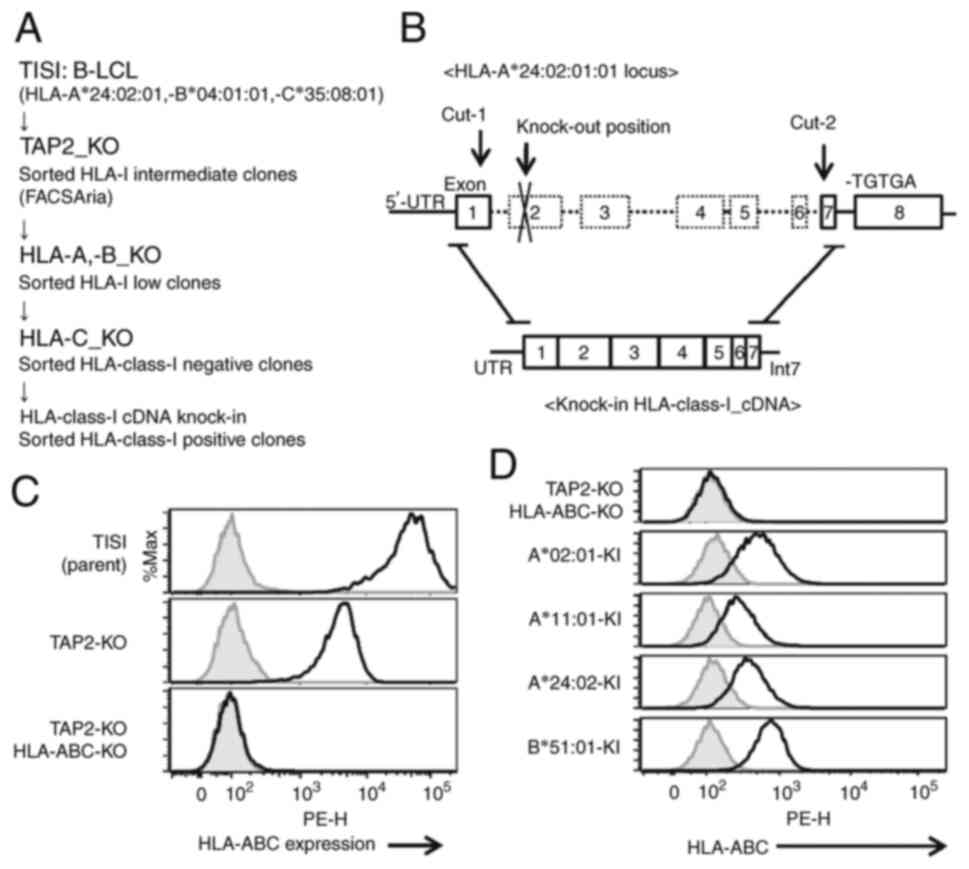
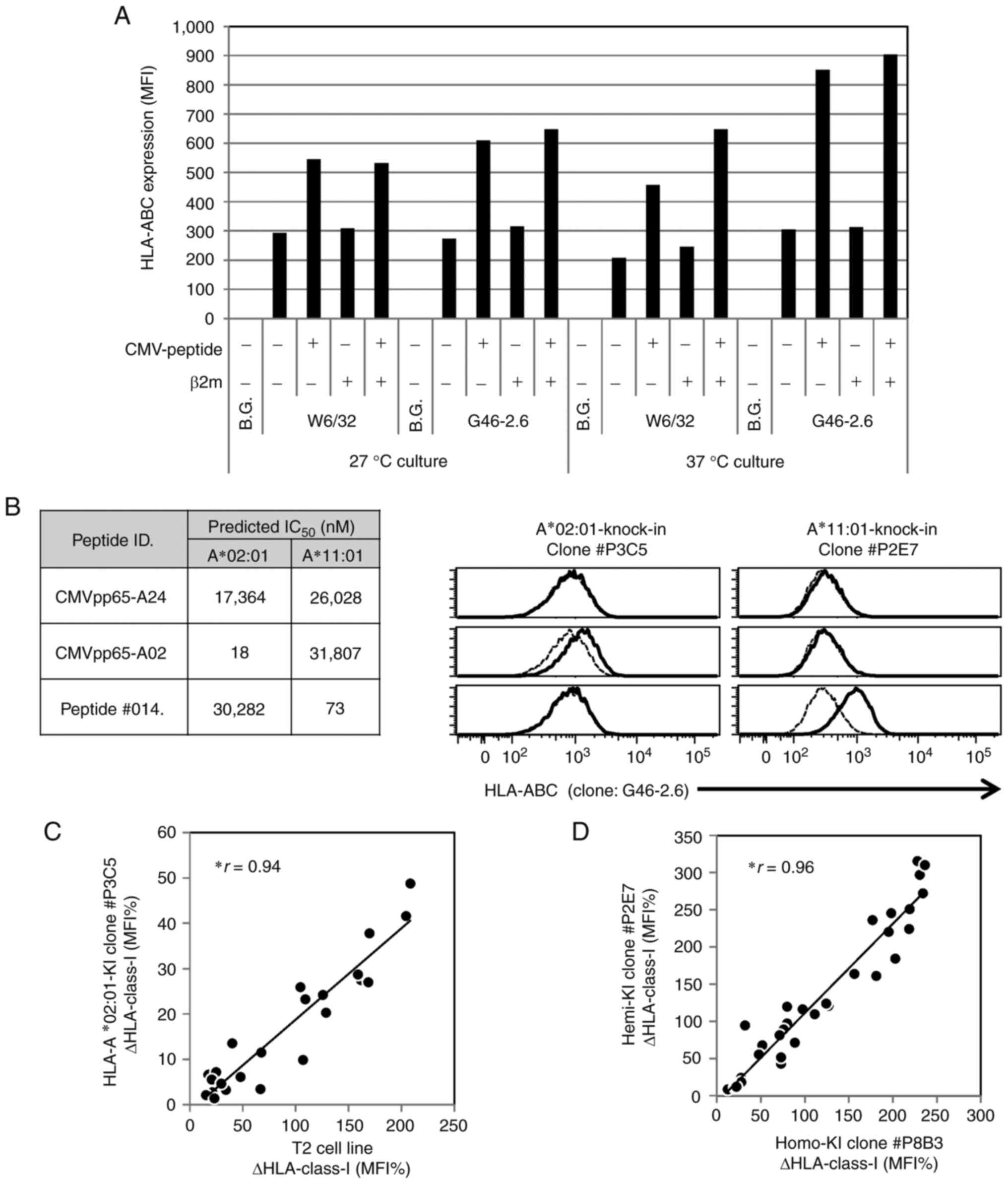
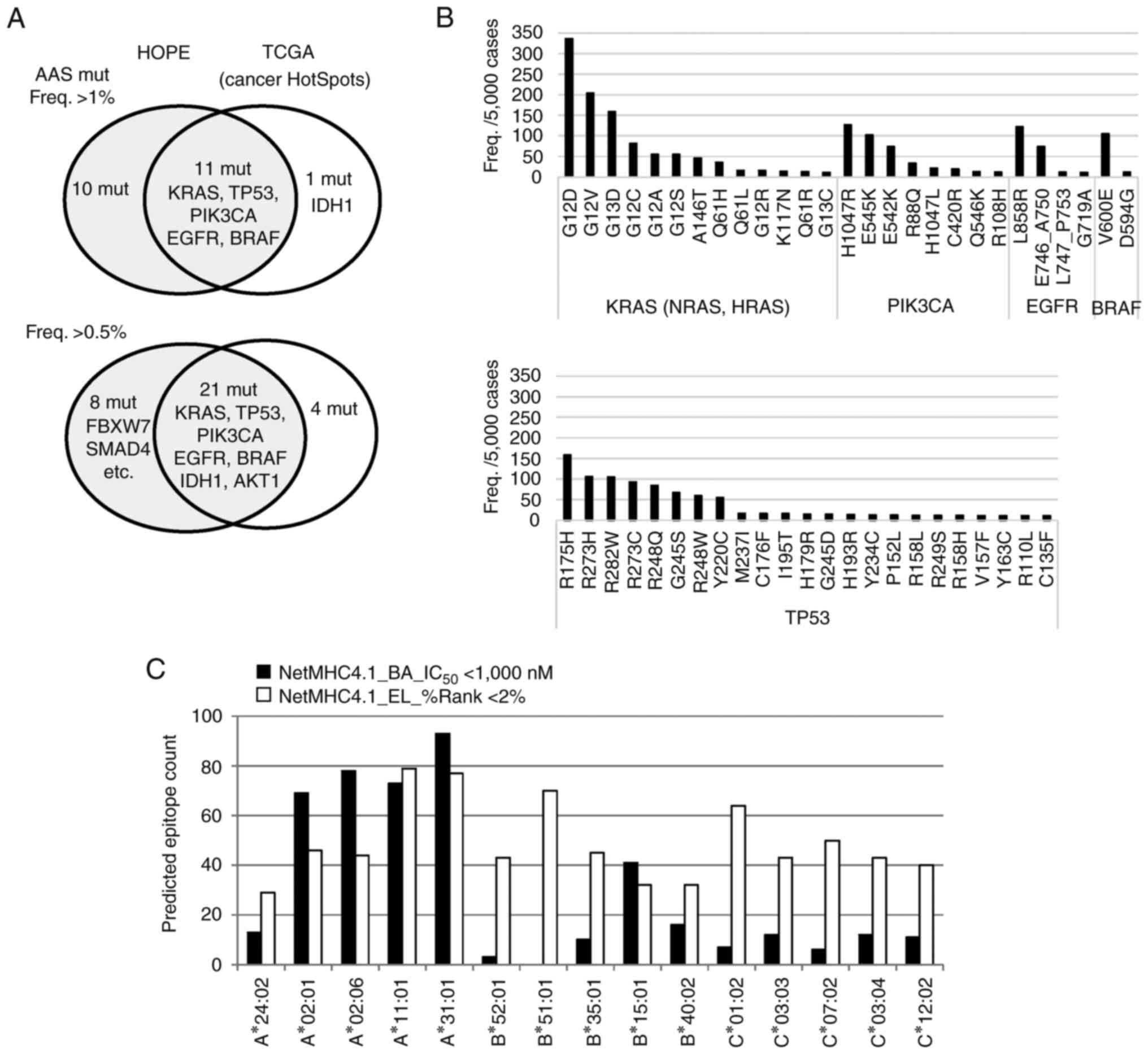
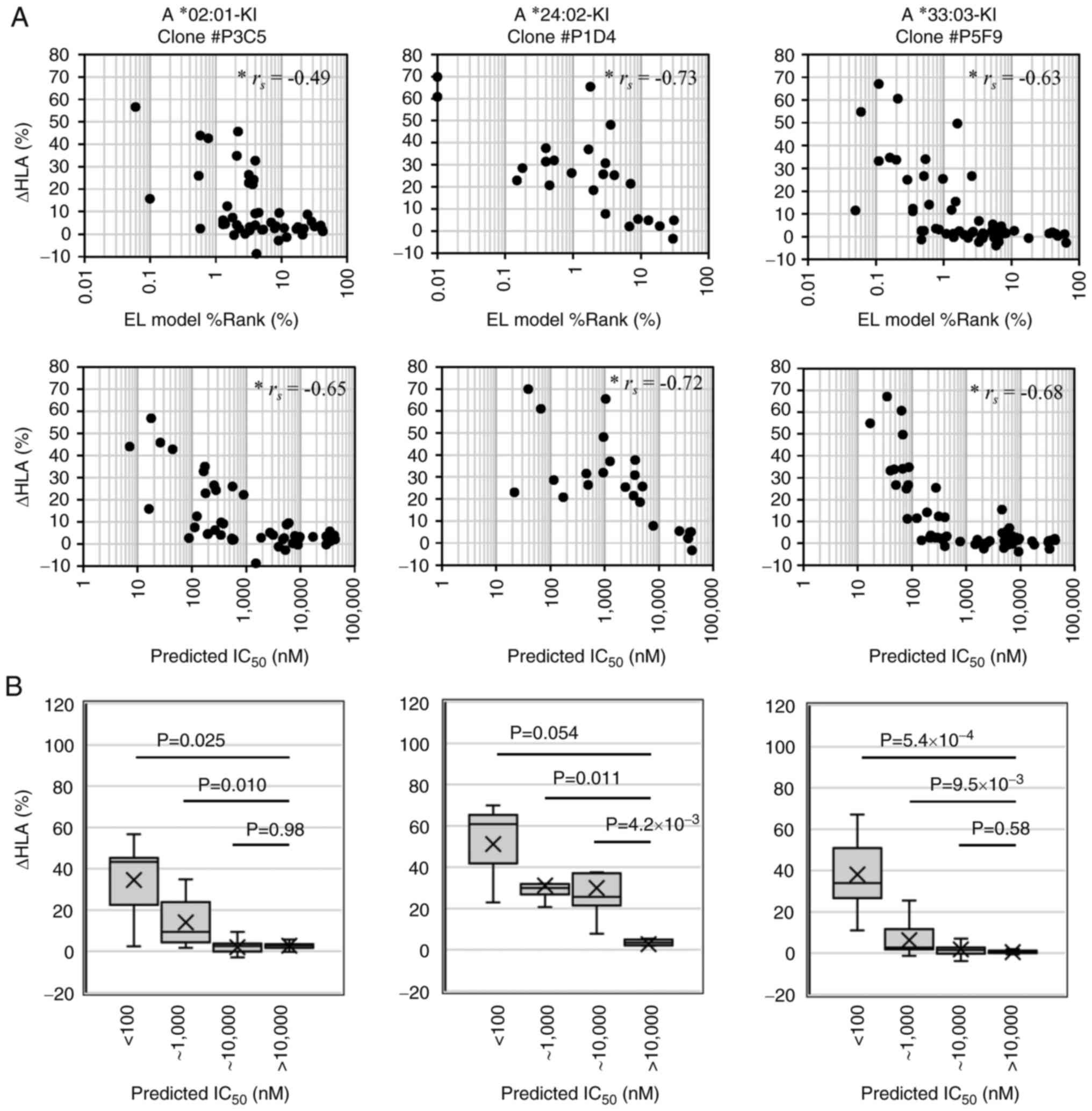
|
1
|
Schumacher TN and Schreiber RD:
Neoantigens in cancer immunotherapy. Science. 348:69–74. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yarchoan M, Hopkins A and Jaffee EM: Tumor
mutational burden and response rate to PD-1 inhibition. N Engl J
Med. 377:2500–2501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Blass E and Ott PA: Advances in the
development of personalized neoantigen-based therapeutic cancer
vaccines. Nat Rev Clin Oncol. 18:215–229. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
De Mattos-Arruda L, Vazquez M, Finotello
F, Lepore R, Porta E, Hundal J, Amengual-Rigo P, Ng CKY, Valencia
A, Carrillo J, et al: Neoantigen prediction and computational
perspectives towards clinical benefit: Recommendations from the
ESMO Precision Medicine Working Group. Ann Oncol. 31:978–990. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McGranahan N, Furness AJ, Rosenthal R,
Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak
NJ, Hiley CT, et al: Clonal neoantigens elicit T cell
immunoreactivity and sensitivity to immune checkpoint blockade.
