Introduction
Recently, with improvements in sequencing
technology, information on human cancer genomes has increased.
Tumor mutation burden (TMB) and mutation-derived neoantigens are
novel findings obtained from cancer genome sequencing (1–3).
TMB-high, microsatellite instability-high (MSI-high) or mismatch
repair-deficient (dMMR) tumors, which leads to a number of
neoantigens, are known predictive markers for the efficacy of
immune checkpoint inhibitor (ICI) therapies (4–6);
pembrolizumab became the first drug approved for the treatment of
cancer selectively according to these biomarkers rather than the
primary tumor site. The advent of cancer immunotherapy has been a
revolution for cancer patients, and indications for ICIs and
combination therapies have expanded to various cancer types,
ranging from melanoma, colorectal cancer, gastric cancer,
hepatocellular carcinoma and others over the past decade (7). However, the use of ICIs, which allows
the immune system to comprehensively target antigens, is known to
cause immune-related adverse effects (8), hyperprogressive disease (9) and an increased risk of early mortality
(10); it has also not benefited
for the patients with low TMB tumors. On the other hand,
tebentafusp, which targets specific cancer antigen epitopes on
human leucocyte antigen (HLA), has also entered clinical use
(11). Therefore, in the
development and selection of immune-oncology therapeutics, the
detection of whether a tumor has a mutation that can be targeted by
the patient's immune system has become a crucial factor.
The majority of neoantigens are derived from
passenger mutations that are not involved in carcinogenesis, and
they are more likely to exert immunogenic effects than to be cancer
driver mutations (12). In general,
driver mutations are considered to be difficult antigens to target,
as they are associated with cancer development and exert
immunomodulatory activity in the tumor microenvironment (13–15).
Nevertheless, researchers have successfully cloned and used driver
mutation-specific tumor-infiltrating T-cell (TIL) or T-cell
receptor (TCR) repertoires for adoptive immunotherapy (16,17).
Small clinical trials of immunotherapy using driver
mutation-derived neoantigens have been performed (18,19),
and moderate antitumor effects have been verified. In particular,
Chen et al (20)
demonstrated that vaccines targeting common driver mutations are
applicable to metastatic cancers, and the combination of vaccines
and immunomodulatory therapies may constitute a promising regimen.
However, the combined diversity of HLAs, antigen-presenting
molecules and mutation-derived epitopes is a major obstacle to the
clinical development of cancer vaccines. Although the accurate
prediction of epitopes is an essential component of personalized
immunotherapy, it is difficult to validate in silico
predicted epitopes as human T-cell clonal diversity cannot be
reproduced in vitro or in vivo.
To confirm predicted antigen epitopes, immunological
and biochemical approaches such as major histocompatibility complex
(MHC) stabilization assays or mass spectrometry (MS)-mediated
identification have been developed based on HLA-B-, HLA-C-null T2
cells (CVCL_2211) (21) or
HLA-class-I-null LCL721.221 cells (CVCL_6263, ATCC CRL-1855), to
which HLA-alleles have been transferred (22–24);
however, HLA-C expression in LCL721.221 cells was not found to be
completely abrogated (25);
therefore, this cell line was discontinued by the American Type
Culture Collection (ATCC). The present study generated antigen
peptide transporter 2 (TAP2)-knockout (KO) and monoallelic
HLA-class-I-expressing B-cell lines derived from the TISI cell line
(CVCL_E851) (26) for MHC
stabilization assays, screened frequent hot spot mutations in 5,143
cancer patients treated at the Shizuoka Cancer Center Hospital and
identified driver mutation-based neoantigen epitope candidates.
Materials and methods
Comprehensive cancer research project
HOPE
A total of 5,143 cancer cases were analyzed in the
High-tech Omics-based Patient Evaluation (HOPE) project, which has
been conducted in Shizuoka Cancer Center since 2014 using
multiomics analyses, such as whole-exome sequencing (WES) and gene
expression profiling (27). For the
WES analysis, somatic mutations were identified by comparing data
on tumors and corresponding blood samples. Total exonic mutations
for each sequenced tumor included single-nucleotide variants and
indel/frameshift mutations. All nonsynonymous mutations detected in
the cases were collected and screened.
Prediction of epitope peptides in
silico
We used the NetMHCpan 4.1 (Immune Epitope Database,
http://www.iedb.org/) to assess HLA presentation
of 8 to 11-mer peptide epitopes from cancer driver gene mutation to
predict affinity (IC50) on the basis of the binding
affinity (BA) model or to predict rank scores (%Rank) on the basis
of the eluted ligand (EL) model.
Cell lines
TISI human B-lymphoblastoid cell line (B-LCL)
(CVCL_E851) with homozygous HLA-class-I loci (26) were supplied by Takara Shuzo Co.,
Ltd. (Otsu, Shiga, Japan), and we verified the HLA types by Sanger
sequencing using SeCore SBT kit (#5300025, One Lambda, Thermo
Fisher Scientific, Waltham, Massachusetts, USA). T2 cells were
purchased from American Type Culture Collection (#CRL-1992, ATCC,
Manassas, VA, USA). These cell lines were maintained in RPMI-1640
medium (#R8758, Sigma-Aldrich, Merck KGaA, Darmstadt, Germany)
supplemented with 10% (v/v) FBS (#10437, Gibco, Thermo Fisher
Scientific, Waltham, Massachusetts, USA) and 1×
penicillin-streptomycin (#151401222, Gibco). Newly constructed
TAP2-KO HLA-ABC monoallelic cell clones were maintained in
RPMI-1640 medium supplemented with 10% (v/v) FBS, 100 units/ml
penicillin, 100 µg/ml streptomycin, 1 mM sodium pyruvate
(#11360070, Gibco), 1× MEM nonessential amino acids (#11140050,
Gibco), and 55 µg/ml 2-mercaptoethanol (#21985-023, Gibco).
Flow cytometry and antibodies
Monoallelic cell cloning was performed using a
PE-labeled anti-HLA-ABC monoclonal antibody (clone: G46-2.6, BD
Biosciences, Franklin Lakes, New Jersey, USA) to stain HLA-ABC KO
or knock-in (KI) cells and a FACSAria cell sorter (BD Biosciences)
for sorting single cells to 96-well plates. For the MHC
stabilization assay, we used PE-labeled anti-HLA-ABC antibody clone
G46-2.6 diluted 1:10 and clone W6/32 (Dako Denmark A/S, Dako North
America, Inc., Carpinteria, California, USA) diluted 10 µg/ml with
FACSCanto flow cytometer (BD Biosciences) for analysis. PE-labeled
anti-mouse Ig polyclonal antibody (#550589, BD Biosciences) was
used as the secondary antibody. FlowJo software ver.8.8.7 (Tomy
digital biology Co., Ltd. Tokyo, Japan) was used for analysis.
Construction of TAP2-deficient and HLA
class I monoallelic cell lines
Using the CRISPR/Cas9 system (Invitrogen, Thermo
Fisher Scientific), TAP2 and HLA-A, HLA-B, and
HLA-C genes were knocked out in the TISI cell line using
synthetic gRNAs (#A35510, Invitrogen) (Table SI). Each synthetic HLA-class-I
allele cDNA (Genewiz, Tokyo, Japan) corresponding to a
TISI-HLA-A*24:02:01:01-deleted site was transcribed to
single-stranded DNA and isolated from double strand using agarose
gel (#50070, Lonza) electrophoresis with ethidium bromide
(#315-90051, Nippongene), and Purified single-stranded DNA was
knocked in the TAP2- and HLA-ABC-KO TISI subclone using a paired
sgRNA. Each KO and KI site was confirmed by PCR and Sanger
sequencing. All sgRNAs and primer pairs are shown in Tables SI and SII.
Reagents and solvents for peptide
synthesis
Fmoc-amino acids were obtained from CEM Corp.
