Introduction
Thyroid cancer (TC) is among the most prevalent
endocrine malignancies globally, with epidemiological data
indicating a 20% annual increase in incidence and a 1.1% annual
mortality rate (1–3). This trend is primarily due to the
rising occurrence and heightened mortality of papillary thyroid
carcinoma (PTC), which, together with follicular thyroid carcinoma
(FTC), comprises differentiated thyroid cancer (DTC), accounting
for >95% of all TC cases (4,5). The
completion of the Human Genome Project in 2003 ushered in the
post-genomic era, characterized by the emergence of functional
genomics, which seeks to uncover the functions and regulatory
mechanisms of the genome (6).
Following the advent of the post-genomic era, medical research
transitioned into the precision medicine era, leveraging
various-omics technologies to elucidate the molecular mechanisms
underpinning human malignancies in numerous publications (7,8). DTC
is fundamentally a genetic disease, with its development being
protracted, multifaceted, staged and intricate, inextricably
connected to multiple molecular and environmental factors (5,8).
Transcriptomics and proteomics involve the comprehensive study of
gene transcription or proteins in living organisms. The vast amount
of data generated by high-throughput transcriptome sequencing
enables the identification of gene-specific characteristics by
analyzing genes correlated with phenotypic information. By
contrast, proteomics, as a functional translation of the genome,
offers direct insight into the mechanisms that instigate pathogenic
effects at the genomic level (9).
Consequently, transcriptomics and proteomics can be employed to
identify molecular markers of DTC, monitor its progression and
select and validate therapeutic targets, demonstrating their
long-term potential in clinical diagnostics and treatment. The
present review examined the crucial transcriptomic and proteomic
data of DTC identified recently, offering novel perspectives for
the advancement of DTC diagnosis and therapy.
Transcriptome analysis of DTC
Recent advancements in transcriptomics-based
sequencing analysis have led to the identification of potential
molecular markers for DTC at various levels, such as pathogenic
risk (10,11), early diagnosis (12–14),
metastasis and progression (15–19),
prognosis prediction (20–25) and therapeutic targets (26–32)
(Table I). This progress has
increased our understanding of the molecular mechanisms underlying
DTC and has helped resolve complex issues. However, only a few of
these molecules have proved useful in clinical settings. The
present review focused on the molecular classification of DTC and
radioactive iodine (131I) refractory (RAIR)-DTC, which
is DTC that does not respond to radioactive iodine.
 | Table I.Application of massively parallel
sequencing methods to analyze the latest partial transcriptome
studies of DTC. |
Table I.
Application of massively parallel
sequencing methods to analyze the latest partial transcriptome
studies of DTC.
| Author, year | Category | Biomarkers | Expression | Regulatory target
or pathway | Significance | Samples | (Refs.) |
|---|
| Comiskey et
al, 2020 | Diagnosis | LRRC34 | - | ranBP1 | Variations in
LRRC34 at 3q26 revealed two independent mechanisms for
predisposition to PTC, one in G protein signaling and the other in
transcriptional control. | TPC1,BCPAP and 293T
cell lines | (10) |
| Corrado et
al, 2021 |
| LGALS3 | - | TTF-1 | Polymorphism of
Pro64His (rs4644) in LGALS3 serves as the risk factor for DTC,
while rs4644 represents the trans-expression quantitative trait
locus for modifying downstream gene transcriptional expression by
modulating TTF-1. | DTC (n=1,142) | (11) |
| Barros-Filho et
al, 2020 | Prognosis | GADD45B | Up | FoxO/GADD45B
axis | GADD45B transcript
is discovered to be the new candidate prognostic biomarker for PTC
cases receiving radioiodine therapy and thyroidectomy. | PTC (n=48) | (16) |
| Guan et al,
2020 |
| ITGA7 | Down | EMT | Downregulating
ITGA7 promotes PTC growth, invasion and migration via EMT. | PTC (n=19), TPC1,
KTC-1 and BCPAP cell lines | (22) |
| Ramírez-Moya et
al, 2021 |
| edit-CDK13 | Up | ADAR1-mediated
A-to-I editing/CDK13 | The ADAR1-dependent
splicing can be mainly regulated through trans-acting elements such
as CDK13, but not the direct cis-element editing, thus, further
affecting splicing in numerous genes. CDK13 editing is the possible
effective mechanism associated with cancer aggressiveness and
deterioration. | PTC (n=6), Cal62
and TPC1 cell lines | (23) |
| Wang et al,
2020 |
| MALAT1 | - | miR-214/MALAT1
rs619586/CTNNB1 | rs619586 SNP (G
allele) within MALAT1 upregulated miR-214 and later downregulated
CTNNB1, which was the possible prognostic biomarker for DTC. | PTC (n=214), FTC
(n=14) | (24) |
| Saqcena et
