Introduction
Lung cancer is not only a life-threatening disease
that causes thousands of deaths (1), but it is also a severe social problem
that causes billions in economic burden every year (2). The 5-year survival rate of lung cancer
is <20%. Non-small cell lung cancer (NSCLC) is the most common
form of lung cancer with lung adenocarcinoma (LUAD) being the most
common subtype of NSCLC. In recent years, programmed
death-1/programmed death-ligand 1 immune checkpoint therapy and
driver gene mutation-oriented targeted therapy have resulted in
marked survival benefits for some patients with LUAD (3). However, the problem of drug resistance
gradually hinders the effect of treatment, and more potential novel
targets are needed for the prediction and treatment of LUAD
(4).
ATP-binding cassette (ABC) subfamily A member 3
(ABCA3) is a gene located on chromosome 16p13.3*-. Studies on ABCA3
have revealed that this gene serves an important role in the
development of malignant disease; Schimanski et al (5) revealed that decreased ABCA3 expression
in breast cancer was associated with poor prognosis. Steinbach
et al (6) showed that ABCA3
was upregulated in childhood acute myeloid leukemia (AML) compared
with in healthy bone marrow, and ABCA3 was identified as the most
likely transporter that causes drug resistance. Although there are
a few studies on the effects of ABCA3 in lung cancer, the exact
role of ABCA3 in LUAD is not clear (7,8). In
the present study, a series of public databases such as The Cancer
Genome Atlas (TCGA) and the Clinical Proteomic Tumor Analysis
Consortium (CPTAC) were analyzed, and cytological and molecular
experiments were performed to explore the role of ABCA3 in the
occurrence and development of LUAD.
Materials and methods
Data analysis from public
databases
mRNA expression data from TCGA were obtained from
the University of Alabama at Birmingham Cancer Data Analysis Portal
(UALCAN; http://ualcan.path.uab.edu/analysis.html) (9) and UCSC XENA browser (https://xenabrowser.net/) (10), and protein expression data from the
CPTAC were also obtained from the University of Alabama at
Birmingham Cancer Data Analysis Portal (https://ualcan.path.uab.edu/analysis.html). Survival
data from TCGA were obtained from UCSC XENA browser and
Kaplan-Meier plotter online analysis database (https://kmplot.com/analysis/index.php?p=service&cancer=lung).
Co-expressed genes were obtained from UALCAN and cBioportal
(http://www.cbioportal.org/index.do)
(11,12). Kyoto Encyclopedia of Genes and
Genomes (KEGG) and Gene Ontology (GO) analyses were performed with
ClueGo V2.5.2 (13) in Cytoscape
software V3.7.0 (14). The
protein-protein interaction network was analyzed using the Search
Tool for the Retrieval of Interacting Genes/Proteins V11.5
(https://cn.string-db.org/) (15).
Cell culture and transfection
The A549 LUAD cell line was obtained from Hebei
Medical University (Shijiazhuang, China), and the cells were
cultured in a 5% CO2 incubator at 37°C. DMEM (Corning,
Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) and penicillin-streptomycin (100 U/ml) (Gibco; Thermo Fisher
Scientific, Inc.) was used for cell culture. During the culture
process, PCR detection was performed once a week to prevent
mycoplasma contamination, and cell passage was performed when the
cell confluence reached 70%. To knock down the expression of ABCA3,
cell transfection was performed when the cell confluence reached
70%. ABCA3 and negative control (NC) small interfering RNA (siRNA)
sequences were synthesized by Sangon Biotech Co., Ltd.
Lipofectamine® RNAiMAX Transfection Reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) was used to transfect siRNA into
the cells. According to the instructions of the transfection
reagent, the siRNA concentration was 50 nM and the transfection
process was performed in a 37°C incubator; the initial incubation
duration was 5 min and the final incubation time was 72 h. The
effect of protein knockdown (KD) was detected by western blotting
72 h after transfection. The siRNA sequences were as follows: NC
forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse:
5′-ACGUGACACGUUCGGAGAATT-3′; and ABCA3 forward,
5′-CGCUGUUCCUCAAGCAGAAAU-3′ and reverse,
5′-UUCUGCUUGAGGAACAGCGAG-3′.
