Introduction
Gastric cancer (GC) ranks as the fifth most commonly
diagnosed cancer and the third most common cause of cancer-related
mortality worldwide (1). Peritoneal
metastasis (PM) is a major cause of the development of recurrence
and distant dissemination in patients with GC (2). The median survival time of patients
with GC and PM is ~four months and such patients can barely survive
beyond five years (3,4). Malignant ascites (MA) is a common
manifestation of advanced GC with PM, which implies a shorter life
expectancy in patients with GC (5).
Among all patients with GC and PM, ~40% of them have MA fluid
(6). Although intraperitoneal
chemotherapy combined with systemic paclitaxel chemotherapy has
shown promising effects on the survival of patients with advanced
GC and PM, the prognoses of these patients are still poor (7).
An imbalance between the production and outflow of
fluid in the abdominal cavity results in ascites, which is
primarily reported in liver cirrhosis and a variety of malignancies
(8), such as ovarian (9) and gastrointestinal cancer with PM
(10). Accumulated ascites in the
abdominal cavity, which are composed of cellular and acellular
components, constitute a unique microenvironment for PM and the
tumor cells suspended in the peritoneal fluid (11). Non-cancer cells in ascites, such as
macrophages (12) and
cancer-associated fibroblasts (13), interact with cancer cells and serve
a pivotal role in the progression of PM. In addition, the acellular
fraction of ascites, such as exosomes (14), metabolites (15), soluble growth factor (16) and chemokines (17) is involved in peritoneal
dissemination, epithelial-to-mesenchymal transition (EMT),
chemoresistance and cell proliferation (14,18).
Previous research indicated that pleural effusion and ascites from
breast, lung and ovarian cancer may induce EMT and manifestation of
cancer stem cell traits via activation of the PI3K/Akt/mTOR pathway
(19). However, little is known
about the effects of MA supernatant on GC cells, which needs
further exploration.
Asparagine synthetase (ASNS), which catalyzes the
synthesis of asparagine and glutamate using aspartic acid and
glutamine, is ubiquitous in mammalian cells (20). Upregulation of the expression of
ASNS is associated with poor prognosis in patients with colorectal
cancer (21), hepatocellular
carcinoma (22) and malignant
gliomas (23), suggesting a
prominent role in cancer progression. In addition, the
downregulation of ASNS expression inhibits the growth of GC
(24). Nevertheless, a consensus
has not been established on the correlation between ASNS expression
and cancer evolution (25,26). Few studies have explored the role of
the ASNS in mediating cancer cell progression associated with
ascites or peritoneal lesions in patients with GC.
The current study investigated the effects of MA on
the proliferation of GC cells. RNA sequencing was used to explore
the differentially expressed genes (DEGs) between MA-treated GC
cells and benign ascites-treated GC cells. ASNS, one of the
DEGs, was assessed its role in proliferation-promoting effects of
MA on GC cells. Notably, MA may initiate activation of the
activating transcription factor 4 (ATF4)-ASNS axis to promote the
proliferation of GC cells.
Materials and methods
Ascites samples
MA fluid was obtained from GC patients with large
volumes (>1,000 ml) of MA, who were admitted to Changhai
Hospital (Shanghai, China) between May 1, 2020 and Jan 31, 2021.
All patients were free from peritonitis, life-threatening
complications and secondary cancers. The clinicopathological
features of the patients are shown in Table I. A sample of benign ascites was
collected from a 46-year-old female patient diagnosed with liver
cirrhosis. Ascites (50–100 ml) were collected from the enrolled
patients who underwent peritoneal paracentesis. Ascites samples
were processed within 24 h after collection. After being dispensed
into 50 ml sterile centrifuge tubes, the samples were centrifuged
at 400 × g at 4°C for 10 min. The cell pellets were then used for
organoid construction and the supernatant was transferred into a
new centrifuge tube after being filtered through a 0.22 µm
sterilizing filter. All ascites supernatants were cryopreserved at
−80°C and heat-inactivated prior to use.
 | Table I.Clinicopathological characteristics
of gastric cancer patients with malignant ascites. |
Table I.
Clinicopathological characteristics
of gastric cancer patients with malignant ascites.
| No. | Sex | Age (years) | Disease | Primary cancer
sites | Pathology type | Clinical stage | Her-2 status |
|---|
| 1 |
Male | 36 | Gastric cancer |
Body | Poorly
differentiated | IV |
Positive |
| 2 |
Male | 61 | Gastric cancer | Antrum | Medium
differentiated | IV |
Negative |
| 3 | Female | 69 | Gastric cancer |
Cardia | Poorly
differentiated | IV | Not detected |
Cell culture
The human GC cell lines, AGS, SNU5 and SNU16, were
purchased from the Cell Bank of Chinese Academy of Science and Bena
Culture Collection. AGS is an adherent cell line, while SNU5 and
SNU16 are semi-adherent and suspended cells respectively. AGS cells
were cultivated in high-glucose DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100
ng/ml streptomycin. SNU5 and SNU16 cells were maintained in
RIPM1640 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS. All cell lines were subjected to STR analysis and the
absence of mycoplasma contamination was confirmed. The three cell
lines were cultivated at 37°C with 5% CO2. Organoids
derived from MA were constructed as previously described (27).
Cell proliferation assay
Cell proliferation was measured using Cell Counting
Kits-8 (CCK-8) according to the instructions of the manufacturer
(Beyotime Institute of Biotechnology). Briefly, 2,000 cells per
well (100 µl) were seeded in a 96-well plate. After incubation with
or without 10% MA for 24, 48 and 72 h, the normalized proliferation
of cells was determined. A total of 10 µl of CCK-8 was added to
each well. After 2 h of incubation, the optical density was
measured at 450 nm.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from 1×106 cells
using RNAiso Plus reagent (Takara Bio, Inc.) according to the
manufacturer's protocol. Reverse transcription was then performed
to obtain cDNA using PrimeScript RT Master Mix (Takara Bio, Inc.)
according to manufacturer's instructions. Next, qPCR was performed
in a total reaction volume of 20 µl to determine the relative mRNA
expression of the target genes using the Powerup SYBR Green Master
Mix (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The parameters of qPCR were as
follows: Initial denaturation at 95°C for 120 sec; followed by 40
cycles of denaturation at 95°C for 10 sec, and annealing and
extension at 60°C for 30 sec. The primers used are listed in
Table SI. Relative mRNA expression
of genes was calculated using the 2−ΔΔCq formula
(28) and β-actin was used as an
internal control. The experiments were performed in triplicate.
Western blotting (WB)
Total protein of GC cells was extracted using RIPA
lysis buffer (Beyotime Institute of Biotechnology) containing
phenylmethylsulfonyl fluoride and phosphatase inhibitors. The
concentration of protein was determined using the Pierce BCA
protein kit (Thermo Fisher Scientific, Inc.). A total of 40 µg of
the total protein was loaded in each gel lane and proteins with
different molecular weights were separated by 10% SDS-PAGE. After
the proteins were transferred onto PVDF membranes, the membranes
were blocked with 5% skimmed milk at 25°C for 1 h. The membranes
were then incubated with ASNS (Santa Cruz Biotechnology, Inc.;
1:800; cat. no. 365809; mouse), ATF4 (Proteintech Group, Inc.;
1:1,000; cat. no. 10835-1-AP; rabbit) or β-actin antibody (Cell
Signaling Technology, Inc.; 1:1,000; cat. no. 3700; mouse) at 4°C
overnight. The membrane was then incubated with the corresponding
HRP-conjugated secondary antibodies (Biosharp Life Sciences; cat.
nos. BL001A and BL003A; 1:5,000; goat) at 25°C for 1 h. Protein
expression was detected by the Amersham imager 680 (Cytiva).
Quantification of the strips was performed using ImageJ software
(National Institutes of Health; version: 1.8.0).