al, 2021 |
| SWI/SNF | Down | - | SWI/SNF complexes
have important effects on maintaining differentiation within TC,
while the loss of such complexes induces resistance to MAPK
inhibitor-based re-differentiation and radioiodine treatments. | Mice thyroid
cells | (25) |
| Augenlicht et
al, 2021 | Treatment | miR-7-5p | Down | miR-7-5p/EGFR/MAPK
or IRS2/PI3K signaling pathways | miR-7-5p suppresses
TC growth via EGFR/MAPK as well as IRS2/PI3K pathways. | Classical variation
of PTC and FTC, LNM, adjacent to normal thyroid tissues. TPC1 and
HTori-3 cell lines | (26) |
| Hou et al,
2021 |
| miR-146b-3p | Up |
miR-146b-3p/MUC20/MET signaling
pathways | Regulating the MET
pathway via miR-146b-3p can possibly target MUC20 and is associated
with DTC dedifferentiation. It induces 131I resistance
along with failure to absorb iodine into DTC cancer foci and cancer
becomes RAIR-DTC. | RAIR-DTC (n=3) and
control group (n=3); WRO cell line | (27) |
| Huang et al,
2022 |
| FTO | Down | FTO/APOE axis | FTO inhibited
expression of APOE through IGF2BP2-mediated m6A modification and
may inhibit glycolytic metabolism in PTC by modulating
IL-6/JAK2/STAT3 signaling pathway, thus, abrogating tumor
growth. | PTC (n=150);
Nthy-ori3-1, TPC1, K1, IHH4 and BCPAP cell lines | (28) |
| Li et al,
2018 |
| TBX3 | Up | TBX3/p57KIP2
(CDKN1C) axis | TBX3 promotes the
proliferation of PTC cells by facilitating PRC2-mediated p57KIP2
repression. | PTC (n=98), K1 and
TPC1 cell lines | (29) |
| Long et al,
2020 |
|
hsa_circ_0007694 | Down |
hsa_circ_0007694/PI3K/AKT/mTOR or Wnt
signaling pathways | The circRNA
hsa_circ_0007694 is down-regulated in PTC and is, therefore, a
potential therapeutic target. | PTC and adjacent to
normal thyroid tissues (n=3) | (31) |
| Ramírez-Moa et
al, 2022 |
| SPTY2D1-AS1 | Down | miR-221 | SPTY2D1-AS1 is the
strong tumor suppressor in vivo and in vitro, which
shows reduced expression within many advanced TC cases and can
block primary miR-221 processing. | PTC (n=8) | (32) |
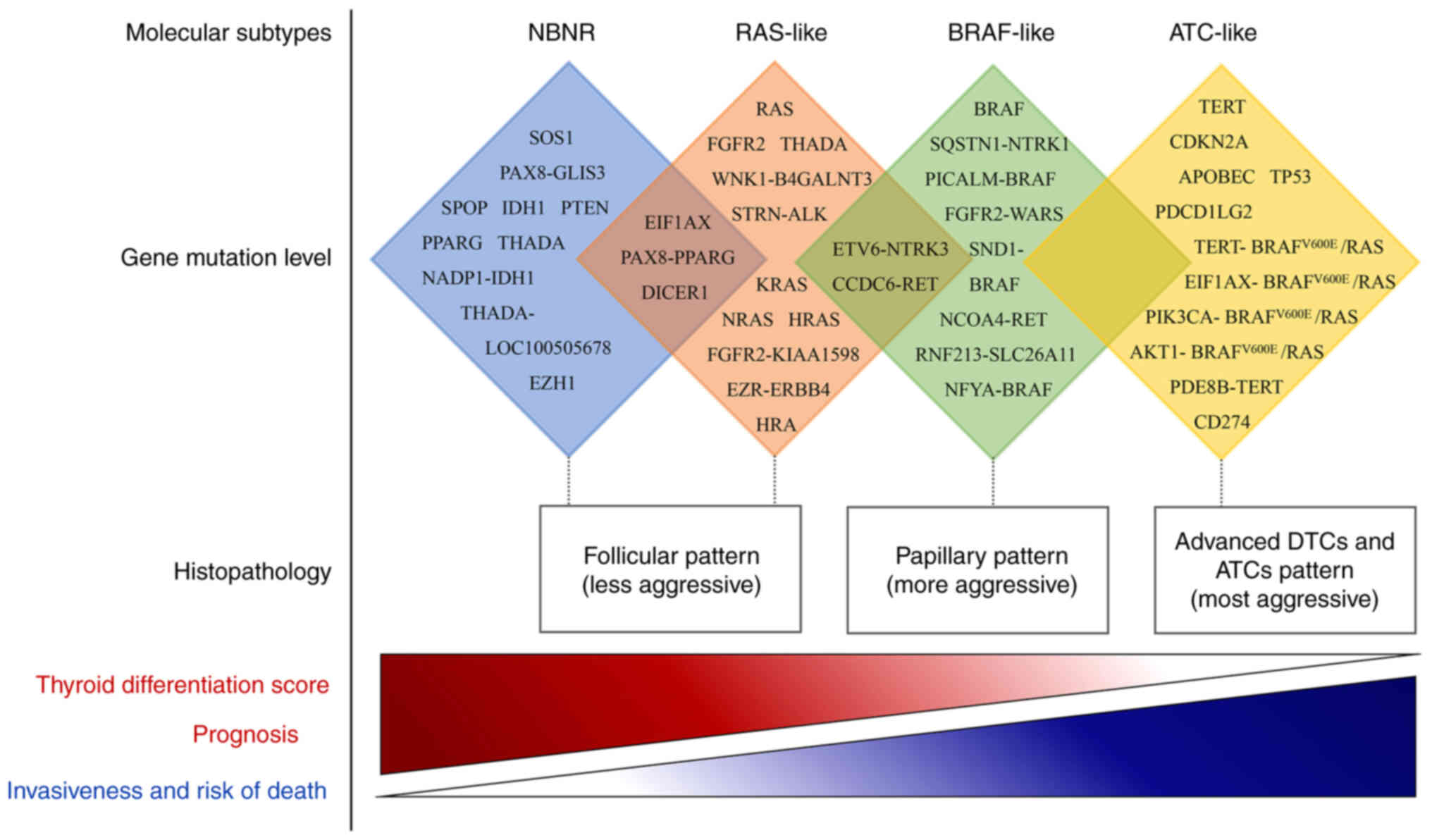
Molecular subtypes
DTC is categorized into distinct subtypes according
to their gene expression levels, revealing differences in
clinicopathological features, intracellular mechanisms and
mutational profiles that contribute to a deeper understanding of
TC. The initial integrative study by the Cancer Genome Atlas (TCGA)
in 2014 (33) classified PTC into
two molecular subtypes: BRAF-like (BVL) and RAS-like (RL), which
are associated with BRAF or RAS mutations, respectively. BVL
exhibits more aggressive clinical features compared with RL. This
research marked the first transcriptome-based classification of PTC
molecular subtypes. The study employed unsupervised clustering
methods on four genomic datasets, each producing distinct subtypes
that supported the overarching separation of BVL-PTC and RL-PTC.
Patients in the RL-PTC group exhibited follicular variant
histology, a relatively low recurrence risk and highly
differentiated tumors associated with younger patients. By
contrast, the different datasets divided the BVL-PTC group into
varying numbers of subtypes that did not overlap, resulting in an
inconsistent lower-level partitioning of BVL-PTC. To investigate
signaling and differentiation in PTC, researchers developed a
BRAFV600E-RAS score (BRS) and a thyroid differentiation
score (TDS). The BRS quantifies the extent to which the gene
expression profile of a given tumor resembles either the
BRAFV600E- or RAS-mutant profiles, providing a
continuous reference scale from most BVL to most RL to examine the
signaling consequences of other, less common mutations. The TDS,
derived from the expression levels of 16 highly correlated thyroid
metabolic and functional genes within this cohort, plays a central
role in TC. Based on the TDS and BRS results, the study (30) found that all BRAF fusions were BVL.
Among the six EIF1AX mutations, four were RL, one was neutral and
one was weakly BVL. All PAX8/peroxisome proliferator activated
receptor γ (PPARG) fusions were RL, in line with their prevalence
in follicular-patterned tumors. Almost all Ret Proto-Oncogene (RET)
fusions were weakly BVL, while non-invasive follicular thyroid
neoplasm (NTRK)1/3 and anaplastic lymphoma receptor tyrosine kinase
(ALK) fusions were predominantly neutral. In conclusion, the
comprehensive multi-platform molecular data and large sample size
in this study offer an opportunity to refine the classification of
PTC into molecular subtypes and correlate them with clinically
relevant parameters. The BRS and TDS measures were utilized to
elucidate the relationships between tumor cluster, histology,
genotype, signaling and differentiation.
A number of studies have supported the notion that
there are three molecular subtypes of DTC: BVL, RL and
non-BRAF-non-RAS (NBNR) (34,35).