Science. 351:1463–1469. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rizzo A, Ricci AD, Di Federico A, Frega G,
Palloni A, Tavolari S and Brandi G: Predictive biomarkers for
checkpoint inhibitor-based immunotherapy in hepatocellular
carcinoma: Where do we stand? Front Oncol. 11:8031332021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Morad G, Helmink BA, Sharma P and Wargo
JA: Hallmarks of response, resistance, and toxicity to immune
checkpoint blockade. Cell. 184:5309–5337. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schneider BJ, Naidoo J, Santomasso BD,
Lacchetti C, Adkins S, Anadkat M, Atkins MB, Brassil KJ, Caterino
JM, Chau I, et al: Management of immune-related adverse events in
patients treated with immune checkpoint inhibitor therapy: ASCO
Guideline Update. J Clin Oncol. 39:4073–4126. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park HJ, Kim KW, Won SE, Yoon S, Chae YK,
Tirumani SH and Ramaiya NH: Definition, incidence, and challenges
for assessment of Hyperprogressive disease during cancer treatment
with immune checkpoint inhibitors: A systematic review and
Meta-analysis. JAMA Netw Open. 4:e2111362021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Viscardi G, Tralongo AC, Massari F,
Lambertini M, Mollica V, Rizzo A, Comito F, Di Liello R, Alfieri S,
Imbimbo M, et al: Comparative assessment of early mortality risk
upon immune checkpoint inhibitors alone or in combination with
other agents across solid malignancies: A systematic review and
meta-analysis. Eur J Cancer. 177:175–185. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dhillon S: Tebentafusp: First approval.
Drugs. 82:703–710. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schumacher TN, Scheper W and Kvistborg P:
Cancer neoantigens. Annu Rev Immunol. 37:173–200. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kortlever RM, Sodir NM, Wilson CH,
Burkhart DL, Pellegrinet L, Brown Swigart L, Littlewood TD and Evan
GI: Myc cooperates with Ras by programming inflammation and immune
suppression. Cell. 171:1301–1315. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Ma JA, Zhang HX, Jiang YN and Luo
WH: Cancer vaccines: Targeting KRAS-driven cancers. Expert Rev
Vaccines. 19:163–173. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takeuchi Y, Tanegashima T, Sato E, Irie T,
Sai A, Itahashi K, Kumagai S, Tada Y, Togashi Y, Koyama S, et al:
Highly immunogenic cancer cells require activation of the WNT
pathway for immunological escape. Sci Immunol. 6:eabc64242021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tran E, Robbins PF, Lu YC, Prickett TD,
Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, et al:
T-Cell transfer therapy targeting mutant KRAS in cancer. N Engl J
Med. 375:2255–2262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chandran SS, Ma J, Klatt MG, Dündar F,
Bandlamudi C, Razavi P, Wen HY, Weigelt B, Zumbo P, Fu SN, et al:
Immunogenicity and therapeutic targeting of a public neoantigen
derived from mutated PIK3CA. Nat Med. 28:946–957. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li F, Deng L, Jackson KR, Talukder AH,
Katailiha AS, Bradley SD, Zou Q, Chen C, Huo C, Chiu Y, et al:
Neoantigen vaccination induces clinical and immunogenic responses
in non-small cell lung cancer patients harboring EGFR mutations. J
Immunother Cancer. 9:e0025312021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Quandt J, Schlude C, Bartoschek M, Will R,
Cid-Arregui A, Schölch S, Reissfelder C, Weitz J, Schneider M,
Wiemann S, et al: Long-peptide vaccination with driver gene
mutations in p53 and Kras induces cancer mutation-specific effector
as well as regulatory T cell responses. Oncoimmunology.