Immunofluorescence
Cells at a density of 3×105 per well were
seeded onto the 6-well plate. AGS cells treated with or without
ascites were fixed using 4% paraformaldehyde for 10 min at 25°C.
After blocking with 5% donkey serum (cat. no. ab7475; Abcam) for 30
min, the cells were incubated with a Ki-67 antibody (Abcam; 1:200;
cat. no. ab15580; mouse) at 4°C overnight. After washing with PBST
three times, the cells were cultured with secondary antibodies
conjugated to Alexa-Fluor 488 (Abcam; 1:1,000; cat. no. ab150113;
goat) at 25°C for 1 h. The cells were subsequently stained with
DAPI at 25°C for 10 min. Finally, images were captured using a
fluorescence microscope (magnification, ×200; Olympus Corporation).
A total of five random fields were selected to capture images for
calculating Ki-67 positive cells.
RNA isolation, library preparation and
RNA sequencing (RNA-seq)
RNA-seq of MA-treated AGS and untreated AGS cells
was performed in duplicate. Total RNA was isolated using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The purity and
quantification of RNA were evaluated using the NanoDrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). RNA integrity
assessment was conducted using the Agilent 2100 Bioanalyzer
(Agilent Technologies, Inc.). Libraries were then constructed using
a TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, Inc.) and the
libraries were sequenced on an Illumina HiSeq X Ten platform
(Illumina, Inc.) by 150 bp paired-end sequencing. Then clean data
were mapped to the human genome (GRCh38) using HISAT2 (http://daehwankimlab.github.io/hisat2/).
Transcriptome sequencing and data analysis were conducted by
Shanghai OE Biotech Co., Ltd. P<0.05 and foldchange >2 or
<0.5 were set as differentially expressed genes (DEGs)
threshold. DEGs were identified using the DESeq R package
(http://www.bioconductor.org/; version:
1.34.1) (29).
Small interfering (si) RNA
transfection
ASNS siRNA, ATF4 siRNA and negative control (NC;
non-targeting siRNA) were designed and constructed by Shanghai
GenePharma Co., Ltd. and the sequences are shown in Table SII. A density of 5×105
cells per well were seeded into 6-well plates to achieve 60–80%
confluence after 12 h of cultivation. RNA iMAX (Thermo Fisher
Scientific, Inc.)-siRNA mix (final siRNA concentration, 50 nM) was
then added to each well. Cells were transfected at 37°C for 8 h.
After 48 h, transfected cancer cells were collected to detect the
silencing effect on the target genes at the mRNA and protein
levels. The siRNA with the highest efficacy was used for further
experiments. The subsequent experiments were performed 48 h after
transfection.
Stably transfected cells
Control vector and ASNS-overexpression
(ASNSoe) lentiviral vector were constructed using
pcSlenti-EF1-EGFP-P2A-puro-CMV-3×FLAG-WPRE. The lentivirus was
constructed and packaged by OBiO Technology (Shanghai) Corp., Ltd.
A total of 40 µl lentivirus (4×108 TU/ml) was added to
each well to infect the targeted cells. GPF fluorescence was
detected using a fluorescence microscope (Olympus Corporation)
after 48 h. Transduced cells were selected using 2 µg/ml puromycin
to obtain stable cell lines. The overexpression of ASNS was then
verified by RT-qPCR and WB.
Chromatin immunoprecipitation (ChIP)
assay
This was conducted according to the manufacturer's
instructions using an EZ-Magna CHIP A/G kit (cat. no. 17-10086;
MilliporeSigma). AGS cells (density,
1.3×105/cm2 in a 75-cm2 cell
culture flask) treated with MA or benign ascites were cross-linked
using formaldehyde at 25°C for 10 min and then lysed with ChIP
lysis buffer. Then nuclear lysates were sonicated (20 kHz frequency
at 4°C for 10 min) on ice to generate 100–500 bp DNA fragments.
After the sonication and centrifugation (10,000 × g at 4°C for 10
min), the supernatant was removed to a new tube with CHIP dilution
buffer (part no. CS200624; MilliporeSigma) containing protease
inhibitors (part no. 20-283; MilliporeSigma). Subsequently, 10 µl
of the supernatant was removed as input control, and the input
control was stored at 4°C and then processed via elution and
cross-link reversal, while the rest of the supernatant was divided
into two aliquots and these two aliquots were immunoprecipitated at
4°C for 2 h using anti-ATF4 (Cell Signaling Technology, Inc.;
dilution, 1:200; cat. no. 11815S; rabbit) or IgG antibody (Cell
Signaling Technology, Inc.; dilution, 1:200; cat. no. 2729S;
rabbit) with magnetic protein A/G beads (part no. CS204457;
MilliporeSigma). The magnetic beads were pelleted and the
supernatants were removed. The bead-antibody/chromatin complexes
were washed with the buffers in the following order: Low salt wash
buffer (part no. CS200625; MilliporeSigma), one wash; high salt
wash buffer (part no. CS200626; MilliporeSigma), one wash; LiCl
wash buffer (part no. CS200627; MilliporeSigma), one wash; and TE
buffer (part no. CS200628; MilliporeSigma), one wash. After elution
and cross-link reversal of protein-DNA complex (including the
immunoprecipitated samples and the input control samples), the DNA
was purified using unique polypropylene spin columns which contains
activated silica membrane filters that can capture DNA and separate
the DNA from proteins in combination with the binding reagent A
(part no. 20-292; MilliporeSigma) and washing reagent B (part no.
20-293; MilliporeSigma). The purified DNA was eluted using elution
reagent C (part no. 20-294; MilliporeSigma) and used to perform
qPCR. The enrichment ratio was assessed by qPCR which was performed
as described for RT-qPCR. Primers for the ASNS promoter are
shown in Table SI.
Statistical analysis
All experiments were performed in triplicate and the
data are presented as mean ± standard deviation. One-way ANOVA and
unpaired t-test were used to compare the differences between
multiple groups or two groups, respectively. Tukey's honestly
significant difference test was used after the ANOVA when comparing
multiple groups. GraphPad Prism version 8 (Dotmatics) was used to
generate graphs and perform statistical analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
MA supernatant promotes the
proliferation of GC cells
The survival and proliferation of GC cells in MA is
a pivotal step in PM formation and progression (30). To evaluate the effects of MA on the
proliferation of GC cells, AGS, SNU16 and SNU5 cells were incubated
with different gradients of ascitic fluid (0, 10, 25, 50 and 100%)
for 72 h. Then, the normalized proliferation of the cells was
detected using the CCK-8. The results indicated that the
proliferation of GC cells was increased after treatment with
gradient ascites compared with the untreated cells (P<0.05;
Fig. 1A). However, no significant
dose-dependent effects of MA were observed during the
proliferation-promoting process. To exclude the different effects
of variable gradient ascites on cell proliferation, cells treated
with 10% ascites supernatant were selected as the treated group
while cells treated with 10% benign ascites were regarded as the
control group. To further verify the promotional effects of MA
fluid on cancer cells, the proliferation of GC cells treated with
10% MA from different patients was evaluated. The results
demonstrated that ascites supernatants from three representative
patients promoted the proliferation of GC cells at 72 h (Fig. 1B). Phase contrast images of cell
lines and organoids derived from MA (MADO) cultured in two
dimensions at 48 h and three dimensions (3D) at 72 h were captured
after treatment with MA or benign ascites (Figs. 1C and S1A). The volumes of the spheres of the
treated group in the 3D culture system were larger than those of
the control group (Fig. S1B).
Furthermore, after treatment with 10% MA or benign ascites for 48
h, the proportion of Ki67+ AGS cells was determined. A
higher proportion of Ki67+ cells was observed in the
MA-treated group than in the untreated group (P<0.01; Fig. 1D). Taken together, these findings
suggested that MA promoted the proliferation of GC cells ex
vivo.