Yoo et al (34) conducted a
study on follicular thyroid tumors, including CV-PTC, FV-PTC,
minimally invasive FTC and FA, using molecular typing by TCGA. In
addition to expanding the RL subtype, they analyzed and proposed
another molecular subtype, NBNR. The BVL subtype consisted of
BRAFV600E and fusion genes, including
phosphatidylinositol binding clathrin assembly protein-BRAF,
nuclear transcription factor y subunit α-BRAF, staphylococcal
nuclease and tudor domain containing 1-BRAF, fibroblast growth
factor receptor (FGFR) 2-tryptophanyl-tRNA synthetase, ETS variant
transcription factor 6 (ETV6)-NTRK3, sequestosome 1-NTRK1,
coiled-coil domain containing (CCDC)6-RET, nuclear receptor
coactivator 4-RET and ring finger protein 213-solute carrier family
(SLC) 26 member 11. The RL subtype consisted of H/K/NRAS and fusion
genes, including Striatin-ALK, Ezrin-Erb-B2 receptor tyrosine
kinase 4, fibroblast growth factor receptor 2-KIAA1598, ETV6-NTRK3
and CCDC6-RET. The NBNR subtype was associated with dicer 1,
ribonuclease III, EIF1AX, isocitrate dehydrogenase [NADP(+)] 1,
phosphatase and tensin homolog, PAX8-PPARG and other driver gene
candidates. The study (32) also
implemented TDS and ERK scores to explore tumor signaling and
differentiation and clinical risk factors were integrated to obtain
a progression model of TC. A similar perspective was obtained in a
study by Song et al (36).
Mutations in SPOP P94R and EZH1 Q571R in nonmalignant thyroid
nodules were associated with BVL transcriptomic characteristics,
further supporting the proposal of a third molecular subtype
(12). Due to its inert biological
behavior and molecular profile, enveloped PTC has been reclassified
as non-invasive follicular thyroid neoplasm (NIFTP), which exhibits
papillary nuclear characteristics (36–39).
This reclassification demonstrates that stratification medicine can
be achieved by reclassifying TC based on its mutational and
transcriptional characteristics, regardless of its histological
classification.
Yoo et al (40) proposed an updated view on the
progression of DTC to advanced DTC and degenerative interstitial
types using previous approaches for anaplastic thyroid cancer
(ATC), poorly differentiated thyroid carcinoma, widely invasive FTC
and metastatic PTC. The results showed that TERT, AKT1, PIK3CA and
EIF1AX were frequently co-mutated with driver genes
(BRAFV600E and RAS) in advanced DTCs similar to ATC,
while tumor suppressors (e.g., TP53 and CDKN2A) were predominantly
altered in ATC. CDKN2A loss was significantly associated with poor
disease-specific survival in patients with ATC or advanced DTC and
upregulation of programmed cell death 1 ligand (PDCD1LG) 1 and
PDCD1LG 2. This led to the proposal of a fourth molecular subtype
of TC, ATC-like (AL) and paved the way for targeted therapy of
highly invasive and fatal TCs.
Since the 2014 publication of TCGA research on PTC
(33), genomic studies of DTC using
more advanced massively parallel sequencing have suggested that
diverse subtypes involve different mutations, which coupled with
their clinical and molecular features, show progressive activation
from NBNR to RL to BVL and then to AL, leading to a series of more
aggressive behaviors (8,14,33,34,40)
(Fig. 1). The molecular subtypes of
DTC correspond to different cell signaling and differentiation
characteristics, allowing for more accurate pathological
classification and diagnosis. However, only a few studies have
investigated this topic and further large-scale validation is
necessary.
RAIR-DTC
RAIR-DTC constitutes ~1/3 of distant DTC metastases,
which are the primary cause of TC mortality, with an average
survival of 3–5 years and a 10-year survival rate of ~10% (41). However, molecular-level
investigations on RAIR-DTC have been scarce due to the complexity
and heterogeneity of RAIR-DTC development, the instability and
irreproducibility of the methods employed and the challenges in
acquiring clinical samples (42–44).
Capdevila et al (45)
questioned the limitations of DNA mutation profiles in stratifying
prognostic factors. They correlated RNA expression patterns and
patient survival in cases enrolled in the DECISION trial,
discovering that although RNA expression patterns were associated
with DNA mutation patterns and tumor histology, they exhibited
considerable differences in multivariate analysis, correlating with
prognosis while the other two factors did not. Notably, the RNA
expression profiles of BVL and RL revealed 7–9% of BRAF or RAS
mutated tumors with ‘unexpected’ expression profiles, suggesting
that these classical driver mutations did not dictate the final RNA
gene expression profile in these instances. Additionally, ~40% of
cases in BVL and RL expression profiles lacked mutations in BRAF or
RAS genes, necessitating the identification of potential
genomic/epigenomic aberrations that might influence the final
expression profile. These findings imply that prognostic biomarkers
associated with DTC may become prevalent in individuals with RAIR
status. Therefore, it is essential to identify genomic/epigenomic
mutations that could potentially affect the eventual expression
patterns, as a genome-only stratified prognostic analysis of
RAIR-DTC is insufficient. This study (45) also has a limitation due to the use
of formalin-fixed and paraffin-embedded (FFPE) samples, which,
although representing most tumor sample materials, lack information
such as TERT promoter mutations in the prognostic genome.
Another study supports the view of the RAI
refractory state. Colombo et al (46) investigated gene/miRNA and molecular
expression profiles of PTC primary foci, synchronous lymph node
metastases (LNM) and RAI-LNM, noting that patient characteristics
with RAI+/D+ (metastatic site with initial RAI uptake and disease
persistence) and RAI-/D+ (metastatic site without RAI uptake and
disease persistence) still possessed driving lesions (particularly
BRAFV600E) of BRAF-/RAS-like subtypes with TDS. These
results suggest that DTC maintains its classification as a
molecular subtype based on transcriptome expression profiles, even
in an RAIR environment. Furthermore, the study found transcriptome
similarity in BRAFV600E mutation samples from primary
and post-RAI LNM, indicating that RAI treatment has minimal
influence on the expression profile of RAIR LNMs compared with
primary tumors. This observation may offer a fresh perspective on
the challenge of sample accessibility in future RAIR-DTC
research.
A recent study utilizing whole-exome sequencing of
matched sixty-six iodine-refractory and ninety-two iodine uptake
PTC samples identified the APOBEC SBS13 mutation nomogram as an
independent predictor of radioiodine resistance in PTC (47). When combined with SBS13 and TERTp
mutations, this nomogram significantly increases the likelihood of
prediction.
In contrast to adult PTC, molecular characterization
of pediatric PTC is crucial for the development of
molecule-targeted therapies for progressive RAIR PTC. A genome-wide
and transcriptomic study comparing 106 RAIR-DTC children with
adults (48) revealed a significant
tumor response with the restoration of radioactive iodine uptake
when exposed to NTRK and RET fusion-targeted drug therapy. The
study also identified age-related drivers of pediatric PTC, thus,
supporting fusion-targeted therapy for RAIR-DTC
re-differentiation.
Research on RAIR-DTC is currently limited and more
molecular studies are necessary to address this bottleneck as
quickly as possible.