Cell behavior experiments
All cell experiments were performed 72 h after
transfection. Cell proliferation was measured using an MTT assay.
DMSO (MilliporeSigma) was used to dissolve the purple formazan and
the absorbance was measured at 490 nm by spectrophotometry. Cell
migration was measured using a wound healing assay, which was
performed in cells cultured in DMEM without FBS. A wound was
generated with a sterile pipette tip when cell confluence reached
80%, the scratched cells were washed with PBS and the width of the
wound was measured every 12 h using an IX71 light microscope
(Olympus Corporation). The relative width of the wound was
calculated as follows: The width of the initial wound at the start
of the assay was considered the baseline value, and the ratio of
the width measured at each time point to the baseline value was
considered the relative width. Cell invasion was measured using a
Transwell assay. The 24-well Transwell chamber (diameter, 6.5 mm;
pore size, 8.0 µm; cat. no. 3422, Corning Inc.) was coated with
Matrigel (Corning Inc.) for 24 h at 37°C, and the cells were seeded
in DMEM containing 5% FBS in the upper chamber (2×105
per chamber per well). DMEM with 20% FBS was placed in the bottom
chamber. Crystal violet (2%) was used to stain the cells under the
bottom of the chamber at room temperature for 10 min, and cells
were observed via an IX71 light microscope (Olympus
Corporation).
Protein extraction and western
blotting
Cell proteins were extracted using RIPA lysis
reagent (Thermo Fisher Scientific, Inc.) with protease and
phosphatase inhibitors. The total protein concentration was
determined using a Pierce™ BCA protein assay kit (Thermo Fisher
Scientific, Inc.). Proteins were stored at −80°C and were then
separated by SDS-PAGE on a 12% gel at a voltage of 120 V for 2 h.
Subsequently, proteins were transferred to PVDF membranes, which
were blocked with 5% BSA (Merck & Co., Inc.) for 2 h, incubated
with primary antibodies for 4 h at room temperature and incubated