7:e15006712018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen F, Zou Z, Du J, Su S, Shao J, Meng F,
Yang J, Xu Q, Ding N, Yang Y, et al: Neoantigen identification
strategies enable personalized immunotherapy in refractory solid
tumors. J Clin Invest. 129:2056–2070. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stuber G, Dillner J, Modrow S, Wolf H,
Székely L, Klein G and Klein E: HLA-A0201 and HLA-B7 binding
peptides in the EBV-encoded EBNA-1, EBNA-2 and BZLF-1 proteins
detected in the MHC class I stabilization assay. Low proportion of
binding motifs for several HLA class I alleles in EBNA-1. Int
Immunol. 7:653–663. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shimizu Y and DeMars R: Production of
human cells expressing individual transferred HLA-A,-B,-C genes
using an HLA-A,-B,-C null human cell line. J Immunol.
142:3320–3328. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Abelin JG, Keskin DB, Sarkizova S,
Hartigan CR, Zhang W, Sidney J, Stevens J, Lane W, Zhang GL,
Eisenhaure TM, et al: Mass spectrometry profiling of HLA-associated
peptidomes in mono-allelic cells enables more accurate epitope
prediction. Immunity. 46:315–326. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kaseke C, Park RJ, Singh NK, Koundakjian
D, Bashirova A, Garcia Beltran WF, Takou Mbah OC, Ma J, Senjobe F,
Urbach JM, et al: HLA class-I-peptide stability mediates CD8+ T
cell immunodominance hierarchies and facilitates HLA-associated
immune control of HIV. Cell Rep. 36:1093782021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Partridge T, Nicastri A, Kliszczak AE,
Yindom LM, Kessler BM, Ternette N and Borrow P: Discrimination
between human leukocyte antigen Class I-Bound and Co-Purified
HIV-Derived peptides in immunopeptidomics workflows. Front Immunol.
9:9122018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Turner TR, Hayhurst JD, Hayward DR,
Bultitude WP, Barker DJ, Robinson J, Madrigal JA, Mayor NP and
Marsh SGE: Single molecule real-time DNA sequencing of HLA genes at
ultra-high resolution from 126 International HLA and Immunogenetics
Workshop cell lines. HLA. 91:88–101. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nagashima T, Yamaguchi K, Urakami K,
Shimoda Y, Ohnami S, Ohshima K, Tanabe T, Naruoka A, Kamada F,
Serizawa M, et al: Japanese version of The Cancer Genome Atlas,
JCGA, established using fresh frozen tumors obtained from 5143
cancer patients. Cancer Sci. 111:687–699. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Montealegre S, Venugopalan V, Fritzsche S,
Kulicke C, Hein Z and Springer S: Dissociation of β2-microglobulin
determines the surface quality control of major histocompatibility
complex class I molecules. FASEB J. 29:2780–2788. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
De Silva AD, Boesteanu A, Song R, Nagy N,
Harhaj E, Harding CV and Joyce S: Thermolabile H-2Kb molecules
expressed by transporter associated with antigen
processing-deficient RMA-S cells are occupied by low-affinity
peptides. J Immunol. 163:4413–4420. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Garstka MA, Fish A, Celie PH, Joosten RP,
Janssen GM, Berlin I, Hoppes R, Stadnik M, Janssen L and Ovaa H:
The first step of peptide selection in antigen presentation by MHC
class I molecules. Proc Natl Acad Sci USA. 112:1505–1510. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Martincorena I, Raine KM, Gerstung M,
Dawson KJ, Haase K, Van Loo P, Davies H, Stratton MR and Campbell
PJ: Universal patterns of selection in cancer and somatic tissues.