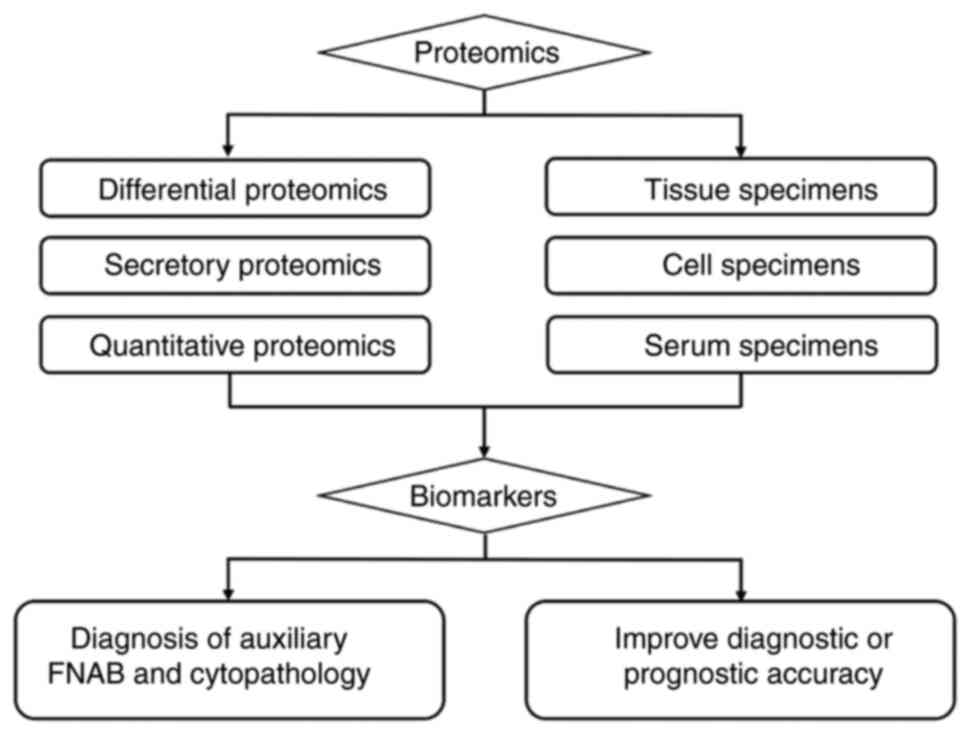
Proteome analysis of DTC
Recent technological advancements in molecular
research have enabled scientists to analyze protein information in
various specimens, such as cells, tissues, cell lines and body
fluids, using different mass spectrometry techniques, thereby
increasing proteome coverage. This has led to the replacement of 2D
gel electrophoresis by mass spectrometry coupled with other
techniques. DTC proteomics research has made significant strides in
improving the accuracy of fine needle aspiration biopsy in
diagnosing indeterminate thyroid nodule cases before surgery, as
well as in predicting diagnosis and prognosis by identifying
biomarkers (Fig. 2; Table II). These advances have facilitated
a better understanding of the pathogenic molecular mechanisms
associated with the occurrence and progression of DTC, thereby
aiding in guiding treatment decisions' (49).
 | Table II.Application of high-throughput
sequencing methods to analyze the latest partial proteomics of
DTC. |
Table II.
Application of high-throughput
sequencing methods to analyze the latest partial proteomics of
DTC.
| Author, year | Category | Biomarkers | Expression | Significance | Samples | (Refs.) |
|---|
| Lai X et al,
2017 | Diagnosis | SUCLG2 | Up | SUCLG2, a
mitochondrial protein upregulated in FC, distinguishes FC from FA
and has a sensitivity of 75% and a specificity of 80% for the
diagnosis of FC. | FA, FC(FFPE, n=54,
52) | (53) |
| Wang Y et
al, 2021 |
| C4A | Down | Low serum levels of
CA4 and PLG in the PTC group could be used as early diagnostic
biomarkers for PTC, with sensitivities of 91.67 and 87.50% and
specificities of 83.33 and 75.00%, respectively. | NG, PTC(serum,
n=29) | (58) |
|
|
| PLG |
|
|
|
|
| Dai J et al,
2020 | Prognosis | SLC27A6 | Up | SLC27A6 was a
specifical upregulated protein in iPTC and may be significantly
associated with the metastasis and staging of PTC. | PTC, iPTC and
adjacent to normal thyroid tissues (n=3) | (62) |
| Zhan, S et
al, 2019 |
| LAMC2 | Up | They played a role
in PTC lymph node metastasis and more importantly, their elevated
mRNA expression in tumorous tissues | PTC and adjacent to
normal thyroid tissues (n =48) | (63) |
|
|
| MYO1G |
| showed a positive
relationship with unfavorable variables, including larger tumor
size, LNM, high AJCC staging, BRAFV600E mutation and
poor prognosis. |
|
|
| Wei X et al,
2018 |
| PDLIM5 | Up | PDLIM5 is highly
expressed in PTC and could promote the migration, invasion and
proliferation of PTC cells through the Ras-ERK pathway. | PTC and adjacent to
normal thyroid tissues (n =75) | (64) |
| Orlandella FM et
al, 2019 |
| JAM-A | Down | Silencing of JAM-A
enhances the proliferation and motility of thyroid cancer cells,
which is closely associated with the phosphorylation of P53 and
GSK3α/β proteins. | PTC, ATC, NT(n=11,
9, 9) | (65) |
| Lin P et al,
2019 |
| ISG15 | Up | ISG15 is highly
expressed in PTMC with lymph node metastasis and thus can be used
as a candidate biomarker for prognosis of lymph node metastatic
PTMC. | PTMC with or
without CLNM (FNAB, n=60) | (72) |
| Krishnan, A et
al, 2020 | Treatment | TFG-RET fusion | Up | Targeting HUWE1 or
DUBs is a promising strategy to prevent RET-induced tumors. | PTC, adjacent to
normal thyroid tissues and LNM | (74) |
Diagnosis of auxiliary fine-needle
aspiration biopsy (FNAB) and cytopathology
The study of biomarkers in DTC tissues poses a
challenge due to the heterogeneous cellular components that can
mask the aberrant expression of tumor proteins. Mass spectrometry
imaging (MSI) can resolve certain limitations by accurately and
locally determining protein levels within tissues (50), which is vital in classifying
indeterminate lesions that cannot be correctly identified by FNAB
and cytopathological examination.
Ucal et al (51) used MSI-based label-free quantitative
methods to investigate the roles of IQGAP proteins, actin
cytoskeletal proteins and energy metabolism alterations in
follicular and classical variants of PTC. Another study (52) evaluated the discriminatory ability
of matrix-assisted laser desorption/ionization (MALDI) combined
with MSI to classify classical variants of PTC using FFPE samples
from CV-PTC, FV-PTC and NIFTP. The signal intensities of S100-A6,
cytoplasmic actin 1 and vimentin were enhanced within FV-PTC, while
60S ribosomal protein L6/L8 and Prelamin A/C were enhanced in NIFTP
and CV-PTC, respectively. The results revealed that the peptide
profiles of NIFTP showed significant differences compared with
invasive FV-PTC, supporting the renaming of non-invasive FV-PTC to
‘non-invasive follicular thyroid neoplasms with papillary nuclear
features’ (NIFTP).
A study using liquid chromatography-tandem mass
spectrometry to explore novel protein biomarkers found that
upregulated mitochondrial proteins, particularly SUVLG2,
differentiate follicular carcinoma (FC) from follicular adenoma
(FA) with a sensitivity and specificity of 75 and 80% respectively
(53). Furthermore, using mass
spectrometry, molecular similarities were observed between seven
thyroid specimens, including the major types of thyroid
malignancies (FV-PTC, CV-PTC, FTC, ATC, medullary thyroid carcinoma
and FA, as well as non-carcinoma thyroid tissue (54,55),
demonstrating the high feasibility of MSI combined with various
techniques in testing cancer types.