with a secondary antibody for 2 h at room temperature. Finally,
electrochemiluminescence was captured by a GE LAS-600 Imaging
system with SuperSignal™ West Femto Maximum Sensitivity Substrate
(Thermo Fisher Scientific, Inc.). The gray value of bands was
semi-quantified using ImageJ software V1.5.0 (National Institutes
of Health). Details of the antibodies involved in this study are
shown in Table I.
 | Table I.Antibody information. |
Table I.
Antibody information.
| Antibody | Brand | Catalog number | Dilution |
|---|
| Primary
antibodies |
|
|
|
|
ABCA3 | Abcam | ab99856 | 1:2,000 |
|
E-cadherin | Cell Signaling
Technology, Inc. | 14472 | 1:1,000 |
|
N-cadherin | Cell Signaling
Technology, Inc. | 13116 | 1:1,500 |
|
Vimentin | Proteintech Group,
Inc. | 60330-1 | 1:5,000 |
|
β-actin | Proteintech Group,
Inc. | 20536-1 | 1:3,000 |
| Secondary
antibodies |
|
|
|
|
Anti-mouse | Proteintech Group,
Inc. | SA00001-1 | 1:5,000 |
|
Anti-rabbit | Proteintech Group,
Inc. | SA00001-2 | 1:5,000 |
Statistical analysis
All assays have been verified with experimental
repeats >3 times. SPSS (v22.0; IBM Corp.) software was used for
statistical analysis. The differences in data between groups were
detected by a t-test, and Z-values that represent standard
deviations from the median across samples for the given cancer type
were presented for the CPTAC database. Gene mRNA levels were
divided into high and low expression groups based on the receiver
operating characteristic (ROC) curve by MedCalc V18 software
(MedCalc Software Ltd.). The survival difference between different
groups was analyzed by Kaplan-Meier survival analysis and log-rank
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Abnormal ABCA3 expression is observed
in TCGA-LUAD dataset
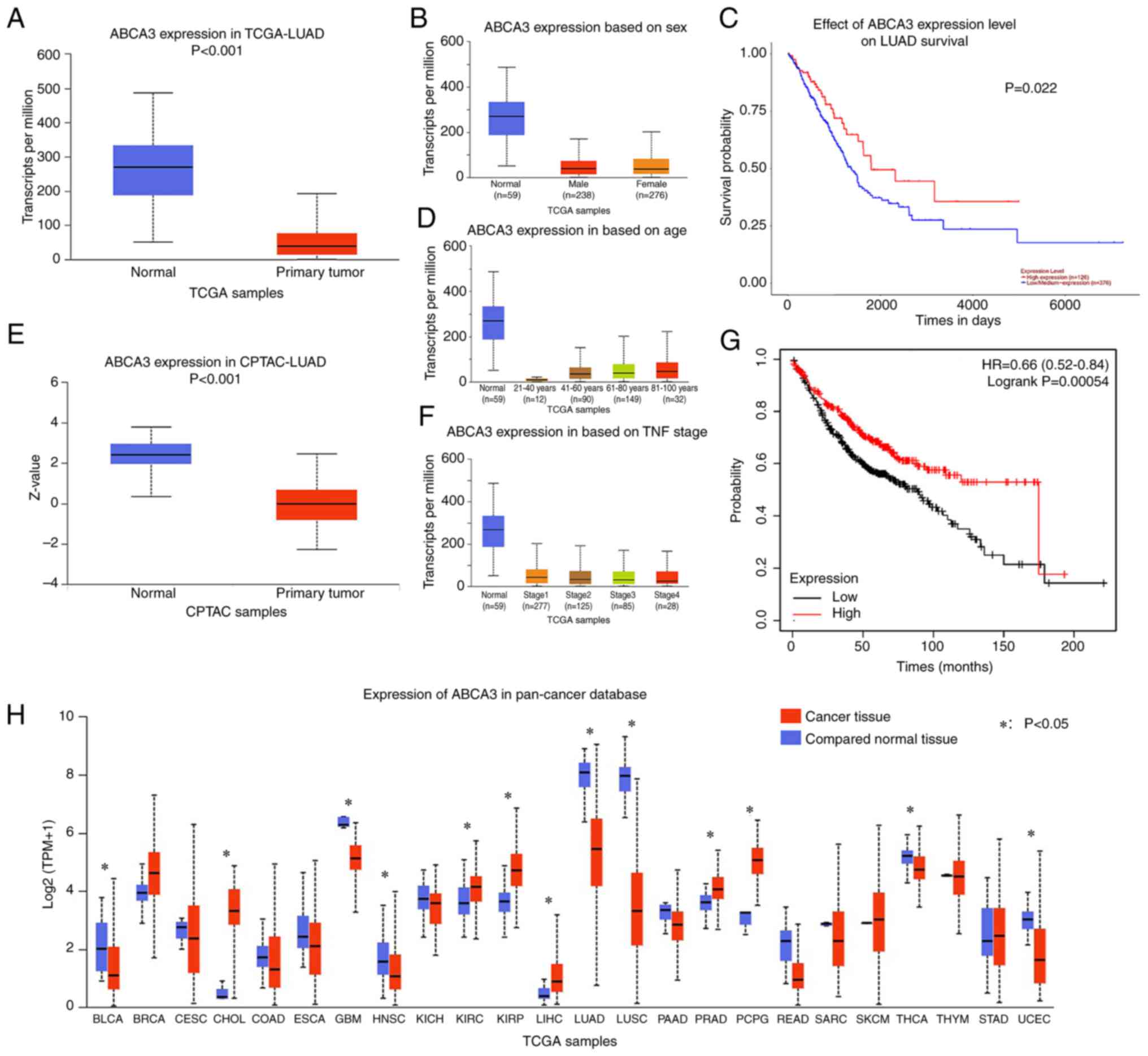
Through analysis of TCGA-LUAD dataset, it was
observed that ABCA3 expression in primary tumor tissues was
significantly reduced compared with that in normal tissues in TCGA
normal lung tissue (Fig. 1A).