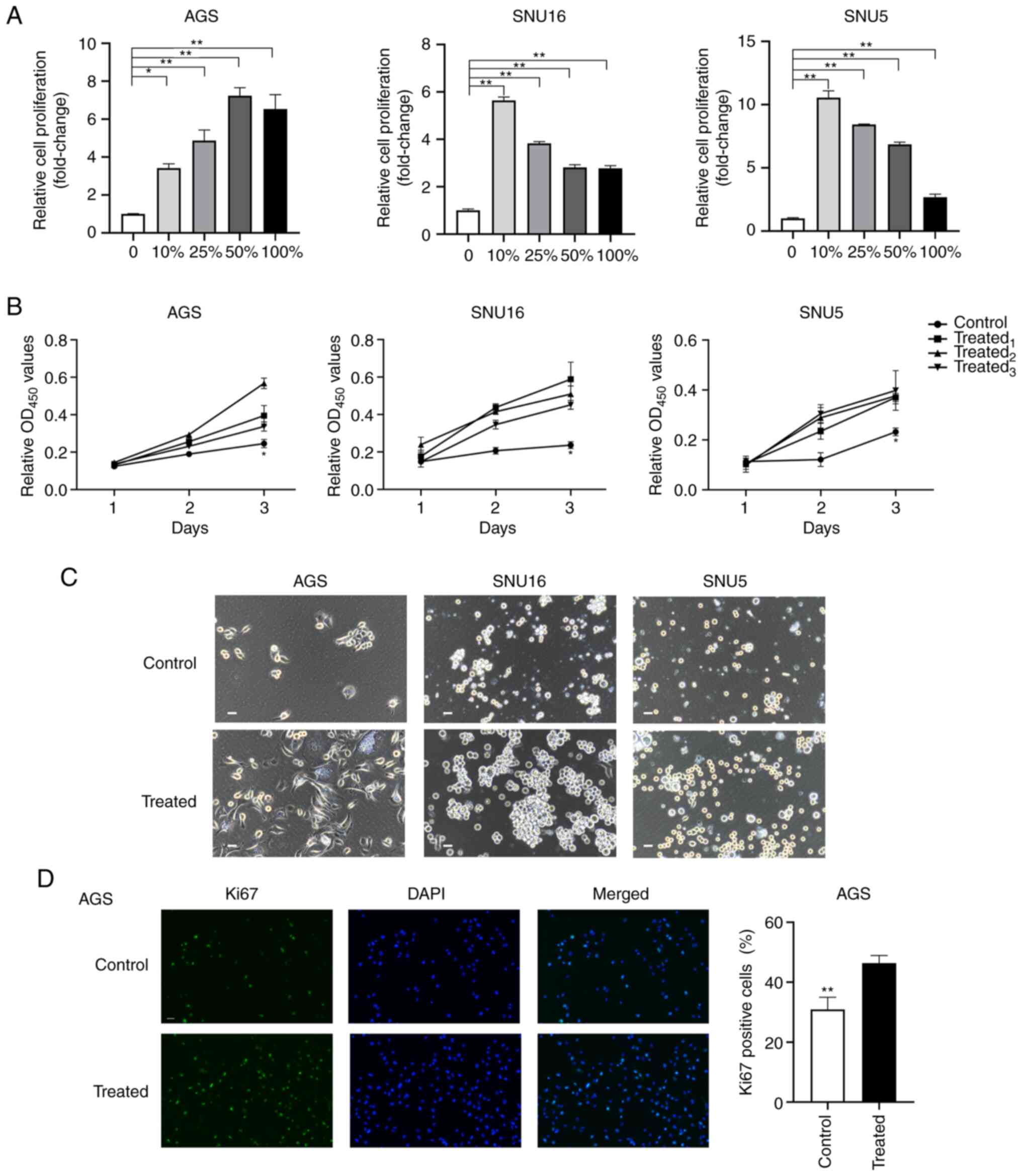 | Figure 1.MA supernatant promotes the
proliferation of gastric cancer cells. (A) Cell proliferation was
assessed by CCK8 after AGS, SNU16 and SNU5 cells were treated with
medium containing 0, 10, 25, 50, 100% gradient MA for 72 h. (B) The
proliferation of AGS and SNU16, SNU5 cells grown in medium with 10%
MA or 10% benign ascites at 24, 48 and 72 h was measured by CCK8
assay. Control: 10% benign ascites treatment; treated1,
treated2 and treated3 represent cells treated
with 10% MA from three different patients with GC. (C) Phase
contrast images of AGS, SNU5, SNU16 cells exposed to medium with or
without MA in two dimensions were captured at 48 h (scale bar, 100
µm; MA was derived from the patient named No.1 in Table I). (D) Immunofluorescence staining
of Ki67 was performed in AGS after being treated with MA or benign
ascites for 24 h (magnification, ×200). The quantification of Ki67
positive cells was shown in the bar graphs. Control: Benign ascites
treatment; treated: Treated with MA which was collected from the
patient named No.1 in Table I. Data
are shown as mean ± standard derivation. P-value was analyzed by
one-way ANOVA or two-tail unpaired Student's t test
(*P<0.05; **P<0.01.). MA, malignant ascites; GC, gastric
cancer. |
MA induces the upregulation of ASNS
expression in GC cells
To further elucidate the underlying molecular
mechanisms mediating the proliferation-promoting effects of MA on
GC cells, transcriptome sequencing was performed to screen for
DEGs. AGS cells, treated with MA (derived from three patients with
GC) or benign ascites for 24 h, were collected for RNA-seq. As
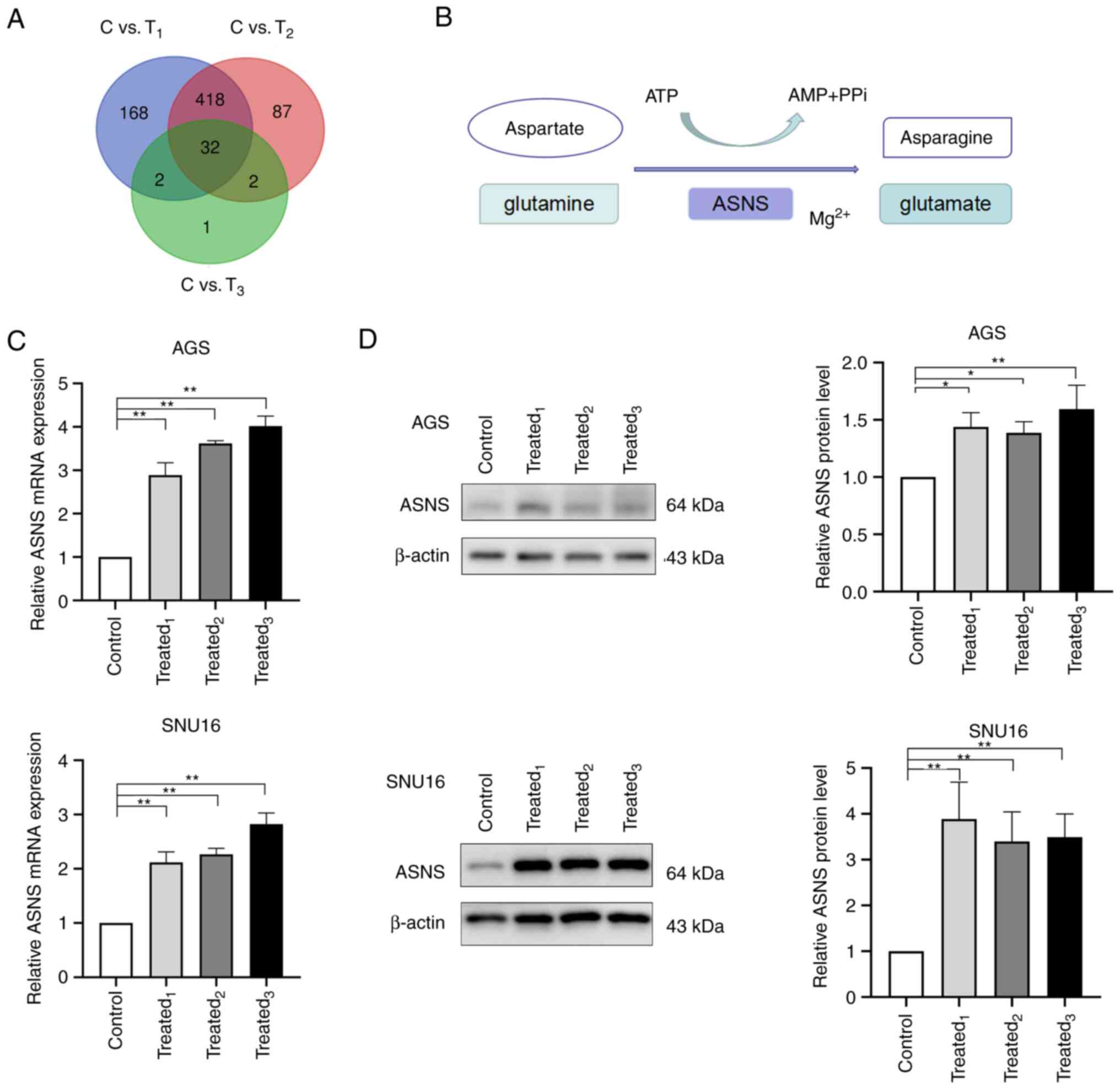
illustrated in the Venn graph shown in Fig. 2A, 32 DEGs were identified. According
to the results of the sequencing, the mRNA levels of 13 genes were
upregulated in the MA-treated groups whereas 19 genes were
downregulated (Table II).
