Onco Targets
Ther. 9:3535–3544. 2016.PubMed/NCBI
|
|
40
|
Yang X, Zang J, Pan X, Yin J, Xiang Q, Yu
J, Gan R and Lei X: miR-503 inhibits proliferation making human
hepatocellular carcinoma cells susceptible to 5-fluorouracil by
targeting EIF4E. Oncol Rep. 37:563–570. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Qiu G, Lin Y, Zhang H and Wu D: miR-139-5p
inhibits epithelial-mesenchymal transition, migration and invasion
of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2.
Biochem Biophys Res Commun. 463:315–321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li J, Fang L, Yu W and Wang Y:
MicroRNA-125b suppresses the migration and invasion of
hepatocellular carcinoma cells by targeting transcriptional
coactivator with PDZ-binding motif. Oncol Lett. 9:1971–1975. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang J, Li Z, Pang Y, Zhou T, Sun J, Cheng
XY and Zheng WV: MicroRNA-139-5p negatively regulates NME1
expression in hepatocellular carcinoma cells. Adv Clin Exp Med.
31:655–670. 2022. View Article : Google Scholar : PubMed/NCBI
|