(Matthews, NC, USA). Fmoc-amino acid-Wang-resins were obtained from
Gyros Protein Technologies Inc. (Tucson, AZ, USA). H-Pro-Barlos
resin and reagents for peptide synthesis,
1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium
3-oxide hexafluorophosphate (HATU), N,N'-diisopropylcarbodiimide
(DIC), Oxyma Pure [Ethyl-2-cyano-2-(hydroxyimino)acetate],
N,N-diisopropylethylamine (DIPEA), trifluoroacetic acid (TFA),
triisopropylsilane (TIPS), and methylene chloride (DCM), were
purchased from Watanabe Chem. IND., LTD. (Hiroshima, Japan).
Piperidine, pyrrolidine, phenol and ethanedithiol (EDT) were
obtained from Fujifilm Wako Pure Chem. Corp. (Tokyo, Japan).
N,N'-Dimethylformamide (DMF), N-methyl-2-pyrrolidone (NMP), HPLC
grade acetonitrile (ACN) and water were obtained from Kanto Chem.
Co. Inc. (Tokyo, Japan).
Peptide synthesis
Neoantigen peptides were synthesized via solid-phase
chemistry using an Fmoc protection strategy. Starting from 0.05
mmol of Fmoc-amino acid-Wang-resin, protected peptide resins were
constructed using 0.5 mmol of Fmoc-amino acids with
tert-butyl-based side-chain-protecting groups on peptide
synthesizers with the peptide synthesizer Tribute (Gyros Protein
Technologies Inc.) at room temperature or with the microwave
peptide synthesizer Liberty PRIME (CEM Corp.) at high temperature
(105°C). As Fmoc removal (deprotection) and peptide
bond formation (coupling) reagents, 20% piperidine/NMP and
HATU/DIPEA for Tribute and 20% pyrrolidine/DMF and DIC/Oxyma for
Liberty PRIME, respectively, were used. The synthesis of peptides
bearing Pro at the C-terminus was carried out starting with
H-Pro-Barlos-resin using a Tribute synthesizer. After final
deprotection of the Nα-Fmoc group of the N-terminal amino acid, the
partially protected peptide resin obtained was washed with DCM and
MeOH and dried in vacuo. Removal of side-chain-protecting
groups and cleavage of the resin were performed by treatment with a
TFA-containing scavenger cocktail
(TFA-phenol-EDT-thioanisole-H2O-TIPS; 90-2-2-2-2-2) at room
temperature for 1.5-3 h. The product was precipitated with ether,
collected by centrifugation, and dried in vacuo. The
resulting crude peptides were purified using a 1525 Binary HPLC
Pump equipped with a 2489UV/VIS detector (Waters Corp., Milford,
MA, USA) with a YMC-Actus Triart C18 column, 5 µm, 20×150 mm (YMC
Corp. Kyoto, Japan). The purity and mass spectrum of the purified
peptides were analyzed using an Alliance e2695 HPLC System equipped
with a 2489UV/VIS detector and an ACQITY QDa mass detector (Waters
Corp.) with an XSelect CSH C18 column, 2.5 µm, 3×75 mm column
(Waters Corp.). All synthetic peptides are shown in Table SIII.
MHC stabilization assay
The TAP2-KO monoallelic HLA class I-expressing TISI
cell lines were precultured with 10,000 U interferon (IFN)-α
(Smiferon300, Sumitomo Pharma, Osaka, Japan) to restore baseline
HLA expression. 1×105 HLA monoallelic cells were
incubated with 25 µM peptide in RPMI 1640 medium with 3 µg/ml
beta-2 microglobulin (#21985023, Sigma-Aldrich), 55 µM
2-melcaptoethanol and 0.1% BSA (#A7906, Sigma-Aldrich) at
37°C with 5% CO2 for 18 h. Cells cultured with
0.5% DMSO without peptide were used as negative controls. Following
incubation, the cells were stained with PE-labeled anti-HLA-ABC
antibody (clone: G46-2.6) diluted 1:10 at RT (23°C) for 60
min, washed 3 times with PBS supplemented with 0.5% BSA and 2 mM
EDTA at 4°C, and fixed with 0.4% paraformaldehyde in PBS
with 0.1% BSA and 2 mM EDTA. HLA class I expression increase (ΔHLA)
was measured using a FACSCanto flow cytometer (BD Biosciences). The
ΔHLA was obtained by calculating the geometric mean of the
fluorescence intensity (MFI) using the following formula:
ΔHLA (% control)={(Sample MFI)-(DMSO cont.
MFI)}/{(DMSO cont. MFI)-(Unstained background MFI)} ×100
Statistical analysis
To determine the significance of differences between
HLA expression levels, Kruskal-Wallis test followed by Steel's
multiple comparison test was performed and P<0.05 was considered
to indicate a statistically significant difference. The
Kruskal-Wallis test, the Steel's multiple comparison test, and
Pearson or Spearman correlation coefficients were calculated using
EZR version 1.55 (Saitama Medical Center, Jichi Medical University,
Saitama, Japan) (28). All means
and standard deviations in the tables are based on three or more
independent experiments.
Results
Generation of TAP2-KO HLA-class-I
monoallelic B-lymphoblastoid cell lines
The MHC stabilization assay was improved with the
use of HLA-partially-null B-lymphoblastoid cell lines to confirm
epitope peptide presentation by HLA molecules (21,22,24).
To perform a more reliable MHC stabilization assay, HLA-A-,
HLA-B-, HLA-C- and TAP2-KO clones derived from the TISI
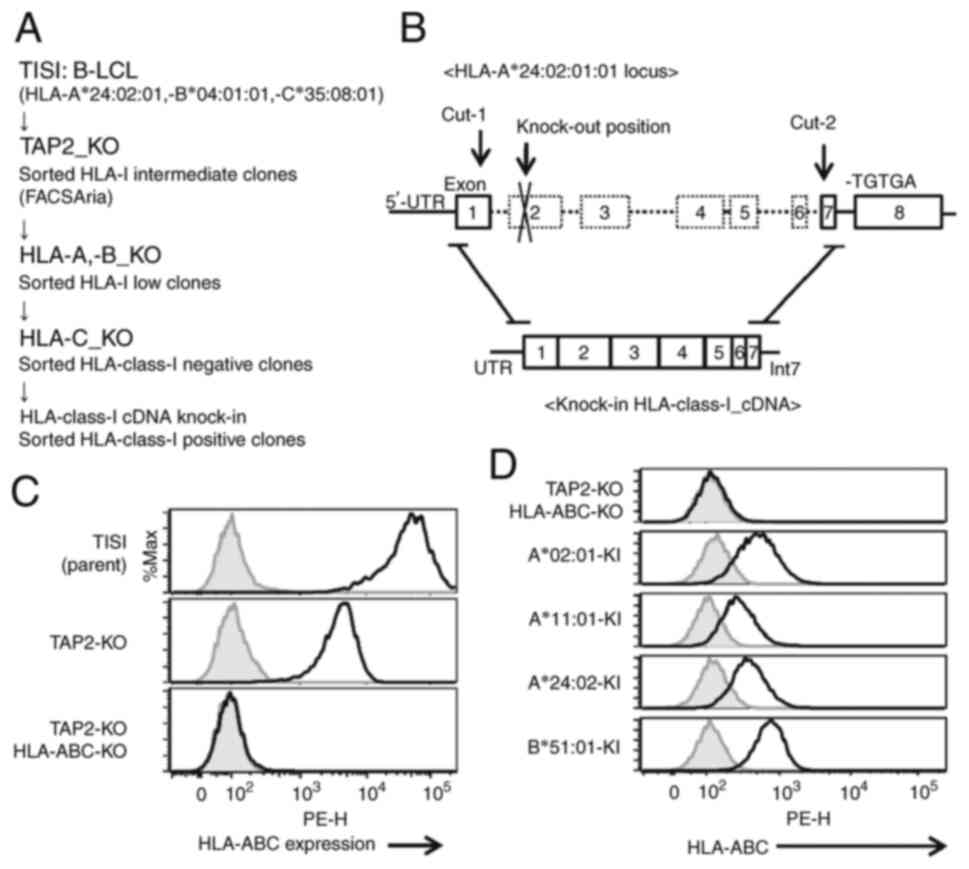
cell line were generated (Fig. S1,
Fig. S2, Fig. S3) and the respective single HLA
alleles were knocked in using the CRISPR/Cas9 system (Fig. S4). TISI cells were selected as they
constitute a B-lymphocyte line with the ability to cross-present
exogenous antigens on HLA molecules and carry homologous
HLA-class-I loci. The HLA allele cDNA was knocked in the loci of
HLA-A*24:02:01 (Fig. 1A and B). HLA
class I expression was not detected in the TAP2, HLA-A, HLA-B and
HLA-C-KO TISI clone (Fig. 1C), and
the cells subjected to HLA class I cDNA knock-in exhibited HLA
class I on their cell surface (Fig.