 | Table II.Differentially expressed genes of
malignant ascites-treated AGS cells compared with the control
group. |
Table II.
Differentially expressed genes of
malignant ascites-treated AGS cells compared with the control
group.
|
|
| Treated1
vs. control | Treated2
vs. control | Treated3
vs. control |
|---|
|
|
|
|
|
|
|---|
| Gene |
Upregulated/Downregulated | Fold change | P-value | Fold change | P-value | Fold change | P-value |
|---|
| ASNS | Upregulated | 5.47 | <0.05 | 5.16 | <0.05 | 2.47 | <0.05 |
| DDIT4 | Upregulated | 5.52 | <0.05 | 4.50 | <0.05 | 2.50 | <0.05 |
| FGF21 | Upregulated | 242.22 | <0.05 | 177.80 | <0.05 | 86.37 | <0.05 |
| INHBE | Upregulated | 25.40 | <0.05 | 20.78 | <0.05 | 3.55 | <0.05 |
| IRAK1BP1 | Upregulated | 2.95 | <0.05 | 2.75 | <0.05 | 2.16 | <0.05 |
| KLHDC7B | Upregulated | 50.10 | <0.05 | 29.50 | <0.05 | 4.51 | <0.05 |
| NUPR1 | Upregulated | 8.41 | <0.05 | 6.87 | <0.05 | 2.43 | <0.05 |
| PCK2 | Upregulated | 3.87 | <0.05 | 3.47 | <0.05 | 2.19 | <0.05 |
| S100P | Upregulated | 8.28 | <0.05 | 6.13 | <0.05 | 2.66 | <0.05 |
| SLC43A1 | Upregulated | 5.32 | <0.05 | 5.34 | <0.05 | 2.66 | <0.05 |
| SPINK1 | Upregulated | 5.02 | <0.05 | 4.52 | <0.05 | 3.06 | <0.05 |
| TGM2 | Upregulated | 2.57 | <0.05 | 2.57 | <0.05 | 2.69 | <0.05 |
| TUBE1 | Upregulated | 4.26 | <0.05 | 3.56 | <0.05 | 2.17 | <0.05 |
| BATF2 | Downregulated | 0.10 | <0.05 | 0.07 | <0.05 | 0.18 | <0.05 |
| CCDC141 | Downregulated | 0.24 | <0.05 | 0.22 | <0.05 | 0.37 | <0.05 |
| CMPK2 | Downregulated | 0.21 | <0.05 | 0.19 | <0.05 | 0.44 | <0.05 |
| CREB3L3 | Downregulated | 0.24 | <0.05 | 0.25 | <0.05 | 0.40 | <0.05 |
| CXCL11 | Downregulated | 0.45 | <0.05 | 0.36 | <0.05 | 0.43 | <0.05 |
| HSD17B2 | Downregulated | 0.18 | <0.05 | 0.23 | <0.05 | 0.38 | <0.05 |
| ISG15 | Downregulated | 0.31 | <0.05 | 0.30 | <0.05 | 0.47 | <0.05 |
| KCNK2 | Downregulated | 0.03 | <0.05 | 0.03 | <0.05 | 0.19 | <0.05 |
| MSMO1 | Downregulated | 0.43 | <0.05 | 0.25 | <0.05 | 0.49 | <0.05 |
| PSG4 | Downregulated | 0.15 | <0.05 | 0.14 | <0.05 | 0.45 | <0.05 |
| REG4 | Downregulated | 0.24 | <0.05 | 0.30 | <0.05 | 0.43 | <0.05 |
| RSAD2 | Downregulated | 0.35 | <0.05 | 0.32 | <0.05 | 0.44 | <0.05 |
| SARM1 | Downregulated | 0.26 | <0.05 | 0.29 | <0.05 | 0.36 | <0.05 |
| SEMA3C | Downregulated | 0.19 | <0.05 | 0.21 | <0.05 | 0.47 | <0.05 |
| SLC7A8 | Downregulated | 0.17 | <0.05 | 0.25 | <0.05 | 0.48 | <0.05 |
| SYNPR | Downregulated | 0.37 | <0.05 | 0.42 | <0.05 | 0.49 | <0.05 |
| SYP | Downregulated | 0.15 | <0.05 | 0.17 | <0.05 | 0.39 | <0.05 |
| TNNC1 | Downregulated | 0.43 | <0.05 | 0.30 | <0.05 | 0.46 | <0.05 |
| TTN | Downregulated | 0.11 | <0.05 | 0.10 | <0.05 | 0.26 | <0.05 |
ASNS was one of the upregulated genes in
MA-treated AGS cells. It is a universally expressed gene in almost
all tissues in humans, with its encoded protein catalyzing the
synthesis of asparagine and glutamate using glutamine and aspartate
in the presence of ATP (Fig. 2B)
(31). The aberrant expression of
ASNS is correlated with cancer progression (32–34).
As few studies have explored the role of ASNS in the
progression of PM in GC, it was selected as the target gene for
further study. To determine whether MA treatment could induce high
expression of ASNS, the mRNA and protein levels of ASNS was
detected in AGS and SNU16 cells after being treated with MA from
three GC patients. Significant upregulation of ASNS expression at
the mRNA level was observed in MA-treated AGS and SNU16 cells when
compared with those in the control group (Fig. 2C). Besides, AGS and SNU16 cells
exposed to a medium containing 10% MA exhibited an increase in ASNS
protein levels (Fig. 2D). The
aforementioned results suggested that GC cells exposed to MA had
enhanced expression of ASNS.
MA exhibits proliferation-promotional
effects on GC cells partially via upregulation of ASNS
expression
Based on the RNA-seq and in vitro
experiments, the findings of the present study implied that MA from
different patients could mediate an increase in ASNS expression in
GC cells. To further determine whether ASNS serves a pivotal role
in mediating the effects of MA on GC cell proliferation, ASNS
knockdown and overexpression were performed. Since the baseline
levels of ASNS in SNU16 cells were higher than those in AGS cells
(P<0.01; Fig. S2A), the AGS
cell line was chosen for overexpression of ASNS. The silencing
efficacy of siRNA (Fig. 3A and B)
and the upregulation of ASNS in ASNSoe
lentivirus-transduced cells (Fig.