Subgroup analysis revealed that similar results could be observed
in all age groups, sexes and Tumor-Node-Metastasis stages (16) across TCGA samples (Fig. 1B-D). In the CPTAC database, a
similar result was observed regarding the protein expression levels
of ABCA3 in LUAD cancer tissues compared with in normal tissues
(Fig. 1E).
Abnormal ABCA3 expression is observed
in a variety of tumors
Pan-cancer analysis was performed to investigate
ABCA3 expression in different types of cancer. The results
indicated that for glioblastoma, head and neck squamous cell
carcinoma, LUAD, lung squamous cell carcinoma (LUSC), thyroid
carcinoma and uterine corpus endometrial carcinoma, ABCA3
expression was significantly lower in tumor tissues than in normal
tissues (Fig. 1F). For some other
types of tumors, such as breast invasive carcinoma,
cholangiocarcinoma, kidney renal clear cell carcinoma, kidney renal
papillary cell carcinoma, liver hepatocellular carcinoma, prostate
adenocarcinoma, and pheochromocytoma and paraganglioma, ABCA3
expression in cancer tissues was higher than that in normal tissues
(Fig. 1F).
Patients with low ABCA3 expression
have a poor OS
Patients in TCGA-LUAD dataset were divided into two
groups based on ABCA3 expression, and the cut-off value was
determined by ROC curve analysis. The results demonstrated that
compared with patients in the high-expression group, patients in