1D). Each clone was confirmed using genomic DNA sequencing.
Validation of the MHC stabilization
assay using T2 cells and the newly generated monoallelic HLA
cells
The principle of the MHC stabilization assay is that
peptides binding to HLA molecules stabilize and increase HLA
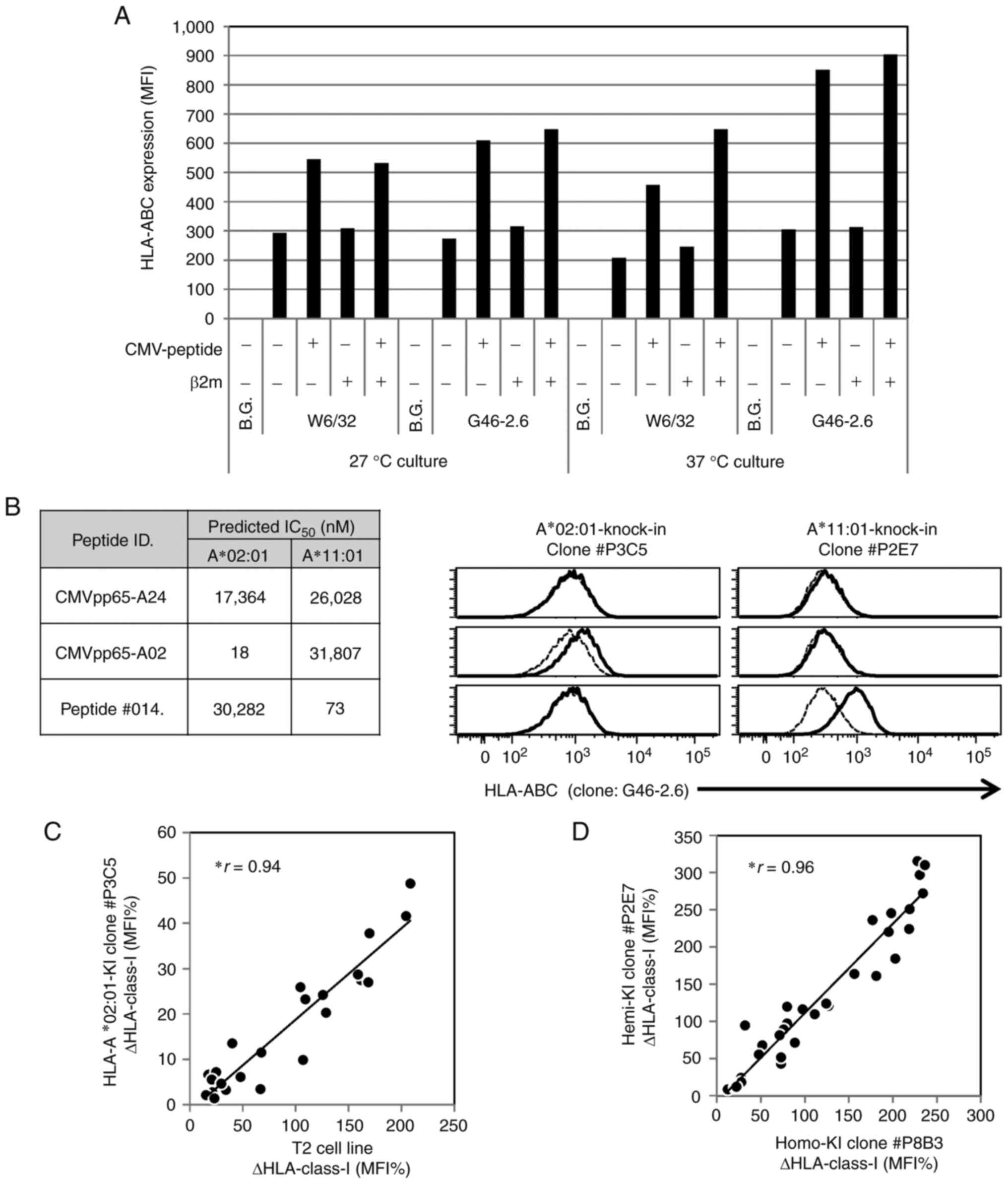
expression levels on the cell surface. The first study was based on
TAP2-deficient and HLA-A*02:01 monoallelic T2 cells and antibody
clone W6/32, which can bind HLA-ABC associated with β-2
microglobulin (β2m) and requires 26°C culture conditions to prevent
β2m dissociation (21,29). On the other hand, antibody clone
G46-2.6 binds to HLA-ABC with or without β2m, which permits
evaluation under physiological 37°C culture conditions, preventing
low-affinity peptide binding at a low temperature (30) or the effect of bovine β2m in FBS
(31). The present study therefore
used clone G46-2.6, which enabled clearer observations. The
addition of β2m to the assay culture supernatant was necessary for
the detection of HLA by antibody clone W6/32, although it exerted a
minimal effect on HLA expression (Figs.
2A and S5). In addition,
interferons in the culture supernatant can easily alter HLA
expression levels. Since HLA expression was extremely low
immediately following knock-in cell cloning, the assays were
performed after culturing the cells with IFN-α for 1 week to
restore the HLA expression levels.
The results revealed increased HLA expression levels
(ΔHLA) in MHC stabilization assays using the in-house-generated
monoallelic HLA knock-in cell lines, although the predicted
peptide/HLA IC50 did not necessarily associate with ΔHLA
(Fig. 2B). To assess the usability
of the novel monoallelic HLA cells, the MHC stabilization assays
were compared between the T2 cells and the monoallelic HLA-A*02:01
knock-in cell line using synthetic peptides (Table SIII). Although the knock-in cell
line exhibited low HLA-A expression levels, it exhibited a good
correlation with the T2 cells in the original method (Fig. 2C). Furthermore, there was a slight
difference between the HLA-A*11:01 homo knock-in and hemi knock-in
cell lines (Fig. 2D).
Screening of amino acid substituted
(AAS) driver mutations in 5,143 cancer patients
The HOPE cohort comprised 5,521 tumor specimens
derived from 5,143 patients treated at the Shizuoka Cancer Center
Hospital between January, 2014 and March, 2019. The major cancer
types were colon (18.4%), lung (16.5%), rectal (13.3%) and stomach
(10.8%) cancers (27). As
cancer-driving gene mutations are high-frequency mutations at
specific sites in the genome due to evolutionary convergence
(32), mutation frequency data were
used to select the driver mutations assessed in the present study.
This approach is consistent with the objective of screening for
mutations that are common in the cancer patient population.