S2B) at the mRNA and protein levels were assessed. Next, the
relative proliferation of GC cells transfected with
siRNA-NC/siRNA-ASNS combined with treatment with or without MA was
determined. The results indicated that the relative proliferation
of AGS cells treated with MA was partially inhibited upon a
decrease in ASNS expression (Fig. 3C). In addition, AGS cells stably
expressing upregulated ASNS levels exhibited higher proliferation
ability (Fig. 3D). In addition, in
MA-treated AGS, the number of Ki67+ cells also decreased
when ASNS expression was knocked down by siRNA, compared
with AGS cells transfected with siRNA-NC (P<0.05; Fig. 3E). The above findings revealed that
MA may promote the proliferation of GC cells partially via elevated
ASNS expression and downregulation of ASNS could, in part, reverse
the effects of MA on GC cell proliferation.
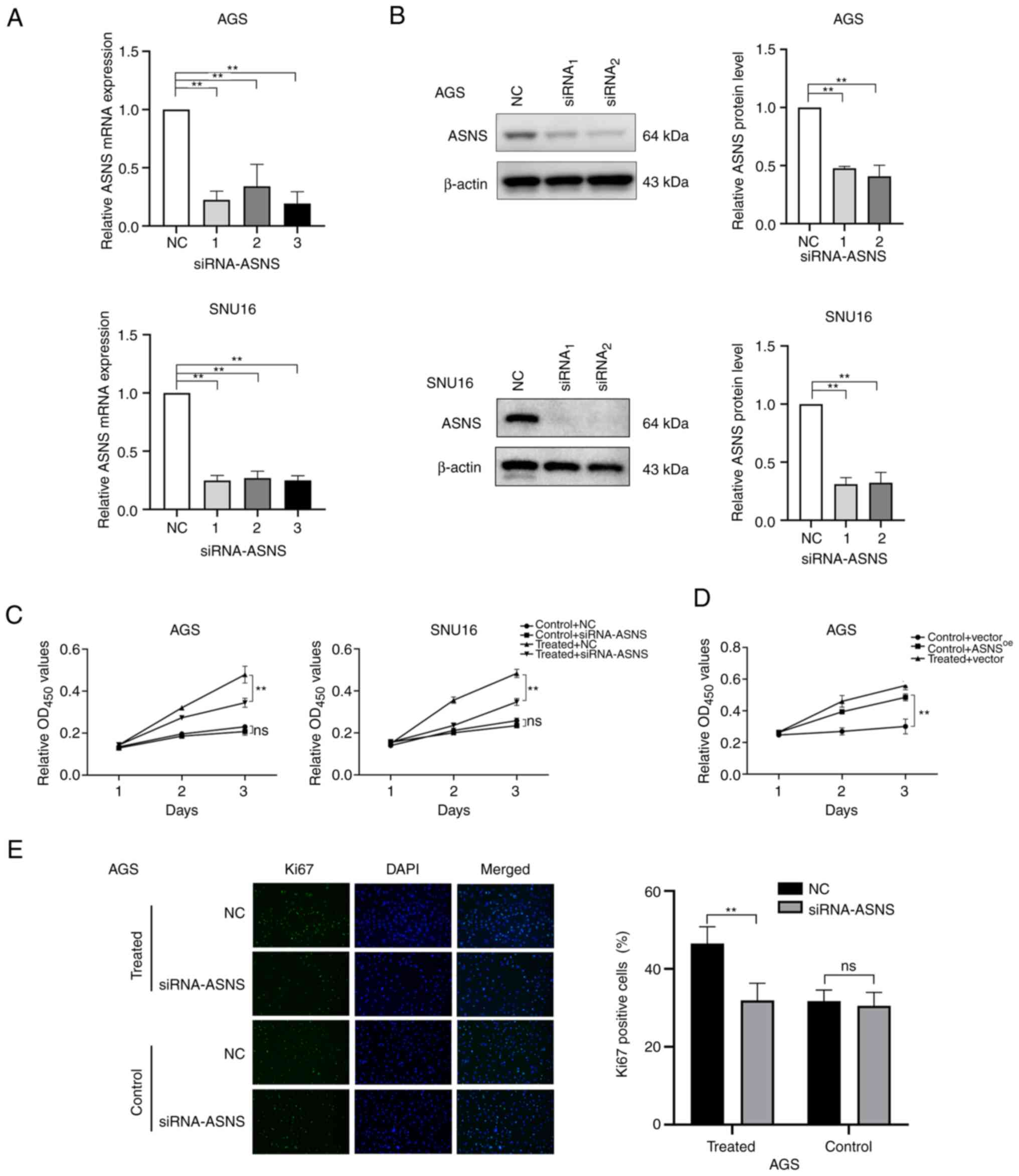 | Figure 3.Enhanced proliferation of GC cells
following MA treatment may be partially mediated by the
upregulation of ASNS. (A) Knockdown efficacy of siRNA on the mRNA
level of ASNS was quantified by reverse transcription-quantitative
PCR. 1, 2, 3 represent three different siRNAs targeting ASNS. (B)
Silencing efficacy of siRNA targeting ASNS was detected by western
blotting at the protein level. (C) After cells were transfected
with siRNA-ASNS or NC, the growth of AGS and SNU16 cells exposed to
medium with or without MA (obtained from the No.1 patient in
Table I) were analyzed using CCK8
on day 1, 2 and 3. (D) The proliferation of AGS cells with or
without MA (from the No.1 patient in Table I) treatment, after being infected
with negative control (vector)/ASNS-overexpression lentivirus
(ASNSoe), was determined by CCK8 at day 1, 2, 3. (E)
Immunofluorescence staining was applied to detect the proportion of
Ki67-positive cells in MA-treated/control AGS with siRNA-ASNS or NC
(magnification, ×200). MA was from the patient named No.1 in
Table I. Control: With benign
ascites treatment; treated: Treated with MA. **P<0.01; ns, no
statistical significance. GC, gastric cancer; MA, malignant
ascites; ASNS, asparagine synthetase; si, small interfering; NC,
negative control. |
Malignant abdominal fluid of patients
with GC promotes the proliferation of cancer cells via the
activated ATF4-ASNS axis
ATF4 is the main transcription factor (TF) that
regulates ASNS (35). Prediction
results presented on JASPAR (http://jaspar.genereg.net/), an online TF database
(36), indicated that ATF4 might
bind to the ASNS promoter region with a relative score of
7.17. In addition, a ChIP-qPCR assay using AGS cells suggested that
ATF4 binds to the promoter of ASNS and that MA treatment
induced enhanced binding activity of ATF4 to the promoter region of
ASNS (Fig. 4A). Considering
the aforementioned results, it was hypothesized that MA might
initiate the upregulation of ASNS via ATF4. To verify this
hypothesis, the expression of ATF4 after MA treatment was detected.
Relative protein levels of ATF4 increased in AGS and SNU16 cells
(Fig. 4B) following treatment with
MA derived from different patients. Then, siRNA (Table SIII) targeting ATF4 was constructed
to knockdown the expression of ATF4. siRNA1 and
siRNA3 targeting ATF4 exhibited the highest
silencing efficacy in AGS and SNU16 cells, respectively (Fig. S2C). After ATF4 expression was
knocked down, the promoting effects of MA on cancer growth were
partially mitigated at 72 h (P<0.01; Fig. 4C) and the proportion of
Ki67-positive cells decreased in MA-treated AGS cells (Fig. 4D). These results suggested that MA
could upregulate ATF4 expression, accelerating the proliferation of
cancer cells.