Serum samples offer a readily available, easily
monitored and less invasive means of detecting biomarkers for
pathological diagnosis in DTC (56). Zhang et al (57) used label-free analysis to identify
and quantify the overall proteome, complete N-glycopeptides and
desialic acid N-glycopeptides within urine and plasma samples of
females with PTC, thus, establishing an integrated, highly
reproducible and rapid approach for exploring potential
non-invasive diagnostic and prognostic biomarkers and therapeutic
targets for PTC. Wang et al (58) discovered that serum-based proteomic
profiles showed significantly lower levels of CA4 and plasminogen
(PLG) in the PTC group compared with the nodular goiter group. C4A
and PLG were identified as excellent diagnostic biomarkers for PTC
cases, with sensitivities of 91.67 and 87.50% and specificities of
83.33 and 75.00%, respectively. Another study combining the
expression of oncogene NRASQ61R within Nthy-ori 3–1 cells (based on
blood and cell lines) with a multi-dimensional proteomics
technology DISER, the combination of 2D-difference gel
electrophoresis (2D-DIGE) and serological proteome analysis
(SERPA), established a comprehensive way to identify
disease-related tumor-associated antigens (TAAs) in thyroid tumors,
enabling the identification of both cell-based and TAA biomarkers.
Among these biomarkers, autoantibodies against CNN3 and PGK1 were
identified as tumor-specific biomarkers of the thyroid envelope
follicular type/RAS-like phenotype (EFP/RLP), which can be used to
differentiate tumors of diverse malignancy grades (59).
MALDI-MSI has shown promise in cytopathology for
diagnosing thyroid nodules, using FNAB needle washes as a sample
source (60). Furthermore, new
diagnostic tools, such as nanoparticle-assisted proteomics
approaches, are constantly being developed (61). Taken together, proteomics appears to
be a very promising alternative for diagnosing thyroid nodules that
are difficult for cytopathologists to identify.
Identification of biomarkers to
improve diagnostic or prognostic accuracy
The progression of DTC is marked by invasion and
metastasis events, particularly LNM in PTC, which heighten the risk
of relapse and result in a dismal prognosis. The discovery of novel
protein biomarkers for improved prediction of DTC prognostic risk
is of utmost importance.
Dai et al (62) employed TMT-based mass spectrometry
and Gene Expression Omnibus and TCGA databases to identify
differential proteins between PTC and invasive PTC phenotype
(iPTC). They discovered that SLC 27 Member 6, a protein acting as a
transporter mediating long-chain fatty acid uptake, was
specifically upregulated in iPTC and may be significantly linked to
PTC metastasis and staging. Similarly, LAMC2 and myosin IG were
found to serve a role in PTC lymph node metastasis and, more
importantly, their increased mRNA expression in tumor tissues
demonstrated a positive correlation with unfavorable variables such
as larger tumor size, LNM, high American Joint Committee on Cancer
(AJCC) staging, BRAFV600E mutation and poor prognosis
(63). Another study first revealed
that PDZ and LIM domain 5 is highly expressed in PTC and could
promote the migration, invasion and proliferation of PTC cells
through the Ras-ERK pathway (64).
Junctional adhesion molecule A, a transmembrane protein involved in
various biological processes, including epithelial-to-mesenchymal
transition, was shown to have its silencing enhance the
proliferation and motility of TC cells, which is closely associated
with the phosphorylation of P53 and GSK3α/β proteins. Each of these
proteins could positively affect their potential as candidate
biomarkers for predicting PTC prognosis (65). Luo et al (66) compared the proteomic profiles of
serum purified exosomes (SPEs) of PTC cases (with and without LNM)
to those of healthy individuals and observed overexpression of
proteins like talin 1, SRC proto-oncogene, non-receptor tyrosine
kinase, calpain small subunit 1 and integrin subunit β 2, which are
associated with tumor cell migration. Furthermore, abnormal
activation of integrin signaling in the SPE of PTC with LNM
promoted the aggressiveness of BHT101 TC cells. These findings
offer insight into the biological activities and features of serum
exosomes during LNM of PTC; thus, aiding in understanding
DTC-related molecular mechanisms.
The global increase in PTC morbidity is primarily
associated with a rise in papillary thyroid microcarcinoma (PTMC)
diagnosis. The need for surgical treatment of PTMC largely depends
on LNM. Jin et al (67)
studied PTMC with and without LNM and screened for proteins
potentially associated with migration and invasion, such as
α-1-antitrypsin, α-actinin-1, carbonic anhydrase 4, high mobility
group protein HMGI-C and hepatocellular carcinoma-derived growth
factor. ISG15 ubiquitin like modifier (ISG15), a type I
interferon-regulated ubiquitin-like molecule that plays crucial
roles in modulating cell growth and cancer progression, has been
reported to be highly expressed in various cancers (68–71).
Similarly, ISG15 was well distinguished between patients with lymph
node metastatic and non-metastatic PTMC. ISG15 knockdown reduced
proliferation and infiltration of human PTC-derived BCPAP cells
in vitro. Additionally, silencing ISG15 inhibited
xenografted tumor growth in immunodeficient nude mice (72).
Targeted therapy remains a primary treatment for
managing advanced TC. However, resistance to multiple drugs
restricts the clinical utility of targeted agents. A study by
Mishall et al (73)
highlight the need for targeting multiple pathways simultaneously.
They developed TC cells resistant to four Src inhibitors
(dasatinib) and proteomic analysis revealed that the MAPK and
AKT/mTOR pathways were critical for resistance to single-drug Src
inhibitors.
Through proteogenomic analysis of matched normal
thyroid tissue, primary tumor foci and LNM in one case, Krishnan
et al (74) discovered a
novel TFG-RET gene fusion. This fusion involves the 5′-terminal of
Trk fusion gene (TFG) exons 1–4 and the 3′-terminal of RET tyrosine
kinase. Further studies have revealed the involvement of HUWE1 in
RET-induced oncogenic conversion of PTC and suggest that targeting
HUWE1 or deubiquitinating enzymes (DUBs) is a promising therapeutic
strategy for preventing RET-induced tumors. Clinical trials
targeting the ubiquitinome are currently underway in other tumors
and appear promising for PTC (75).
Proteomics has proven to be an efficient approach to
identify potential biomarkers for DTC, but further research is
needed to expand our understanding of DTC at the molecular level
beyond the two aspects discussed earlier.
Discussion
DTC is the most prevalent type of TC worldwide and
early detection remains challenging due to the lack of reliable and
specific markers. Thus, identifying biomarkers that can aid in
better diagnostic stratification, prognosis prediction and precise
treatment is crucial. Recent research has focused on exploring the
molecular mechanisms of DTC at the transcript and protein abundance
levels. With the advancement of high-resolution histology
technologies, even small changes in transcriptional, translational
and post-translational modifications have been uncovered, which
were previously undetectable using genomic or proteomic approaches.