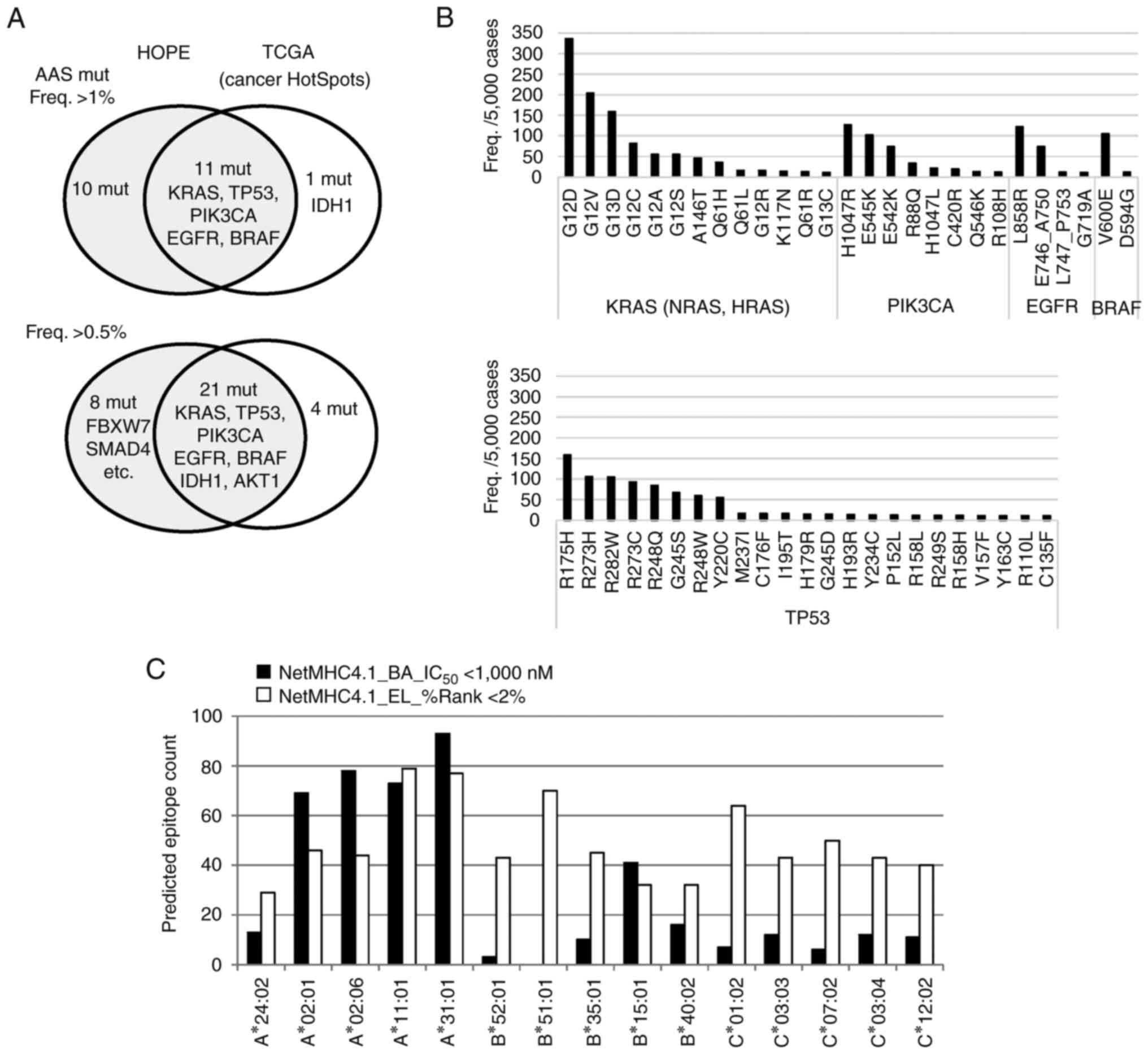
The results revealed 21 AAS mutations with >1%
frequency in the HOPE cohort, and these mutations were found in
only five cancer driver genes (Fig.
3A). The highly frequent mutations mostly overlapped with those
in The Cancer Genome Atlas (TCGA) Cancer HotSpots (33). In the Japanese population, the KRAS
G12S and G13D mutants were the most common, whereas KRAS G12R was
less common. EGFR mutations were also more common in the Japanese
population; however, as the resected primary tumors were the main
samples, the EGFR T790M mutation associated with drug resistance
was found in only 1 patient in this cohort (Table SIV). The eight most frequent TP53
AAS mutations were prominent at six positions. TP53 is a
tumor suppressor gene; however, these dominant-negative mutations
appear to be high-frequency targets as the mutants function as
oncogenes (Fig. 3B).
The present study first evaluated the frequent
mutation-derived AAS epitope presentation on HLA molecules through
the NetMHCpan4.1 binding-affinity (BA) model and the eluted-ligand
(EL) model to predict binding affinities (IC50) and rank
scores (%Rank). In the Japanese dominant HLA alleles (34), a significant number of peptides were
predicted to have a high affinity for HLA-A and a smaller number
for HLA-B and HLA-C (Fig. 3C).
Correlation of the in silico
prediction of AAS epitopes with MHC stabilization assays using the
newly generated cell lines
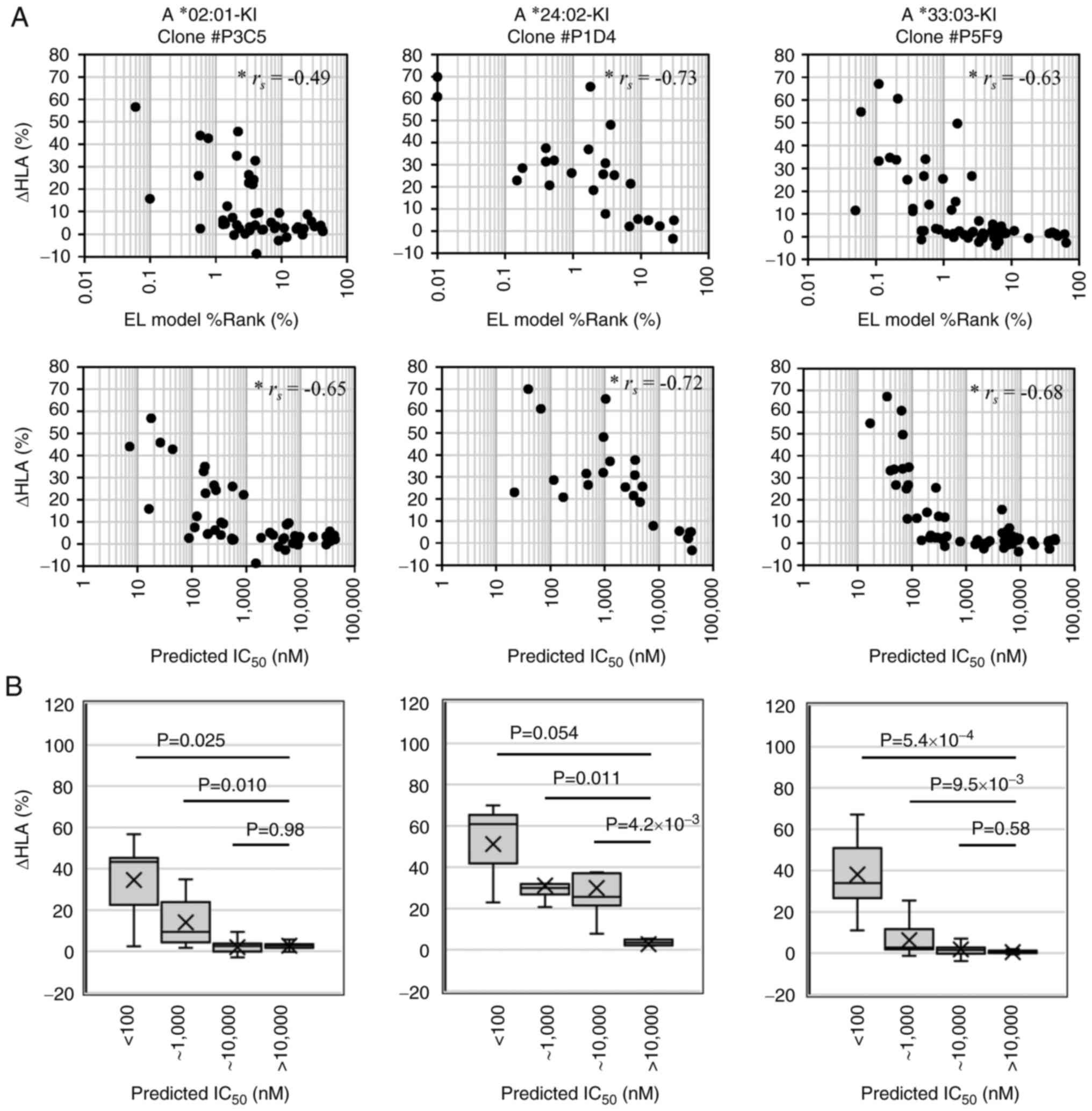
The present study compared in silico
predictions and in vitro assays of the epitopes derived from
driver mutations with >0.2% frequency. A total of 138 epitope
peptides were synthesized with a predicted affinity
(IC50) <500 nM or IC50 <1,000 nM and
%Rank <1% for the 10 most frequently appearing HLA-A, -B and -C
alleles in the Japanese population, and two peptides derived from
cytomegalovirus (Table SIII). For
each HLA allele, synthesized peptides with predicted
IC50 <10,000 nM were used for the assays, and six to
eight peptides with predicted IC50 >10,000 nM were
used as negative controls. All assay results are presented in
Table SV.