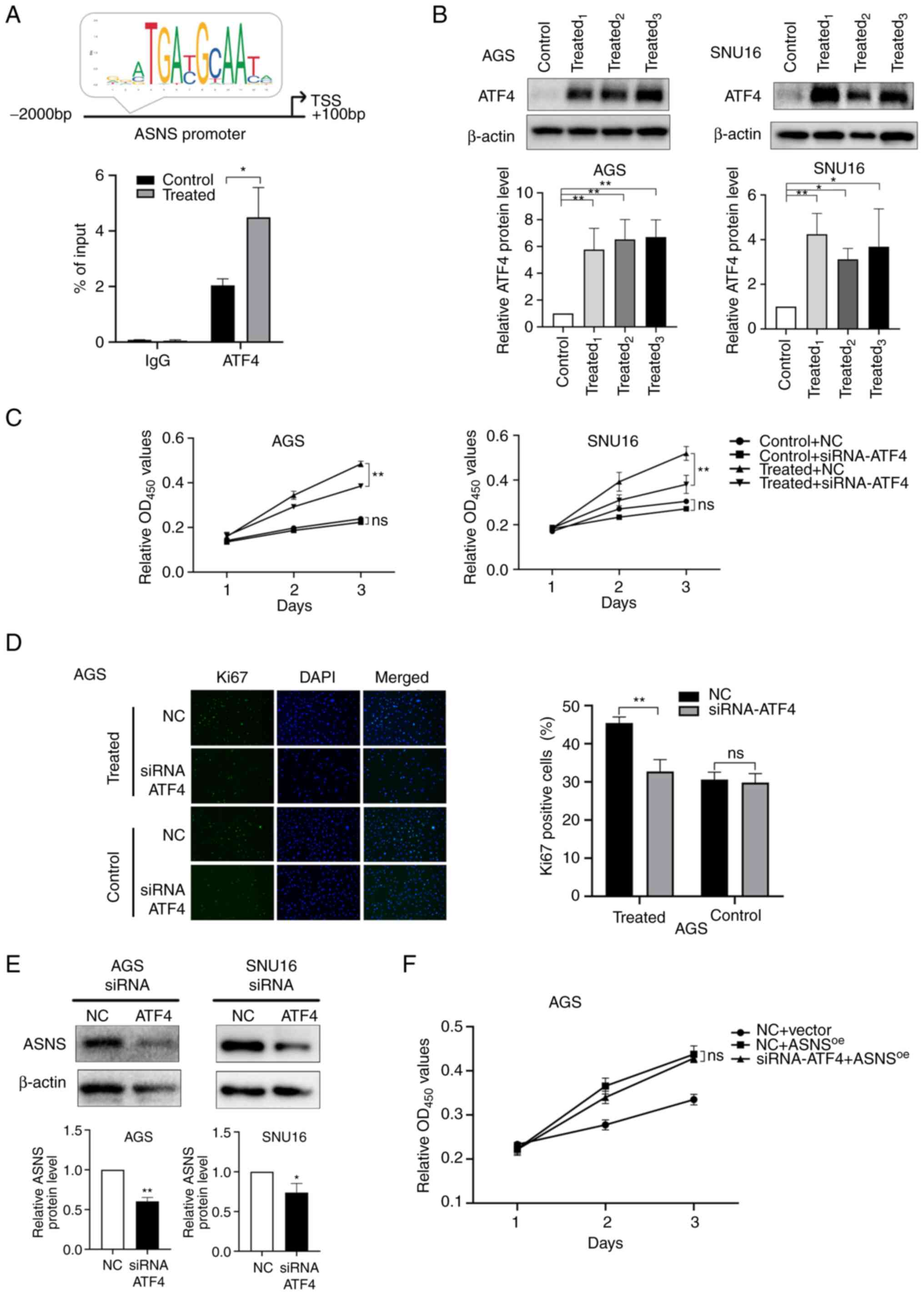 | Figure 4.MA promotes the growth of GC cells
partially via the activation of ATF4-ASNS axis. (A) Top: Predictive
binding site of ATF4 at the promoter region of ASNS by JASPAR
database and (bottom) chromatin immunoprecipitation assay of ATF4
on ASNS promoter in AGS cells treated with malignant ascites or
benign ascites for 48 h. (B) The protein levels of ATF4 in AGS and
SNU16 cells and the quantification were measured by western
blotting after cells were treated with 10% benign ascites or 10% MA
for 48 h. Control: Treated with benign ascites;
treated1, treated2 and treated3:
Cells treated with MA from three different GC patients. (C) Cell
viability of AGS and SNU16 cells, after cells were transfected with
siRNA-NC/siRNA-ATF4, were detected on day 1, 2 and 3 by CCK8 when
cells were treated with benign ascites (control) or MA (treated).
(D) Ki67-positive cells were counted in MA-treated/control AGS
after siRNA-ATF4 or NC was transfected (magnification, ×200). MA
used in (C) and (D) was from the No.1 patient in Table I. (E) ASNS expression at the protein
level in AGS and SNU16 was detected by western blotting when ATF4
was downregulated using siRNA-ATF4. (F) In negative control
lentivirus (vector) or ASNS-overexpressed lentivirus
(ASNSoe) transduced AGS, the proliferation of cells
treated with siRNA-ATF4/NC were recorded at 24, 48 and 72 h by
CCK-8 assay (ns, no statistical significance; *P<0.05;
**P<0.01). MA, malignant ascites; GC, gastric cancer; ATF4,
activating transcription factor 4; ASNS, asparagine synthetase; si,
small interfering; NC, negative control. |
To further elucidate whether ascites promotes cell
proliferation by influencing the expression of ASNS via the
upregulation of ATF4, the expression of ASNS after ATF4 silencing
was determined. The results showed that ASNS levels were
simultaneously downregulated when effective siRNA-ATF4 was applied
(Fig. 4E). In addition, for AGS
cells, no significant differences were observed in proliferation
between siRNA-NC and siRNA-ATF4 groups when ASNS was overexpressed
(Fig. 4F), suggesting that
ASNS upregulation could reverse the inhibitory effects of
siRNA-ATF4 on cell proliferation. Taken together, these results
indicated that MA might enhance the proliferation of GC cells via
activation of the ATF4-ASNS axis.
Discussion
MA fluid is often observed in advanced stage and
invasive cancers which supports the view that MA is involved in
disease progression (37).
According to clinical data, the management of MA in patients with
advanced GC would improve their prognoses (6). Survival and proliferation in the
peritoneal cavity are critical steps for GC cells to form
peritoneal metastases after cancer cells detach from the primary
cancer sites (38). The tumor
microenvironment has been a hotspot in cancer research for decades
(39–41). MA is a special microenvironment for
detached GC cells and the metastasis sites of the peritoneum and
has garnered significant interest recently. Previous research has
implied that components such as growth factors (IL6 and VEGF)
(19), soluble proteins (16), lipids (15) and microRNAs (14) may accelerate the progression of PM.
However, the precise mechanisms involved in the process by which MA
influences the biological behaviors of cancer cells remain to be
elucidated. In the present study, the results suggested that the
proliferation of AGS and SNU16 cells treated with MA from different
patients with GC (stage IV with PM) was significantly enhanced
compared with that of the cells treated with benign ascites. This
finding is consistent with prior studies that demonstrated that
pleural fluid or MA can promote the proliferation of cancer cells
(42,43). In the present study, no significant
dose-dependent promotion effects of MA on the proliferation of
cancer cells were observed, which may be attributed to the
exhaustion of nutrients in the culture system. Therefore, a more
complex and refined model may be needed for an improved
representation of the actual conditions in vivo.
Most previous studies on pleural effusion or ascites
have primarily explored the influence of one component or a certain
type of substance on the progression of cancer (14,16,19).
The present study aimed to elucidate the potential universal
mechanisms underlying the effects of different MA on GC cell
proliferation. It was found that MA treatment induced the
upregulation of ASNS levels in GC cells, which serves a role
in mediating the proliferation-promoting effects of MA on GC cells.
In addition, the upstream TF ATF4, a main regulator of ASNS,
was upregulated at the protein level following MA treatment.
Finally, the findings indicated that the increased proliferation of
MA-treated cells may be partially attributed to the activation of
the ATF4-ASNS axis.
Previous research on ASNS has mainly focused on
hematological malignancies (44).