Cell. 171:1029–1041. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang MT, Bhattarai TS, Schram AM, Bielski
CM, Donoghue MTA, Jonsson P, Chakravarty D, Phillips S, Kandoth C,
Penson A, et al: Accelerating discovery of functional mutant
alleles in cancer. Cancer Discov. 8:174–183. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ikeda N, Kojima H, Nishikawa M, Hayashi K,
Futagami T, Tsujino T, Kusunoki Y, Fujii N, Suegami S, Miyazaki Y,
et al: Determination of HLA-A, -C, -B, -DRB1 allele and haplotype
frequency in Japanese population based on family study. Tissue
Antigens. 85:252–259. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hongo A, Kanaseki T, Tokita S, Kochin V,
Miyamoto S, Hashino Y, Codd A, Kawai N, Nakatsugawa M, Hirohashi Y,
et al: Upstream position of proline defines Peptide-HLA class I
repertoire formation and CD8 + T cell responses. J Immunol.
202:2849–2855. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Trujillo JA, Croft NP, Dudek NL,
Channappanavar R, Theodossis A, Webb AI, Dunstone MA, Illing PT,
Butler NS, Fett C, et al: The cellular redox environment alters
antigen presentation. J Biol Chem. 289:27979–27991. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Haque M, Hawes JW and Blum JS:
Cysteinylation of MHC class II ligands: Peptide endocytosis and
reduction within APC influences T cell recognition. J Immunol.
166:4543–4551. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kantoff PW, Higano CS, Shore ND, Berger
ER, Small EJ, Penson DF, Redfern CH, Ferrari AC, Dreicer R, Sims
RB, et al: Sipuleucel-T immunotherapy for castration-resistant
prostate cancer. N Engl J Med. 363:411–422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yi M, Zheng X, Niu M, Zhu S, Ge H and Wu
K: Combination strategies with PD-1/PD-L1 blockade: Current
advances and future directions. Mol Cancer. 21:282022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Laureano RS, Sprooten J, Vanmeerbeerk I,
Borras DM, Govaerts J, Naulaerts S, Berneman ZN, Beuselinck B, Bol
KF, Borst J, et al: Trial watch: Dendritic cell (DC)-based
immunotherapy for cancer. Oncoimmunology. 11:20963632022.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Douglass J, Hsiue EH, Mog BJ, Hwang MS,
DiNapoli SR, Pearlman AH, Miller MS, Wright KM, Azurmendi PA, Wang
Q, et al: Bispecific antibodies targeting mutant RAS neoantigens.
Sci Immunol. 6:eabd55152021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hsiue EH, Wright KM, Douglass J, Hwang MS,
Mog BJ, Pearlman AH, Paul S, DiNapoli SR, Konig MF, Wang Q, et al:
Targeting a neoantigen derived from a common TP53 mutation.
Science. 371:eabc86972021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Blees A, Januliene D, Hofmann T, Koller N,
Schmidt C, Trowitzsch S, Moeller A and Tampé R: Structure of the
human MHC-I peptide-loading complex. Nature. 551:525–528. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Harndahl M, Rasmussen M, Roder G, Dalgaard
Pedersen I, Sørensen M, Nielsen M and Buus S: Peptide-MHC class I
stability is a better predictor than peptide affinity of CTL
immunogenicity. Eur J Immunol. 42:1405–1416. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Parkhurst MR, Robbins PF, Tran E, Prickett
TD, Gartner JJ, Jia L, Ivey G, Li YF, El-Gamil M, Lalani A, et al:
Unique neoantigens arise from somatic mutations in patients with
gastrointestinal cancers. Cancer Discov. 9:1022–1035. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Turajlic S, Litchfield K, Xu H, Rosenthal
R, McGranahan N, Reading JL, Wong YNS, Rowan A, Kanu N, Al Bakir M,
et al: Insertion-and-deletion-derived tumour-specific neoantigens
and the immunogenic phenotype: A pan-cancer analysis. Lancet Oncol.