As was expected, ΔHLA correlated with the predicted
scores and affinities, particularly for HLA-A*33:03 with the
peptide predicted IC50 <1,000 nM (Figs. 4 and 5). The BA model IC50 <1,000
nM or the EL model %Rank <1% are often used as cut-off criteria
in the epitope predictions; however, for HLA-A*24:02 and
HLA-A*11:01, peptides with predicted IC50 >1,000 nM
or with %Rank ranging from 2 to 4% were found to increase HLA
expression. Of note, two 9-mer epitopes derived from TP53
driver mutations exhibited extremely high responses to HLA-A*11:01
despite predicted IC50 >5,000 nM, which was more
notable than the two 10-mer epitopes with higher predicted
affinities (Fig. 5); thus, these
mutations are potential new target candidates. Furthermore, the
majority of the peptides, including the KRAS G12 or G13 mutation
and rich in valine were presented on HLA-A*11:01 regardless of the
variant of the mutation.
Candidate epitopes based on driver
mutations identified through the MHC stabilization assay
The epitopes candidates were screened on the basis
of the following criteria (Fig. 6):
BA_IC50 <1,000 nM, EL_%Rank <1%, no proline in the
three forward positions to interfere with peptide processing
(35), cysteine content <2
molecules (36,37) and ΔHLA >20% in the MHC
stabilization assay with the synthetic peptides. Finally, 27
candidate epitopes were identified in the present study (Table I).
 | Table I.Candidate 27 epitopes of driver
mutations |
Table I.
Candidate 27 epitopes of driver
mutations
|
|
|
|
| HLA |
|
|---|
|
|
|
|
|
|
|
|---|
|
|
|
|
|
|
|
| ΔHLAc, % |
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| No. | AAS mutation | Freq. a |
Sequenceb | Allele | BA_IC50,
nM | EL_Rank, %Rank | Average | ±SD | Estimated
patientd, % |
|---|
| 1 | KRAS_G12D | 336 | VVGADGVGK | A*11:01 | 172 | 0.35 | 203 | 62 | 1.14 |
| 2 | KRAS_G12D | 336 | VVVGADGVGK | A*11:01 | 194 | 0.59 | 236 | 69 | 1.14 |
| 3 | KRAS_G12V | 204 | VVGAVGVGK | A*11:01 | 39 | 0.10 | 230 | 77 | 0.69 |
| 4 | KRAS_G12V | 204 | VVVGAVGVGK | A*11:01 | 69 | 0.35 | 228 | 69 | 0.69 |
| 5 | KRAS_G13D | 159 | VVVGAGDVGK | A*11:01 | 213 | 0.59 | 195 | 59 | 0.54 |
| 6 | KRAS_G13D | 159 | VVGAGDVGK | A*11:01 | 314 | 0.60 | 181 | 43 | 0.54 |
| 7 | KRAS_G12C | 82 | VVGACGVGK | A*11:01 | 73 | 0.52 | 234 | 73 | 0.28 |
| 8 | KRAS_G12C | 82 | VVVGACGVGK | A*11:01 | 122 | 0.59 | 123 | 30 | 0.28 |
| 9 | KRAS_G12A | 55 | VVGAAGVGK | A*11:01 | 60 | 0.18 | 218 | 65 | 0.19 |
| 10 | KRAS_G12S | 55 | VVGASGVGK | A*11:01 | 51 | 0.15 | 219 | 67 | 0.19 |
| 11 | TP53_R175H | 159 | VVRHCPHHER | A*33:03 | 79 | 0.29 | 25 | 8 | 0.45 |
| 12 | TP53_R175H | 159 | EVVRHCPHHER | A*33:03 | 88 | 0.16 | 35 | 3 | 0.45 |
| 13 | TP53_R248Q | 85 | SSCMGGMNQR | A*11:01 | 171 | 0.49 | 89 | 10 | 0.29 |
| 14 | TP53_G245S | 67 | SSCMGSMNRR | A*11:01 | 145 | 0.78 | 28 | 6 | 0.23 |
| 15 | TP53_G245S | 67 | SSCMGSMNR | A*11:01 | 95 | 0.54 | 107 | 31 | 0.23 |
| 16 | TP53_R248W | 60 | NWRPILTII | A*24:02 | 945 | 0.53 | 32 | 6 | 0.71 |
| 17 | TP53_Y220C | 55 | VVPCEPPEV | A*02:01 | 564 | 0.55 | 26 | 7 | 0.23 |
| 18 | PIK3CA_H1047R | 127 | FMKQMNDAR | A*33:03 | 68 | 0.54 | 34 | 13 | 0.36 |
| 19 | PIK3CA_H1047R | 127 | YFMKQMNDAR | A*33:03 | 51 | 0.51 | 27 | 10 | 0.36 |
| 20 | PIK3CA_H1047R | 127 | EYFMKQMNDAR | A*33:03 | 64 | 0.21 | 61 | 15 | 0.36 |
| 21 | PIK3CA_E545K | 102 | STRDPLSEITK | A*11:01 | 92 | 0.04 | 80 | 33 | 0.35 |
| 22 | PIK3CA_E542K | 74 | AISTRDPLSK | A*11:01 | 99 | 0.25 | 80 | 9 | 0.25 |
| 23 | PIK3CA_E542K | 74 | ISTRDPLSK | A*11:01 | 333 | 0.48 | 124 | 30 | 0.25 |
| 24 | PIK3CA_E542K | 74 | KAISTRDPLSK | A*11:01 | 256 | 0.22 | 127 | 29 | 0.25 |
| 25 | EGFR_L858R | 122 | KITDFGRAK | A*11:01 | 101 | 0.19 | 52 | 11 | 0.41 |
| 26 |
EGFR_E746_A750del | 74 | AIKTSPKANK | A*11:01 | 423 | 0.54 | 73 | 13 | 0.25 |
| 27 | BRAF_V600E | 105 | KIGDFGLATEK | A*11:01 | 108 | 0.28 | 98 | 4 | 0.36 |
Discussion
Although, the cancer antigen targeted DC vaccine was
once approved by the FDA (38), the
paradigm shift in immuno-oncology represented by T-cell-targeted
immune checkpoint blockade (ICB) therapy has now made the cancer
vaccines a niche. The ICB is highly effective for TMB-high or
MSI-high tumors, and numerous clinical trials combining ICI and
chemotherapy are underway (39).
However, approaches targeting a specific cancer antigen have
continued. A considerable number of cancer vaccine studies based on
neoantigen and/or dendritic cells have demonstrated more than
marginal antitumor effects; however, these peptides and vaccines
exhibited more potent and more persistent antitumor effects when
used in combination with immunomodulatory chemotherapy (20,40).