the low-expression group had a significantly shorter overall
survival (OS) time (Fig. 1G and
H).
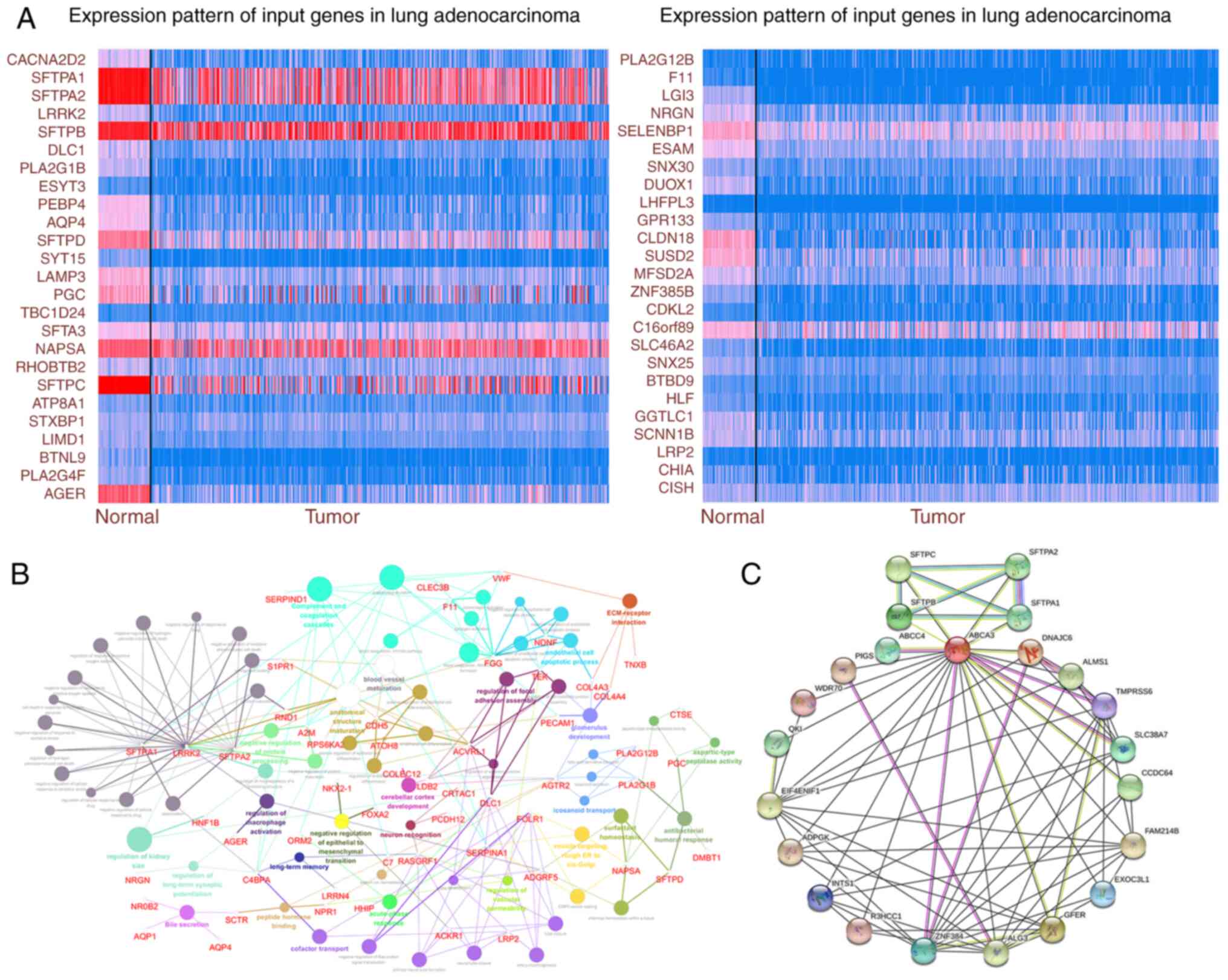
ABCA3 may participate in multiple
pathophysiological activities
To explore the possible pathological role of ABCA3
in LUAD, genes co-expressed with ABCA3 were identified using TCGA
database. The expression of some significantly correlated
co-expressed genes with a Pearson's correlation score of >0.4 in
TCGA is shown in Fig. 2A. KEGG
analysis was performed for ABCA3 and its co-expressed genes. The
results showed that these genes were enriched in certain
pathological pathways, such as ‘extracellular matrix-receptor
interaction’, ‘regulation of cellular response to drug’
(GO:2001038; GO:2001024), ‘regulation of endothelial cell apoptotic
process’ (GO:2000351; GO:2000352), ‘regulation of cell-substrate
junction assembly’ (GO:0090109), ‘endothelial cell apoptotic
process’ (GO:0072577), ‘endothelial cell differentiation’
(GO:0045446; GO:0045601), ‘positive regulation of epithelial cell
differentiation’ (GO:0045603) and ‘negative regulation of
epithelial-mesenchymal transition’ (GO:0010719) (EMT; Fig. 2B). Furthermore, the protein-protein
interaction network of ABCA3 was explored (Fig. 2C), which indicated that there were
multiple interaction relationships between ABCA3 and various
proteins.