18:1009–1021. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Snyder A, Makarov V, Merghoub T, Yuan J,
Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et
al: Genetic basis for clinical response to CTLA-4 blockade in
melanoma. N Engl J Med. 371:2189–2199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Forde PM, Chaft JE, Smith KN, Anagnostou
V, Cottrell TR, Hellmann MD, Zahurak M, Yang SC, Jones DR,
Broderick S, et al: Neoadjuvant PD-1 Blockade in Resectable Lung
Cancer. N Engl J Med. 378:1976–1986. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Anagnostou V, Smith KN, Forde PM, Niknafs
N, Bhattacharya R, White J, Zhang T, Adleff V, Phallen J, Wali N,
et al: Evolution of neoantigen landscape during immune checkpoint
blockade in non-small cell lung cancer. Cancer Discov. 7:264–276.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nejo T, Matsushita H, Karasaki T, Nomura
M, Saito K, Tanaka S, Takayanagi S, Hana T, Takahashi S, Kitagawa
Y, et al: Reduced neoantigen expression revealed by longitudinal
multiomics as a possible immune evasion mechanism in glioma. Cancer
Immunol Res. 7:1148–1161. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Priestley P, Baber J, Lolkema MP, Steeghs
N, de Bruijn E, Shale C, Duyvesteyn K, Haidari S, van Hoeck A,
Onstenk W, et al: Pan-cancer whole-genome analyses of metastatic
solid tumours. Nature. 575:210–216. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Reiter JG, Makohon-Moore AP, Gerold JM,
Heyde A, Attiyeh MA, Kohutek ZA, Tokheim CJ, Brown A, DeBlasio RM,
Niyazov J, et al: Minimal functional driver gene heterogeneity
among untreated metastases. Science. 361:1033–1037. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Marty R, Kaabinejadian S, Rossell D,
Slifker MJ, van de Haar J, Engin HB, de Prisco N, Ideker T,
Hildebrand WH, Font-Burgada J and Carter H: MHC-I genotype
restricts the oncogenic mutational landscape. Cell. 171:1272–1283.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Malekzadeh P, Yossef R, Cafri G, Paria BC,
Lowery FJ, Jafferji M, Good ML, Sachs A, Copeland AR, Kim SP, et
al: Antigen experienced T cells from peripheral blood recognize p53
neoantigens. Clin Cancer Res. 26:1267–1276. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kim SP, Vale NR, Zacharakis N, Krishna S,
Yu Z, Gasmi B, Gartner JJ, Sindiri S, Malekzadeh P, Deniger DC, et
al: Adoptive cellular therapy with autologous tumor-infiltrating
lymphocytes and T-cell Receptor-Engineered T cells targeting common
p53 neoantigens in human solid tumors. Cancer Immunol Res.
10:932–946. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sugio K, Uramoto H, Ono K, Oyama T,
Hanagiri T, Sugaya M, Ichiki Y, So T, Nakata S, Morita M and
Yasumoto K: Mutations within the tyrosine kinase domain of EGFR
gene specifically occur in lung adenocarcinoma patients with a low
exposure of tobacco smoking. Br J Cancer. 94:896–903. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yoshida S, Kato TM, Sato Y, Umekage M,
Ichisaka T, Tsukahara M, Takasu N and Yamanaka S: A clinical-grade
HLA haplobank of human induced pluripotent stem cells matching
approximately 40% of the Japanese population. Med. 4:51–66. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xu H, Wang B, Ono M, Kagita A, Fujii K,
Sasakawa N, Ueda T, Gee P, Nishikawa M, Nomura M, et al: Targeted
disruption of HLA genes via CRISPR-Cas9 generates iPSCs with
enhanced immune compatibility. Cell Stem Cell. 24:566–578.e7. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Harada N, Fukaya S, Wada H, Goto R, Osada
T, Gomori A, Ikizawa K, Sakuragi M and Oda N: Generation of a novel
HLA Class I transgenic mouse model carrying a knock-in mutation at
the b2-Microglobulin locus. J Immunol. 198:516–527. 2017.
View Article : Google Scholar : PubMed/NCBI
|