Moreover, bi-specific antibody targeting the cancer-associated
antigen epitope, restricted by a HLA allele, was approved by the
FDA (11), and similar agents
targeting driver mutations are also under development (41,42).
Cancer driver mutation-targeted immunotherapies are
crucial in the field of clinical oncology, and validating the
immunogenicity of neoantigen epitopes is critical, yet difficult in
preclinical studies. Therefore, candidate cancer antigens
identified via in silico prediction are being evaluated in
immune cell-based assays. To confirm epitope peptide presentation
on HLA molecules, methods such as isotope-labeled peptide binding
to purified HLA molecules, MHC stabilization assays or MS-based
assays have been performed. However, simple in vitro
affinity measurements are not considered reliable, as peptide
loading on HLA molecules is based on a peptide-loading complex in
the endoplasmic reticulum (43).
Moreover, the expression of multiple HLA-class-I alleles renders
epitope prediction unreliable. The MHC stabilization assay with a
single-HLA-expressing cell line is an efficient immunological tool
with which to evaluate the CTL induction activity of antigen
peptides (44). Research using HLA
allele knock-in cells generated from the HLA-A- and HLA-B-null
LCL721.221 cell line has reported large numbers of autoantigen
epitopes with high affinity for HLA molecules based on analysis
using high-throughput mass spectrometry systems (23). Furthermore, Kaseke et al
(24) knocked out the TAP1
gene to generate cells that can be used for MHC stabilization
assays in evaluating viral antigens. The present study successfully
generated the novel TAP2-KO and completely HLA-ABC null
clones, as well as the HLA-A or HLA-B monoallelic clones with the
B-lymphoblastoid TISI cell line using the CRISPR/Cas9 system to
ensure the reliability of the MHC stabilization assay (Fig. 1). These monoallelic HLA-expressing
cell lines were used to evaluate candidate epitopes for use as
cancer vaccines.
Since the development of next-generation sequencing
allowed for the identification of neoantigens derived from
mutations, studies have been conducted to discover and characterize
neoantigens (1,45,46).
As a result, a number of neoantigens that may contribute to the
prediction of a favorable response to ICB therapy have been
identified thus far, and the majority of these neoantigens have
been demonstrated to be products of passenger mutations (47,48).
Passenger mutations are characterized as being i) diverse and
abundant, but not conserved; and ii) likely to exhibit a decreased
expression in recurrent cancers following ICB treatment and to not
produce persistent cancer antigens (49,50).
By contrast, driver mutations are exclusive and highly persistent,
and maintained even at recurrent or metastatic tumor sites, and
their functional inhibitions suppress tumor growth (51,52).
Based on these advantages of driver mutations, small clinical
trials of immunotherapy have been performed using driver
mutation-derived neoantigen peptides or mRNA vaccines with or
without ICB therapy (3), and the
study (53) has reported that
highly immunogenic mutations tend to be less likely to appear, even
in oncogenes. However, KRAS- or p53-driver mutation-specific
T-cells or TCR repertoires have been recently identified and
utilized in patients with metastatic cancer as therapeutics, and a
certain degree of efficacy has been observed (16,54,55).
Therefore, frequent driver mutations are potential therapeutic
targets. The present study screened frequent amino acid-substituted
somatic mutations from the HOPE cohort. KRAS and EGFR
mutations were more common in the HOPE cohort than in the TCGA
cohort, apart from KRAS-G12R. (Table SIV). This finding is consistent
with the fact that approximately half of the cases were colon,
rectal and lung cancers, and EGFR mutations are common among
Japanese non-smoking female patient with lung adenocarcinoma
(56).
In the cancer patients who visited the Shizuoka
Cancer Center Hospital, driver mutation-derived 27 candidate
epitopes for the top four most frequent HLA-A alleles (Table I) were obtained by the procedure
illustrated in Fig. 6 with some
notable results. First, multiple targets were identified in
HLA-A*11:01 and HLA-A*33:03, and the most frequent KRAS G12 and G13
mutation-derived hydrophobic epitopes were included among these
targets. The phenotypic frequency of the A*11:01 allele was 17% in
the Japanese population and ~12% in the European Caucasian
population; the majority of tumors with KRAS mutations were
targetable in these populations. Second, when comparing the in
silico-predicted IC50s with the HLA stabilization assay
results, the epitope prediction for HLA-A*24:02 was underestimated
compared to that for A*02:01, and that for A*33:03 was
overestimated (Fig. 4 and Table SV). Third, strong antigen
presentation was observed at two epitopes derived from TP53
mutations (Fig. 5) that were
excluded from the candidate list (Table
I), due to their predicted very low affinity, and this suggests
that there may be some high-affinity epitopes among the peptides
with low predicted affinities. HLA-A*24:02 is the most frequently
expressed allele in East Asian populations, including Japanese.
Although there is only one HLA-A*24:02-binding epitope included in
the target list (Table I),
experimental evaluation is likely to uncover additional
mutation-derived potential epitopes. It may be possible to expand
the number of driver mutation-derived targets. The accumulation of
immunocytological data using such completely HLA monoallelic cells
may be useful to improve the prediction of epitopes presented on
HLA. More accurate prediction and validation of immune targetable
driver mutations would complement the development of novel
therapeutics through personalized procedure.
The HLA multiplex and polymorphisms also cause
graft-vs.-host disease and that is a major obstacle in cellular
immunotherapy and regenerative medicine; HLA haplobanks, HLA
monoallelic gene expression and HLA knock-in mice may further
facilitate research and development (57–59).
For clinical applications, the potential of antigenic epitopes
needs to be carefully confirmed by CTL induction and/or humanized
mouse models. In the future, the authors aim to perform a cancer
immunotherapy study targeting these epitopes against solid
tumors.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from JSPS KAKENHI
(grant no. 20K07649), Japan.
Availability of data and materials
The somatic gene alterations in the HOPE cohort are
available from the National Bioscience Database Center Human
Database (Research ID, hum0127) as VCF or TSV format files
(https://humandbs.biosciencedbc.jp/en/). The raw data
supporting the conclusions of this article are available from the
corresponding author upon reasonable request.
Authors' contributions
AI and YA were major contributors to the conception
and design of the study, as well as in the drafting of the
manuscript and were responsible for completing the study. TN, KUr,
YS, KO, AS, YO, MTe, KUe, TM, YH, SY, HKa, TS, MTa, HKe and KY
performed patient data acquisition and analysis, and TN, KO and TS
contributed to these analyses and interpretations. NS, AK and YK
contributed to the synthesis and supply of the peptides. AI
performed the development of methodology, generated the genome
editing cell lines and acquired the MHC stabilization assay data.
AI and YA confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The experimental clinical research protocols of the
present study were approved by the Institutional Review Board at
the Shizuoka Cancer Center (Authorization no. 25–33). Written
informed consent was obtained from all patients enrolled in the
present HOPE project study. All experiments using clinical samples
were performed in accordance with the Japanese ethical guidelines
for human genome/gene analysis (Ministry of Health, Labour and
Welfare 2017, http://www.mhlw.go.jp/stf/seisakunitsuite/bunya/hokabunya/kenkyujigyou/i-kenkyu/index.html).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
MHC
|
major histocompatibility complex
|
|
HLA
|
human leukocyte antigen
|
|
TCR
|
T-cell receptor
|
|
AAS
|
amino acid substitution
|
|
TCGA
|
The Cancer Genome Atlas
|
|
ICI
|
immune checkpoint inhibitor
|
|
ICB
|
immune checkpoint blockade
|
References
|
1
|
Schumacher TN and Schreiber RD:
Neoantigens in cancer immunotherapy. Science. 348:69–74. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yarchoan M, Hopkins A and Jaffee EM: Tumor
mutational burden and response rate to PD-1 inhibition. N Engl J
Med. 377:2500–2501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Blass E and Ott PA: Advances in the
development of personalized neoantigen-based therapeutic cancer
vaccines. Nat Rev Clin Oncol. 18:215–229. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
De Mattos-Arruda L, Vazquez M, Finotello
F, Lepore R, Porta E, Hundal J, Amengual-Rigo P, Ng CKY, Valencia
A, Carrillo J, et al: Neoantigen prediction and computational
perspectives towards clinical benefit: Recommendations from the
ESMO Precision Medicine Working Group. Ann Oncol. 31:978–990. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McGranahan N, Furness AJ, Rosenthal R,
Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak
NJ, Hiley CT, et al: Clonal neoantigens elicit T cell
immunoreactivity and sensitivity to immune checkpoint blockade.