As acute lymphoblastic leukemia (ALL) tumor cells lack ASNS,
L-asparaginase, which catalyzes the conversion of asparagine into
aspartate, exhibits anti-tumor activity for ALL cell proliferation
relying on exogenous asparagine (45). In the present study, MA promoted the
upregulation of ASNS, which resulted in increased levels of
asparagine and L-asparaginase, did not show antitumor effects in
MA-treated cancer cells (data not shown). This is consistent with
previous conclusions that upregulated ASNS can confer resistance to
L-asparaginase (46,47). In addition, to the best of the
authors' knowledge, no clinical drugs targeting ASNS have yet been
developed. A recent study suggested that inhibition of the
production of aspartate can sensitize ASNShigh lymphoma
to L-asparaginase (48). This
conclusion may provide new insights into the treatment of GC with
PM and MA.
The present study revealed a previously unknown
mechanism of the proliferation-promoting effects of MA on GC cells
which may be involved in the progression of PM. However, the
substance in MA that specifically affects the proliferation of
cancer cells via the activated ATF4-ASNS axis remains unclear.
Owing to the heterogeneity and complexity of MA, which contains a
variety of proteins, polypeptides, lipids and some small molecules,
the authors are planning to use mass spectrometry and
chemokine/cytokine arrays to explore the active substances in MA in
a future study. In addition, previous research has demonstrated
that the molecular components and cell contents of ascites change
continuously during the course of disease, implying that a changing
environment for cancer cells has emerged in MA (11). Therefore, ascites from patients with
different stages, subtypes or even ascitic fluids derived from one
patient at different times may exert different effects on the
biological behavior of cancer cells. In addition, the number of
ascites samples used in the present study was limited, so more
representative samples are needed in a future study to further
support our conclusions.
Of patients with cancer, ~10% develop MA during the
course of the disease, which is primarily observed in patients with
ovarian, pancreatic and gastric cancer (49). The effect of MA on tumor progression
has not been thoroughly elucidated and the underlying molecular
mechanisms remain obscure. In the present study, it was
demonstrated that the MA supernatant from patients with GC promoted
the proliferation of cancer cells. MA fluid may activate the
ATF4-ASNS axis, which facilitates the enhanced proliferation of GC
cells. These findings imply that, for advanced GC patients with PM
and MA, successful management of MA may slow down the progression
of PM and inhibition of the production of ASNS may be a potential
target for treating GC with MA.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Natural
Science Foundation of China (grant no. 82072707), the Scientific
Research Program of the Shanghai Municipal Commission of Science
and Technology (grant no. 20Y11909400) and the Changhai Hospital
234 Project (grant nos. 2019YXK019 and 2020YXK029).
Availability of data and materials
The RNA-seq datasets generated and/or analyzed
during the current study are available in the Genome Sequence
Archive for Human repository, https://ngdc.cncb.ac.cn/gsa-human/s/VUMxD15b. The
other datasets used and/or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
XZ and WL contributed to the conception, supervision
of the experiments and revision of the manuscript. YJ designed,
performed the experiments, conducted the data analysis and wrote
the manuscript. XP performed experiments, revised the article and
conducted data interpretation. YW, ZH, LC, MW, YZ and JL collected
the clinical samples, clinicopathological information and conducted
the experiments. XZ and YJ confirmed the authenticity of all the
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was performed in accordance with the
Declaration of Helsinki. All the enrolled patients signed informed
consents to provide ascitic biospecimen, which was approved by the
ethics committee of Navy Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wei J, Wu ND and Liu BR: Regional but
fatal: Intraperitoneal metastasis in gastric cancer. World J
Gastroenterol. 22:7478–7485. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thomassen I, van Gestel YR, van Ramshorst
B, Luyer MD, Bosscha K, Nienhuijs SW, Lemmens VE and de Hingh IH:
Peritoneal carcinomatosis of gastric origin: a population-based
study on incidence, survival and risk factors. Int J Cancer.
134:622–628. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saladino E, Fleres F, Mazzeo C, Pruiti V,
Scollica M, Rossitto M, Cucinotta E and Macri A: The role of
prophylactic hyperthermic intraperitoneal chemotherapy in the
management of serosal involved gastric cancer. Anticancer Res.
34:2019–2022. 2014.PubMed/NCBI
|
|
5
|
Chau I, Norman AR, Cunningham D, Waters
JS, Oates J and Ross PJ: Multivariate prognostic factor analysis in
locally advanced and metastatic esophago-gastric cancer-pooled
analysis from three multicenter, randomized, controlled trials
using individual patient data. J Clin Oncol. 22:2395–2403. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maeda H, Kobayashi M and Sakamoto J:
Evaluation and treatment of malignant ascites secondary to gastric
cancer. World J Gastroenterol. 21:10936–10947. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ishigami H, Fujiwara Y, Fukushima R,
Nashimoto A, Yabusaki H, Imano M, Imamoto H, Kodera Y, Uenosono Y,
Amagai K, et al: Phase III trial comparing intraperitoneal and
intravenous paclitaxel plus S-1 Versus Cisplatin Plus S-1 in
patients with gastric cancer with peritoneal metastasis: PHOENIX-GC
trial. J Clin Oncol. 36:1922–1929. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alshuwaykh O, Cheung A, Goel A, Kwong A,
Dhanasekaran R, Ghaziani TT, Ahmed A, Daugherty T, Dronamraju D,
Kumari R, et al: Clinical characteristics and outcomes in those
with primary extrahepatic malignancy and malignant ascites. BMC
Gastroenterol. 22:4102022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ahmed N and Stenvers KL: Getting to know
ovarian cancer ascites: Opportunities for targeted therapy-based
translational research. Front Oncol. 3:2562013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kaufman JL: Care of patients with ascites.
N Engl J Med. 330:1827author reply 1828. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim S, Kim B and Song YS: Ascites
modulates cancer cell behavior, contributing to tumor heterogeneity
in ovarian cancer. Cancer Sci. 107:1173–1178. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ito M, Nakano M, Ariyama H, Yamaguchi K,
Tanaka R, Semba Y, Sugio T, Miyawaki K, Kikushige Y, Mizuno S, et
al: Macrophages are primed to transdifferentiate into fibroblasts
in malignant ascites and pleural effusions. Cancer Lett.
532:2155972022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vokurka M, Lacina L, Brabek J, Kolar M, Ng
YZ and Smetana K Jr: Cancer-associated fibroblasts influence the
biological properties of malignant tumours via paracrine secretion
and exosome production. Int J Mol Sci. 23:9642022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu Y, Qi C, Liu X, Zhang C, Gao J, Wu Y,
Yang J, Zhao Q, Li J, Wang X and Shen L: Malignant ascites-derived
exosomes promote peritoneal tumor cell dissemination and reveal a
distinct miRNA signature in advanced gastric cancer. Cancer Lett.
457:142–150. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xuan Y, Wang H, Yung MM, Chen F, Chan WS,
Chan YS, Tsui SK, Ngan HY, Chan KK and Chan DW: SCD1/FADS2 fatty
acid desaturases equipoise lipid metabolic activity and
redox-driven ferroptosis in ascites-derived ovarian cancer cells.
Theranostics. 12:3534–3552. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Al-Marzouki L, Stavrakos VS, Pal S,
Giannias B, Bourdeau F, Rayes R, Bertos N, Najmeh S, Spicer JD,
Cools-Lartigue J, et al: Soluble factors in malignant ascites
promote the metastatic adhesion of gastric adenocarcinoma cells.
Gastric Cancer. 26:55–68. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Milliken D, Scotton C, Raju S, Balkwill F
and Wilson J: Analysis of chemokines and chemokine receptor
expression in ovarian cancer ascites. Clin Cancer Res. 8:1108–1114.
2002.PubMed/NCBI
|
|
18
|
Puiffe ML, Le Page C, Filali-Mouhim A,
Zietarska M, Ouellet V, Tonin PN, Chevrette M, Provencher DM and
Mes-Masson AM: Characterization of ovarian cancer ascites on cell
invasion, proliferation, spheroid formation, and gene expression in
an in vitro model of epithelial ovarian cancer. Neoplasia.