These research findings can serve as a foundation for future
studies aimed at translating these results into clinical practice,
meeting the demand for clinical stratification of DTC management in
the era of precision medicine.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the NSFC program of
Prof. GZ: 177Lu-CNP-iRGD Nanoprobes for Tracing and Targeting
Cervical Lymph Node Metastasis of Thyroid Papillary Carcinoma
(grant no. 81860313).
Availability of data and materials
Data sharing is not applicable to this article, as
no data sets were generated or analyzed during the current
study.
Authors' contributions
SY conceived, designed, wrote and revised the
manuscript. RH, DF and JF were involved in the design and revision
of the manuscript. GZ reviewed and revised the manuscript. All
authors have read and approved the final manuscript. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Du L, Zhao Z, Zheng R, Li H, Zhang S, Li
R, Wei W and He J: Epidemiology of thyroid cancer: Incidence and
mortality in China, 2015. Front Oncol. 10:17022020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Global Burden of Disease Cancer
Collaboration, . Fitzmaurice C, Dicker D, Pain A, Hamavid H,
Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, et
al: The global burden of cancer 2013. JAMA Oncol. 1:505–527. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lim H, Devesa SS, Sosa JA, Check D and
Kitahara CM: Trends in thyroid cancer incidence and mortality in
the United States, 1974–2013. JAMA. 317:1338–1348. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim J, Gosnell JE and Roman SA: Geographic
influences in the global rise of thyroid cancer. Nat Rev
Endocrinol. 16:17–29. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bilder RM: Phenomics: Building scaffolds
for biological hypotheses in the post-genomic era. Biol Psychiatry.
63:439–440. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hyman DM, Taylor BS and Baselga J:
Implementing genome-driven oncology. Cell. 168:584–599. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Song YS and Park YJ: Genomic
characterization of differentiated thyroid carcinoma. Endocrinol
Metab (Seoul). 34:1–10. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kumar D, Bansal G, Narang A, Basak T,
Abbas T and Dash D: Integrating transcriptome and proteome
profiling: Strategies and applications. Proteomics. 16:2533–2544.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Comiskey DF Jr, He H, Liyanarachchi S,
Sheikh MS, Genutis LK, Hendrickson IV, Yu L, Brock PL and de la
Chapelle A: Variants in LRRC34 reveal distinct mechanisms for
predisposition to papillary thyroid carcinoma. J Med Genet.
57:519–527. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Corrado A, Aceto R, Silvestri R, Dell'Anno
I, Ricci B, Miglietta S, Romei C, Giovannoni R, Poliseno L,
Evangelista M, et al: Pro64His (rs4644) polymorphism within
galectin-3 is a risk factor of differentiated thyroid carcinoma and
affects the transcriptome of thyrocytes engineered via CRISPR/Cas9
system. Thyroid. 31:1056–1066. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ye L, Zhou X, Huang F, Wang W, Qi Y, Xu H,
Yang S, Shen L, Fei X, Xie J, et al: The genetic landscape of
benign thyroid nodules revealed by whole exome and transcriptome
sequencing. Nat Commun. 8:155332017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song YS, Kang BH, Lee S, Yoo SK, Choi YS,
Park J, Park DY, Lee KE, Seo JS and Park YJ: Genomic and
transcriptomic characteristics according to size of papillary
thyroid microcarcinoma. Cancers (Basel). 12:13452020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He H, Liyanarachchi S, Li W, Comiskey DF
Jr, Yan P, Bundschuh R, Turkoglu AM, Brock P, Ringel MD and de la
Chapelle A: Transcriptome analysis discloses dysregulated genes in
normal appearing tumor-adjacent thyroid tissues from patients with
papillary thyroid carcinoma. Sci Rep. 11:141262021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Morillo-Bernal J, Fernández LP and
Santisteban P: FOXE1 regulates migration and invasion in thyroid
cancer cells and targets ZEB1. Endocr Relat Cancer. 27:137–151.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barros-Filho MC, de Mello JBH, Marchi FA,
Pinto CAL, da Silva IC, Damasceno PKF, Soares MBP, Kowalski LP and
Rogatto SR: GADD45B transcript is a prognostic marker in papillary
thyroid carcinoma patients treated with total thyroidectomy and
radioiodine therapy. Front Endocrinol (Lausanne). 11:2692020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma B, Jiang H, Wen D, Hu J, Han L, Liu W,
Xu W, Shi X, Wei W, Liao T, et al: Transcriptome analyses identify
a metabolic gene signature indicative of dedifferentiation of
papillary thyroid cancer. J Clin Endocrinol Metab. 104:3713–3725.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Credendino SC, Moccia C, Amendola E,
D'Avino G, Di Guida L, Clery E, Greco A, Bellevicine C, Brunetti A,
De Felice M and De Vita G: FOXE1 gene dosage affects thyroid cancer
histology and differentiation in vivo. Int J Mol Sci. 22:252020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao B, Huang Z, Zhu X, Cai H, Huang Y,
Zhang X, Zhang Z, Lu H, An C, Niu L and Li Z: Clinical significance
of the expression of co-stimulatory molecule B7-H3 in papillary
thyroid carcinoma. Front Cell Dev Biol. 10:8192362022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Akyay OZ, Gov E, Kenar H, Arga KY, Selek
A, Tarkun İ, Canturk Z, Cetinarslan B, Gurbuz Y and Sahin B:
Mapping the molecular basis and markers of papillary thyroid
carcinoma progression and metastasis using global transcriptome and
microRNA profiling. OMICS. 24:148–159. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Q, Wang P, Sun C, Wang C and Sun Y:
Integrative analysis of methylation and transcriptome identified
epigenetically regulated lncRNAs with prognostic relevance for
thyroid cancer. Front Bioeng Biotechnol. 7:4392020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guan Y, Bhandari A, Xia E, Kong L, Zhang X
and Wang O: Downregulating integrin subunit alpha 7 (ITGA7)
promotes proliferation, invasion, and migration of papillary
thyroid carcinoma cells through regulating
epithelial-to-mesenchymal transition. Acta Biochim Biophys Sin
(Shanghai). 52:116–124. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ramírez-Moya J, Miliotis C, Baker AR,
Gregory RI, Slack FJ and Santisteban P: An ADAR1-dependent RNA
editing event in the cyclin-dependent kinase CDK13 promotes thyroid
cancer hallmarks. Mol Cancer. 20:1152021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang ML and Liu JX: MALAT1 rs619586
polymorphism functions as a prognostic biomarker in the management
of differentiated thyroid carcinoma. J Cell Physiol. 235:1700–1710.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Saqcena M, Leandro-Garcia LJ, Maag J,
Tchekmedyian V, Krishnamoorthy GP, Tamarapu PP, Tiedje V, Reuter V,
Knauf JA, de Stanchina E, et al: SWI/SNF Complex Mutations Promote
Thyroid Tumor Progression and Insensitivity to Redifferentiation
Therapies. Cancer Discov. 11:1158–1175. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Augenlicht A, Saiselet M,
Decaussin-Petrucci M, Andry G, Dumont JE and Maenhaut C: MiR-7-5p
inhibits thyroid cell proliferation by targeting the EGFR/MAPK and
IRS2/PI3K signaling pathways. Oncotarget. 12:1587–1599. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hou S, Xie X, Zhao J, Wu C, Li N, Meng Z,
Cai C and Tan J: Downregulation of miR-146b-3p inhibits
proliferation and migration and modulates the expression and
location of sodium/iodide symporter in dedifferentiated thyroid
cancer by potentially targeting MUC20. Front Oncol. 10:5663652021.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang J, Sun W, Wang Z, Lv C, Zhang T,
Zhang D, Dong W, Shao L, He L, Ji X, et al: FTO suppresses
glycolysis and growth of papillary thyroid cancer via decreasing
stability of APOE mRNA in an N6-methyladenosine-dependent manner. J
Exp Clin Cancer Res. 41:422022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li X, Ruan X, Zhang P, Yu Y, Gao M, Yuan
S, Zhao Z, Yang J and Zhao L: TBX3 promotes proliferation of
papillary thyroid carcinoma cells through facilitating
PRC2-mediated p57KIP2 repression. Oncogene.