Science. 351:1463–1469. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rizzo A, Ricci AD, Di Federico A, Frega G,
Palloni A, Tavolari S and Brandi G: Predictive biomarkers for
checkpoint inhibitor-based immunotherapy in hepatocellular
carcinoma: Where do we stand? Front Oncol. 11:8031332021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Morad G, Helmink BA, Sharma P and Wargo
JA: Hallmarks of response, resistance, and toxicity to immune
checkpoint blockade. Cell. 184:5309–5337. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schneider BJ, Naidoo J, Santomasso BD,
Lacchetti C, Adkins S, Anadkat M, Atkins MB, Brassil KJ, Caterino
JM, Chau I, et al: Management of immune-related adverse events in
patients treated with immune checkpoint inhibitor therapy: ASCO
Guideline Update. J Clin Oncol. 39:4073–4126. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park HJ, Kim KW, Won SE, Yoon S, Chae YK,
Tirumani SH and Ramaiya NH: Definition, incidence, and challenges
for assessment of Hyperprogressive disease during cancer treatment
with immune checkpoint inhibitors: A systematic review and
Meta-analysis. JAMA Netw Open. 4:e2111362021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Viscardi G, Tralongo AC, Massari F,
Lambertini M, Mollica V, Rizzo A, Comito F, Di Liello R, Alfieri S,
Imbimbo M, et al: Comparative assessment of early mortality risk
upon immune checkpoint inhibitors alone or in combination with
other agents across solid malignancies: A systematic review and
meta-analysis. Eur J Cancer. 177:175–185. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dhillon S: Tebentafusp: First approval.
Drugs. 82:703–710. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schumacher TN, Scheper W and Kvistborg P:
Cancer neoantigens. Annu Rev Immunol. 37:173–200. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kortlever RM, Sodir NM, Wilson CH,
Burkhart DL, Pellegrinet L, Brown Swigart L, Littlewood TD and Evan
GI: Myc cooperates with Ras by programming inflammation and immune
suppression. Cell. 171:1301–1315. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Ma JA, Zhang HX, Jiang YN and Luo
WH: Cancer vaccines: Targeting KRAS-driven cancers. Expert Rev
Vaccines. 19:163–173. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takeuchi Y, Tanegashima T, Sato E, Irie T,
Sai A, Itahashi K, Kumagai S, Tada Y, Togashi Y, Koyama S, et al:
Highly immunogenic cancer cells require activation of the WNT
pathway for immunological escape. Sci Immunol. 6:eabc64242021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tran E, Robbins PF, Lu YC, Prickett TD,
Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, et al:
T-Cell transfer therapy targeting mutant KRAS in cancer. N Engl J
Med. 375:2255–2262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chandran SS, Ma J, Klatt MG, Dündar F,
Bandlamudi C, Razavi P, Wen HY, Weigelt B, Zumbo P, Fu SN, et al:
Immunogenicity and therapeutic targeting of a public neoantigen
derived from mutated PIK3CA. Nat Med. 28:946–957. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li F, Deng L, Jackson KR, Talukder AH,
Katailiha AS, Bradley SD, Zou Q, Chen C, Huo C, Chiu Y, et al:
Neoantigen vaccination induces clinical and immunogenic responses
in non-small cell lung cancer patients harboring EGFR mutations. J
Immunother Cancer. 9:e0025312021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Quandt J, Schlude C, Bartoschek M, Will R,
Cid-Arregui A, Schölch S, Reissfelder C, Weitz J, Schneider M,
Wiemann S, et al: Long-peptide vaccination with driver gene
mutations in p53 and Kras induces cancer mutation-specific effector
as well as regulatory T cell responses. Oncoimmunology.
7:e15006712018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen F, Zou Z, Du J, Su S, Shao J, Meng F,
Yang J, Xu Q, Ding N, Yang Y, et al: Neoantigen identification
strategies enable personalized immunotherapy in refractory solid
tumors. J Clin Invest. 129:2056–2070. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stuber G, Dillner J, Modrow S, Wolf H,
Székely L, Klein G and Klein E: HLA-A0201 and HLA-B7 binding
peptides in the EBV-encoded EBNA-1, EBNA-2 and BZLF-1 proteins
detected in the MHC class I stabilization assay. Low proportion of
binding motifs for several HLA class I alleles in EBNA-1. Int
Immunol. 7:653–663. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shimizu Y and DeMars R: Production of
human cells expressing individual transferred HLA-A,-B,-C genes
using an HLA-A,-B,-C null human cell line. J Immunol.
142:3320–3328. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Abelin JG, Keskin DB, Sarkizova S,
Hartigan CR, Zhang W, Sidney J, Stevens J, Lane W, Zhang GL,
Eisenhaure TM, et al: Mass spectrometry profiling of HLA-associated
peptidomes in mono-allelic cells enables more accurate epitope
prediction. Immunity. 46:315–326. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kaseke C, Park RJ, Singh NK, Koundakjian
D, Bashirova A, Garcia Beltran WF, Takou Mbah OC, Ma J, Senjobe F,
Urbach JM, et al: HLA class-I-peptide stability mediates CD8+ T
cell immunodominance hierarchies and facilitates HLA-associated
immune control of HIV. Cell Rep. 36:1093782021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Partridge T, Nicastri A, Kliszczak AE,
Yindom LM, Kessler BM, Ternette N and Borrow P: Discrimination
between human leukocyte antigen Class I-Bound and Co-Purified
HIV-Derived peptides in immunopeptidomics workflows. Front Immunol.
9:9122018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Turner TR, Hayhurst JD, Hayward DR,
Bultitude WP, Barker DJ, Robinson J, Madrigal JA, Mayor NP and
Marsh SGE: Single molecule real-time DNA sequencing of HLA genes at
ultra-high resolution from 126 International HLA and Immunogenetics
Workshop cell lines. HLA. 91:88–101. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nagashima T, Yamaguchi K, Urakami K,
Shimoda Y, Ohnami S, Ohshima K, Tanabe T, Naruoka A, Kamada F,
Serizawa M, et al: Japanese version of The Cancer Genome Atlas,
JCGA, established using fresh frozen tumors obtained from 5143
cancer patients. Cancer Sci. 111:687–699. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Montealegre S, Venugopalan V, Fritzsche S,
Kulicke C, Hein Z and Springer S: Dissociation of β2-microglobulin
determines the surface quality control of major histocompatibility
complex class I molecules. FASEB J. 29:2780–2788. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
De Silva AD, Boesteanu A, Song R, Nagy N,
Harhaj E, Harding CV and Joyce S: Thermolabile H-2Kb molecules
expressed by transporter associated with antigen
processing-deficient RMA-S cells are occupied by low-affinity
peptides. J Immunol. 163:4413–4420. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Garstka MA, Fish A, Celie PH, Joosten RP,
Janssen GM, Berlin I, Hoppes R, Stadnik M, Janssen L and Ovaa H:
The first step of peptide selection in antigen presentation by MHC
class I molecules. Proc Natl Acad Sci USA. 112:1505–1510. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Martincorena I, Raine KM, Gerstung M,
Dawson KJ, Haase K, Van Loo P, Davies H, Stratton MR and Campbell
PJ: Universal patterns of selection in cancer and somatic tissues.