9:820–829. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yin T, Wang G, He S, Shen G, Su C, Zhang
Y, Wei X, Ye T, Li L, Yang S, et al: Malignant pleural effusion and
ascites induce epithelial-mesenchymal transition and cancer
stem-like cell properties via the vascular endothelial growth
factor (VEGF)/Phosphatidylinositol 3-Kinase (PI3K)/Akt/Mechanistic
target of rapamycin (mTOR) pathway. J Biol Chem. 291:26750–26761.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lomelino CL, Andring JT, Mckenna R and
Kilberg MS: Asparagine synthetase: Function, structure, and role in
disease. J Biol Chem. 292:19952–19958. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shen X, Cai Y, Lu L, Huang H, Yan H, Paty
PB, Muca E, Ahuja N, Zhang Y, Johnson CH and Khan SA: Asparagine
metabolism in tumors is linked to poor survival in females with
colorectal cancer: A cohort study. Metabolites. 12:642022.
View Article : Google Scholar
|
|
22
|
Zhang B, Dong LW, Tan YX, Zhang J, Pan YF,
Yang C, Li MH, Ding ZW, Liu LJ, Jiang TY, et al: Asparagine
synthetase is an independent predictor of surgical survival and a
potential therapeutic target in hepatocellular carcinoma. Br J
Cancer. 109:14–23. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Panosyan EH, Lasky JL, Lin HJ, Lai A, Hai
Y, Guo X, Quinn M, Nelson SF, Cloughesy TF and Nghiemphu PL:
Clinical aggressiveness of malignant gliomas is linked to augmented
metabolism of amino acids. J Neurooncol. 128:57–66. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu Q, Wang X, Wang L, Zheng J, Wang J and
Wang B: Knockdown of asparagine synthetase (ASNS) suppresses cell
proliferation and inhibits tumor growth in gastric cancer cells.
Scand J Gastroenterol. 51:1220–1226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Toda K, Kawada K, Iwamoto M, Inamoto S,
Sasazuki T, Shirasawa S, Hasegawa S and Sakai Y: Metabolic
alterations caused by KRAS mutations in colorectal cancer
contribute to cell adaptation to glutamine depletion by
upregulation of asparagine synthetase. Neoplasia. 18:654–665. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin CY, Sheu MJ, Li CF, Lee SW, Lin LC,
Wang YF and Chen SH: Deficiency in asparagine synthetase expression
in rectal cancers receiving concurrent chemoradiotherapy: Negative
prognostic impact and therapeutic relevance. Tumour Biol.
35:6823–6830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Xu H, Zhang L, Song L, Feng D, Peng
X, Wu M, Zou Y, Wang B, Zhan L, et al: Malignant ascites-derived
organoid (MADO) cultures for gastric cancer in vitro modelling and
drug screening. J Cancer Res Clin Oncol. 145:2637–2647. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
R Core Team (2019), . A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: Available from:. http://www.R-project.org/
|
|
30
|
Kanda M and Kodera Y: Molecular mechanisms
of peritoneal dissemination in gastric cancer. World J
Gastroenterol. 22:6829–6840. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chiu M, Taurino G, Bianchi MG, Kilberg MS
and Bussolati O: Asparagine synthetase in cancer: beyond acute
lymphoblastic leukemia. Front Oncol. 9:14802019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu Y, Lv F, Zhu X, Wu Y and Shen X: Loss
of asparagine synthetase suppresses the growth of human lung cancer
cells by arresting cell cycle at G0/G1 phase. Cancer Gene Ther.
23:287–294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cai DJ, Zhang ZY, Bu Y, Li L, Deng YZ, Sun
LQ, Hu CP and Li M: Asparagine synthetase regulates lung-cancer
metastasis by stabilizing the β-catenin complex and modulating
mitochondrial response. Cell Death Dis. 13:5662022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nishikawa G, Kawada K, Hanada K, Maekawa
H, Itatani Y, Miyoshi H, Taketo MM and Obama K: Targeting
asparagine synthetase in tumorgenicity using patient-derived
tumor-initiating cells. Cells. 11:32732022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gwinn DM, Lee AG, Briones-Martin-Del-Campo
M, Conn CS, Simpson DR, Scott AI, Le A, Cowan TM, Ruggero D and
Sweet-Cordero EA: Oncogenic KRAS regulates amino acid homeostasis
and asparagine biosynthesis via ATF4 and alters sensitivity to
L-Asparaginase. Cancer Cell. 33:91–107. e62018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sandelin A, Alkema W, Engström P,
Wasserman WW and Lenhard B: JASPAR: An open-access database for
eukaryotic transcription factor binding profiles. Nucleic Acids
Res. 32((Database issue)): D91–D94. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng LN, Wen F, Xu P and Zhang S:
Prognostic significance of malignant ascites in gastric cancer
patients with peritoneal metastasis: A systemic review and
meta-analysis. World J Clin Cases. 7:3247–3258. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Z, Chen JQ, Liu JL and Tian L: Issues
on peritoneal metastasis of gastric cancer: an update. World J Surg
Oncol. 17:2152019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Anderson NM and Simon MC: The tumor
microenvironment. Curr Biol. 30:R921–R925. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiao Y and Yu D: Tumor microenvironment as
a therapeutic target in cancer. Pharmacol Ther. 221:1077532021.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Oya Y, Hayakawa Y and Koike K: Tumor
microenvironment in gastric cancers. Cancer Sci. 111:2696–2707.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Asciak R, Kanellakis NI, Yao X, Abd Hamid
M, Mercer RM, Hassan M, Bedawi EO, Dobson M, Fsadni P, Montefort S,
et al: Pleural Fluid Has Pro-Growth biological properties which
enable cancer cell proliferation. Front Oncol. 11:6583952021.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cheah HM, Lansley SM, Varano Della
Vergiliana JF, Tan AL, Thomas R, Leong SL, Creaney J and Lee YC:
Malignant pleural fluid from mesothelioma has potent biological
activities. Respirology. 22:192–199. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Krejci O, Starkova J, Otova B, Madzo J,
Kalinova M, Hrusak O and Trka J: Upregulation of asparagine
synthetase fails to avert cell cycle arrest induced by
L-asparaginase in TEL/AML1-positive leukaemic cells. Leukemia.
18:434–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stams WA, den Boer ML, Holleman A, Appel
IM, Beverloo HB, van Wering ER, Janka-Schaub GE, Evans WE and
Pieters R: Asparagine synthetase expression is linked with
L-asparaginase resistance in TEL-AML1-negative but not
TEL-AML1-positive pediatric acute lymphoblastic leukemia. Blood.
105:4223–4225. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zwaan CM, Kaspers GJ, Pieters R, Hählen K,
Janka-Schaub GE, van Zantwijk CH, Huismans DR, de Vries E, Rots MG,
Peters GJ, et al: Different drug sensitivity profiles of acute
myeloid and lymphoblastic leukemia and normal peripheral blood
mononuclear cells in children with and without Down syndrome.
Blood. 99:245–251. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Aslanian AM, Fletcher BS and Kilberg MS:
Asparagine synthetase expression alone is sufficient to induce
l-asparaginase resistance in MOLT-4 human leukaemia cells. Biochem
J. 357((Pt 1)): 321–328. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Srivastava S, Jiang J, Misra J, Seim G,
Staschke KA, Zhong M, Zhou L, Liu Y, Chen C, Davé U, et al:
Asparagine bioavailability regulates the translation of MYC
oncogene. Oncogene. 41:4855–4865. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Garrison RN, Kaelin LD, Galloway RH and
Heuser LS: Malignant ascites. Clinical and experimental
observations. Ann Surg. 203:644–651. 1986. View Article : Google Scholar : PubMed/NCBI
|