37:2773–2792. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liao Y, Hua Y, Li Y, Zhang C, Yu W, Guo P,
Zou K, Li W, Sun Y, Wang R, et al: CRSP8 promotes thyroid cancer
progression by antagonizing IKKα-induced cell differentiation. Cell
Death Differ. 28:1347–1363. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Long MY, Chen JW, Zhu Y, Luo DY, Lin SJ,
Peng XZ, Tan LP and Li HH: Comprehensive circular RNA profiling
reveals the regulatory role of circRNA_0007694 in papillary thyroid
carcinoma. Am J Transl Res. 12:1362–1378. 2020.PubMed/NCBI
|
|
32
|
Ramírez-Moya J, Wert-Lamas L, Acuña-Ruíz
A, Fletcher A, Wert-Carvajal C, McCabe CJ, Santisteban P and
Riesco-Eizaguirre G: Identification of an interactome network
between lncRNAs and miRNAs in thyroid cancer reveals SPTY2D1-AS1 as
a new tumor suppressor. Sci Rep. 12:77062022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cancer Genome Atlas Research Network, .
Integrated genomic characterization of papillary thyroid carcinoma.
Cell. 159:676–690. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yoo SK, Lee S, Kim SJ, Jee HG, Kim BA, Cho
H, Song YS, Cho SW, Won JK, Shin JY, et al: Comprehensive analysis
of the transcriptional and mutational landscape of follicular and
papillary thyroid cancers. PLoS Genet. 12:e10062392016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Costa V, Esposito R, Ziviello C, Sepe R,
Bim LV, Cacciola NA, Decaussin-Petrucci M, Pallante P, Fusco A and
Ciccodicola A: New somatic mutations and WNK1-B4GALNT3 gene fusion
in papillary thyroid carcinoma. Oncotarget. 6:11242–11251. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Song YS, Won JK, Yoo SK, Jung KC, Kim MJ,
Kim SJ, Cho SW, Lee KE, Yi KH, Seo JS and Park YJ: Comprehensive
transcriptomic and genomic profiling of subtypes of follicular
variant of papillary thyroid carcinoma. Thyroid. 28:1468–1478.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Johnson DN, Furtado LV, Long BC, Zhen CJ,
Wurst M, Mujacic I, Kadri S, Segal JP, Antic T and Cipriani NA:
Noninvasive follicular thyroid neoplasms with papillary-like
nuclear features are genetically and biologically similar to
adenomatous nodules and distinct from papillary thyroid carcinomas
with extensive follicular growth. Arch Pathol Lab Med. 142:838–850.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nikiforov YE, Seethala RR, Tallini G,
Baloch ZW, Basolo F, Thompson LD, Barletta JA, Wenig BM, Al Ghuzlan
A, Kakudo K, et al: Nomenclature revision for encapsulated
follicular variant of papillary thyroid carcinoma: A paradigm shift
to reduce overtreatment of indolent tumors. JAMA Oncol.
2:1023–1029. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pool C, Walter V, Bann D, Goldenberg D,
Broach J, Hennessy M, Cottrill E, Washburn E, Williams N, Crist H,
et al: Molecular characterization of tumors meeting diagnostic
criteria for the non-invasive follicular thyroid neoplasm with
papillary-like nuclear features (NIFTP). Virchows Arch.
474:341–351. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yoo SK, Song YS, Lee EK, Hwang J, Kim HH,
Jung G, Kim YA, Kim SJ, Cho SW, Won JK, et al: Integrative analysis
of genomic and transcriptomic characteristics associated with
progression of aggressive thyroid cancer. Nat Commun. 10:27642019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Berdelou A, Lamartina L, Klain M,
Leboulleux S and Schlumberger M: Treatment of refractory thyroid
cancer. Endocr Relat Cancer. 25:R209–R223. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Goossens N, Nakagawa S, Sun X and Hoshida
Y: Cancer biomarker discovery and validation. Transl Cancer Res.
4:256–269. 2015.PubMed/NCBI
|
|
43
|
Bossuyt PM: Where are all the new
omics-based tests? Clin Chem. 60:1256–1257. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Boutros PC: The path to routine use of
genomic biomarkers in the cancer clinic. Genome Res. 25:1508–1513.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Capdevila J, Matos I, Mancuso FM, Iglesias
C, Nuciforo P, Zafon C, Palmer HG, Ogbah Z, Muiños L, Hernando J,
et al: Identification of expression profiles defining distinct
prognostic subsets of radioactive-iodine refractory differentiated
thyroid cancer from the DECISION trial. Mol Cancer Ther.
19:312–317. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Colombo C, Minna E, Gargiuli C, Muzza M,
Dugo M, De Cecco L, Pogliaghi G, Tosi D, Bulfamante G, Greco A, et
al: The molecular and gene/miRNA expression profiles of radioiodine
resistant papillary thyroid cancer. J Exp Clin Cancer Res.
39:2452020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Siraj S, Masoodi T, Siraj AK, Azam S,
Qadri Z, Parvathareddy SK, Bu R, Siddiqui KS, Al-Sobhi SS, AlDawish
M and Al-Kuraya KS: APOBEC SBS13 mutational signature-A novel
predictor of radioactive iodine refractory papillary thyroid
carcinoma. Cancers (Basel). 14:15842022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Franco AT, Ricarte-Filho JC, Laetsch TW
and Bauer AJ: Oncogene-specific inhibition in the treatment of
advanced pediatric thyroid cancer. J Clin Invest. 131:e1526962021.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Boufraqech M and Nilubol N: Multi-omics
signatures and translational potential to improve thyroid cancer
patient outcome. Cancers (Basel). 11:19882019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mainini V, Lalowski M, Gotsopoulos A,
Bitsika V, Baumann M and Magni F: MALDI-imaging mass spectrometry
on tissues. Methods Mol Biol. 1243:139–164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ucal Y, Eravci M, Tokat F, Duren M, Ince U
and Ozpinar A: Proteomic analysis reveals differential protein
expression in variants of papillary thyroid carcinoma. EuPA Open
Proteom. 17:1–6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ucal Y, Tokat F, Duren M, Ince U and
Ozpinar A: Peptide profile differences of noninvasive follicular
thyroid neoplasm with papillary-like nuclear features, encapsulated
follicular variant, and classical papillary thyroid carcinoma: An
application of matrix-assisted laser desorption/ionization mass
spectrometry imaging. Thyroid. 29:1125–1137. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lai X, Umbricht CB, Fisher K, Bishop J,
Shi Q and Chen S: Identification of novel biomarker and therapeutic
target candidates for diagnosis and treatment of follicular
carcinoma. J Proteomics. 166:59–67. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gawin M, Wojakowska A, Pietrowska M,
Marczak Ł, Chekan M, Jelonek K, Lange D, Jaksik R, Gruca A and
Widłak P: Proteome profiles of different types of thyroid cancers.