Cell. 171:1029–1041. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang MT, Bhattarai TS, Schram AM, Bielski
CM, Donoghue MTA, Jonsson P, Chakravarty D, Phillips S, Kandoth C,
Penson A, et al: Accelerating discovery of functional mutant
alleles in cancer. Cancer Discov. 8:174–183. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ikeda N, Kojima H, Nishikawa M, Hayashi K,
Futagami T, Tsujino T, Kusunoki Y, Fujii N, Suegami S, Miyazaki Y,
et al: Determination of HLA-A, -C, -B, -DRB1 allele and haplotype
frequency in Japanese population based on family study. Tissue
Antigens. 85:252–259. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hongo A, Kanaseki T, Tokita S, Kochin V,
Miyamoto S, Hashino Y, Codd A, Kawai N, Nakatsugawa M, Hirohashi Y,
et al: Upstream position of proline defines Peptide-HLA class I
repertoire formation and CD8 + T cell responses. J Immunol.
202:2849–2855. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Trujillo JA, Croft NP, Dudek NL,
Channappanavar R, Theodossis A, Webb AI, Dunstone MA, Illing PT,
Butler NS, Fett C, et al: The cellular redox environment alters
antigen presentation. J Biol Chem. 289:27979–27991. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Haque M, Hawes JW and Blum JS:
Cysteinylation of MHC class II ligands: Peptide endocytosis and
reduction within APC influences T cell recognition. J Immunol.
166:4543–4551. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kantoff PW, Higano CS, Shore ND, Berger
ER, Small EJ, Penson DF, Redfern CH, Ferrari AC, Dreicer R, Sims
RB, et al: Sipuleucel-T immunotherapy for castration-resistant
prostate cancer. N Engl J Med. 363:411–422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yi M, Zheng X, Niu M, Zhu S, Ge H and Wu
K: Combination strategies with PD-1/PD-L1 blockade: Current
advances and future directions. Mol Cancer. 21:282022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Laureano RS, Sprooten J, Vanmeerbeerk I,
Borras DM, Govaerts J, Naulaerts S, Berneman ZN, Beuselinck B, Bol
KF, Borst J, et al: Trial watch: Dendritic cell (DC)-based
immunotherapy for cancer. Oncoimmunology. 11:20963632022.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Douglass J, Hsiue EH, Mog BJ, Hwang MS,
DiNapoli SR, Pearlman AH, Miller MS, Wright KM, Azurmendi PA, Wang
Q, et al: Bispecific antibodies targeting mutant RAS neoantigens.
Sci Immunol. 6:eabd55152021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hsiue EH, Wright KM, Douglass J, Hwang MS,
Mog BJ, Pearlman AH, Paul S, DiNapoli SR, Konig MF, Wang Q, et al:
Targeting a neoantigen derived from a common TP53 mutation.
Science. 371:eabc86972021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Blees A, Januliene D, Hofmann T, Koller N,
Schmidt C, Trowitzsch S, Moeller A and Tampé R: Structure of the
human MHC-I peptide-loading complex. Nature. 551:525–528. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Harndahl M, Rasmussen M, Roder G, Dalgaard
Pedersen I, Sørensen M, Nielsen M and Buus S: Peptide-MHC class I
stability is a better predictor than peptide affinity of CTL
immunogenicity. Eur J Immunol. 42:1405–1416. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Parkhurst MR, Robbins PF, Tran E, Prickett
TD, Gartner JJ, Jia L, Ivey G, Li YF, El-Gamil M, Lalani A, et al:
Unique neoantigens arise from somatic mutations in patients with
gastrointestinal cancers. Cancer Discov. 9:1022–1035. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Turajlic S, Litchfield K, Xu H, Rosenthal
R, McGranahan N, Reading JL, Wong YNS, Rowan A, Kanu N, Al Bakir M,
et al: Insertion-and-deletion-derived tumour-specific neoantigens
and the immunogenic phenotype: A pan-cancer analysis. Lancet Oncol.
18:1009–1021. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Snyder A, Makarov V, Merghoub T, Yuan J,
Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et
al: Genetic basis for clinical response to CTLA-4 blockade in
melanoma. N Engl J Med. 371:2189–2199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Forde PM, Chaft JE, Smith KN, Anagnostou
V, Cottrell TR, Hellmann MD, Zahurak M, Yang SC, Jones DR,
Broderick S, et al: Neoadjuvant PD-1 Blockade in Resectable Lung
Cancer. N Engl J Med. 378:1976–1986. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Anagnostou V, Smith KN, Forde PM, Niknafs
N, Bhattacharya R, White J, Zhang T, Adleff V, Phallen J, Wali N,
et al: Evolution of neoantigen landscape during immune checkpoint
blockade in non-small cell lung cancer. Cancer Discov. 7:264–276.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nejo T, Matsushita H, Karasaki T, Nomura
M, Saito K, Tanaka S, Takayanagi S, Hana T, Takahashi S, Kitagawa
Y, et al: Reduced neoantigen expression revealed by longitudinal
multiomics as a possible immune evasion mechanism in glioma. Cancer
Immunol Res. 7:1148–1161. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Priestley P, Baber J, Lolkema MP, Steeghs
N, de Bruijn E, Shale C, Duyvesteyn K, Haidari S, van Hoeck A,
Onstenk W, et al: Pan-cancer whole-genome analyses of metastatic
solid tumours. Nature. 575:210–216. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Reiter JG, Makohon-Moore AP, Gerold JM,
Heyde A, Attiyeh MA, Kohutek ZA, Tokheim CJ, Brown A, DeBlasio RM,
Niyazov J, et al: Minimal functional driver gene heterogeneity
among untreated metastases. Science. 361:1033–1037. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Marty R, Kaabinejadian S, Rossell D,
Slifker MJ, van de Haar J, Engin HB, de Prisco N, Ideker T,
Hildebrand WH, Font-Burgada J and Carter H: MHC-I genotype
restricts the oncogenic mutational landscape. Cell. 171:1272–1283.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Malekzadeh P, Yossef R, Cafri G, Paria BC,
Lowery FJ, Jafferji M, Good ML, Sachs A, Copeland AR, Kim SP, et
al: Antigen experienced T cells from peripheral blood recognize p53
neoantigens. Clin Cancer Res. 26:1267–1276. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kim SP, Vale NR, Zacharakis N, Krishna S,
Yu Z, Gasmi B, Gartner JJ, Sindiri S, Malekzadeh P, Deniger DC, et
al: Adoptive cellular therapy with autologous tumor-infiltrating
lymphocytes and T-cell Receptor-Engineered T cells targeting common
p53 neoantigens in human solid tumors. Cancer Immunol Res.
10:932–946. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sugio K, Uramoto H, Ono K, Oyama T,
Hanagiri T, Sugaya M, Ichiki Y, So T, Nakata S, Morita M and
Yasumoto K: Mutations within the tyrosine kinase domain of EGFR
gene specifically occur in lung adenocarcinoma patients with a low
exposure of tobacco smoking. Br J Cancer. 94:896–903. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yoshida S, Kato TM, Sato Y, Umekage M,
Ichisaka T, Tsukahara M, Takasu N and Yamanaka S: A clinical-grade
HLA haplobank of human induced pluripotent stem cells matching
approximately 40% of the Japanese population. Med. 4:51–66. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xu H, Wang B, Ono M, Kagita A, Fujii K,
Sasakawa N, Ueda T, Gee P, Nishikawa M, Nomura M, et al: Targeted
disruption of HLA genes via CRISPR-Cas9 generates iPSCs with
enhanced immune compatibility. Cell Stem Cell. 24:566–578.e7. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Harada N, Fukaya S, Wada H, Goto R, Osada
T, Gomori A, Ikizawa K, Sakuragi M and Oda N: Generation of a novel
HLA Class I transgenic mouse model carrying a knock-in mutation at
the b2-Microglobulin locus. J Immunol. 198:516–527. 2017.
View Article : Google Scholar : PubMed/NCBI
|