Mol Cell Endocrinol. 472:68–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kurczyk A, Gawin M, Chekan M, Wilk A,
Łakomiec K, Mrukwa G, Frątczak K, Polanska J, Fujarewicz K,
Pietrowska M and Widlak P: Classification of thyroid tumors based
on mass spectrometry imaging of tissue microarrays; a single-pixel
approach. Int J Mol Sci. 21:62892020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Farrokhi Yekta R, Arefi Oskouie A, Rezaei
Tavirani M, Mohajeri-Tehrani MR and Soroush AR: Decreased
apolipoprotein A4 and increased complement component 3 as potential
markers for papillary thyroid carcinoma: A proteomic study. Int J
Biol Markers. 33:455–462. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang Y, Zhao W, Zhao Y, Mao Y, Su T,
Zhong Y, Wang S, Zhai R, Cheng J, Fang X, et al: Comparative
glycoproteomic profiling of human body fluid between healthy
controls and patients with papillary thyroid carcinoma. J Proteome
Res. 19:2539–2552. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang Y, Zhou S, Wang D, Wei T, Zhu J and
Li Z: Complement C4-A and plasminogen as potential biomarkers for
prediction of papillary thyroid carcinoma. Front Endocrinol
(Lausanne). 12:7376382021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Belousov PV, Afanasyeva MA, Gubernatorova
EO, Bogolyubova AV, Uvarova AN, Putlyaeva LV, Ramanauskaite EM,
Kopylov AT, Demin DE, Tatosyan KA, et al: Multi-dimensional
immunoproteomics coupled with in vitro recapitulation of oncogenic
NRASQ61R identifies diagnostically relevant autoantibody
biomarkers in thyroid neoplasia. Cancer Lett. 467:96–106. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Capitoli G, Piga I, Galimberti S, Leni D,
Pincelli AI, Garancini M, Clerici F, Mahajneh A, Brambilla V, Smith
A, et al: MALDI-MSI as a complementary diagnostic tool in
cytopathology: A pilot study for the characterization of thyroid
nodules. Cancers (Basel). 11:13772019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
García-Vence M, Chantada-Vázquez MDP,
Cameselle-Teijeiro JM, Bravo SB and Núñez C: A novel nanoproteomic
approach for the identification of molecular targets associated
with thyroid tumors. Nanomaterials (Basel). 10:23702020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Dai J, Yu X, Han Y, Chai L, Liao Y, Zhong
P, Xie R, Sun X, Huang Q, Wang J, et al: TMT-labeling proteomics of
papillary thyroid carcinoma reveal invasive biomarkers. J Cancer.
11:6122–6132. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhan S, Wang T, Wang M, Li J and Ge W:
In-depth proteomics analysis to identify biomarkers of papillary
thyroid cancer patients older than 45 years with different degrees
of lymph node metastases. Proteomics Clin Appl. 13:e19000302019.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wei X, Zhang Y, Yu S, Li S, Jiang W, Zhu
Y, Xu Y, Yang C, Tian G, Mi J, et al: PDLIM5 identified by
label-free quantitative proteomics as a potential novel biomarker
of papillary thyroid carcinoma. Biochem Biophys Res Commun.
499:338–344. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Orlandella FM, Mariniello RM, Iervolino
PLC, Auletta L, De Stefano AE, Ugolini C, Greco A, Mirabelli P,
Pane K, Franzese M, et al: Junctional adhesion molecule-A is
down-regulated in anaplastic thyroid carcinomas and reduces cancer
cell aggressiveness by modulating p53 and GSK3 α/β pathways. Mol
Carcinog. 58:1181–1193. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Luo D, Zhan S, Xia W, Huang L, Ge W and
Wang T: Proteomics study of serum exosomes from papillary thyroid
cancer patients. Endocr Relat Cancer. 25:879–891. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Jin S, Bao W, Yang YT, Fu Q, Bai Y and Liu
Y: Proteomic analysis of the papillary thyroid microcarcinoma. Ann
Endocrinol (Paris). 80:293–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Mondaza-Hernandez JL, Moura DS,
Lopez-Alvarez M, Sanchez-Bustos P, Blanco-Alcaina E,
Castilla-Ramirez C, Collini P, Merino-Garcia J, Zamora J,
Carrillo-Garcia J, et al: ISG15 as a prognostic biomarker in
solitary fibrous tumour. Cell Mol Life Sci. 79:4342022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kariri YA, Alsaleem M, Joseph C, Alsaeed
S, Aljohani A, Shiino S, Mohammed OJ, Toss MS, Green AR and Rakha
EA: The prognostic significance of interferon-stimulated gene 15
(ISG15) in invasive breast cancer. Breast Cancer Res Treat.
185:293–305. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang Q, Wang J, Qiao H, Huyan L, Liu B,
Li C, Jiang J, Zhao F, Wang H and Yan J: ISG15 is downregulated by
KLF12 and implicated in maintenance of cancer stem cell-like
features in cisplatin-resistant ovarian cancer. J Cell Mol Med.
25:4395–4407. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Qu T, Zhang W, Yan C, Ren D, Wang Y, Guo
Y, Guo Q, Wang J, Liu L, Han L, et al: ISG15 targets glycosylated
PD-L1 and promotes its degradation to enhance antitumor immune
effects in lung adenocarcinoma. J Transl Med. 21:3412023.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lin P, Yao Z, Sun Y, Li W, Liu Y, Liang K,
Liu Y, Qin J, Hou X and Chen L: Deciphering novel biomarkers of
lymph node metastasis of thyroid papillary microcarcinoma using
proteomic analysis of ultrasound-guided fine-needle aspiration
biopsy samples. J Proteomics. 204:1034142019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Mishall KM, Beadnell TC, Kuenzi BM,
Klimczak DM, Superti-Furga G, Rix U and Schweppe RE: Sustained
activation of the AKT/mTOR and MAP kinase pathways mediate
resistance to the Src inhibitor, dasatinib, in thyroid cancer.
Oncotarget. 8:103014–103031. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Krishnan A, Berthelet J, Renaud E,
Rosigkeit S, Distler U, Stawiski E, Wang J, Modrusan Z, Fiedler M,
Bienz M, et al: Proteogenomics analysis unveils a TFG-RET gene
fusion and druggable targets in papillary thyroid carcinomas. Nat
Commun. 11:20562020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Rajalingam K and Dikic I: SnapShot:
Expanding the ubiquitin code. Cell. 164:1074–1074.e1. 2016.
View Article : Google Scholar : PubMed/NCBI
|