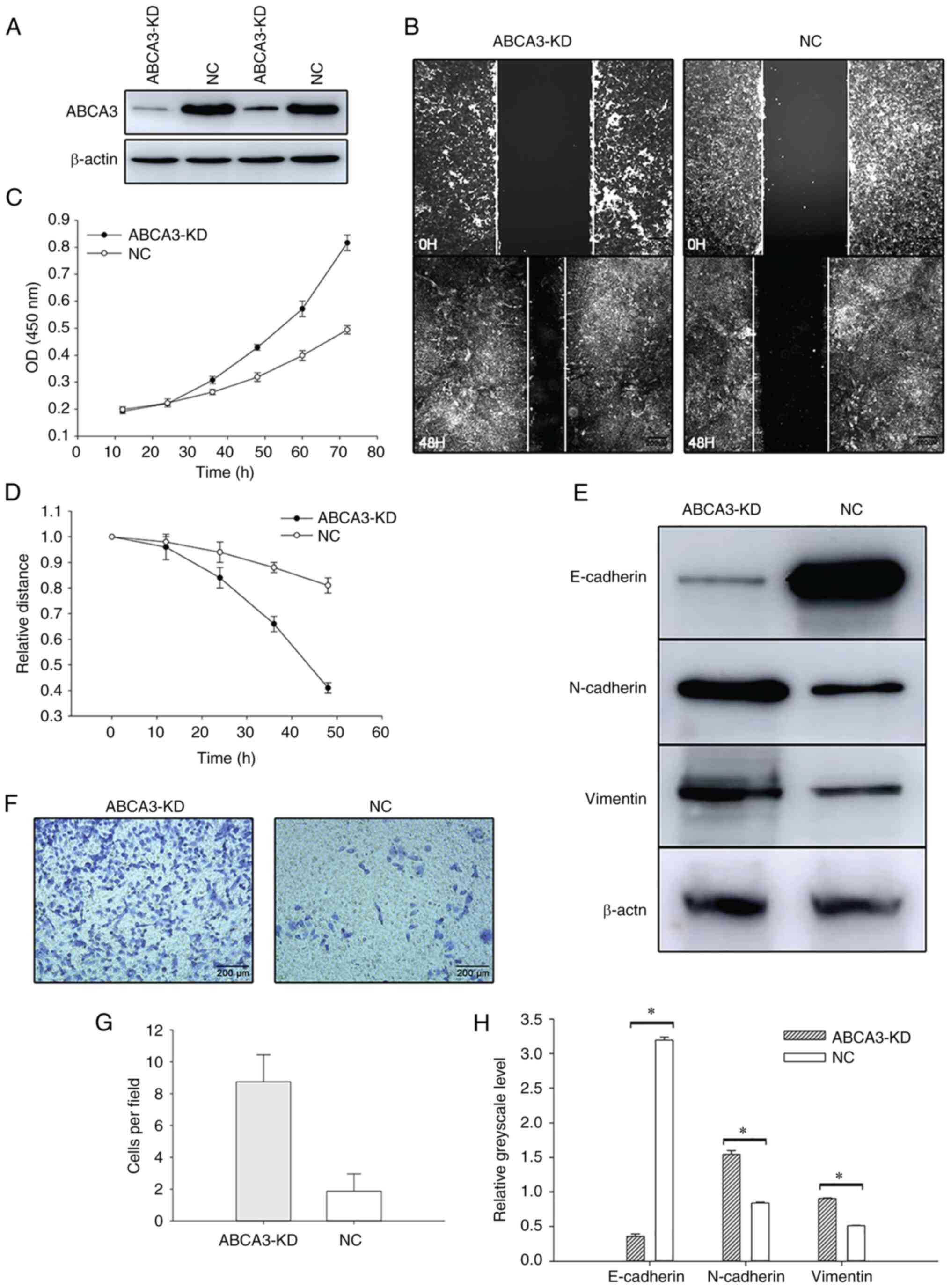
Malignant behavior of tumor cells is
enhanced when ABCA3 expression is knocked down
To investigate the role of ABCA3 in LUAD, the A549
cell line was cultured for further research. siRNA was transfected
into the A549 cell line to knock down ABCA3 expression, and the MTT
assay showed that after siRNA transfection, the proliferation of
A549 cells was increased (Fig. 3A and
B). Wound healing assays showed that the wound healing speed in
ABCA3-KD cells was faster than that in normal A549 cells (Fig. 3C and D). In the Transwell assay,
compared with A549 cells transfected with a negative control, more
ABCA3-KD cells passed through the Transwell membrane, which
indicated that the invasion was significantly enhanced after
transfection (Fig. 3E and F).
EMT is activated after KD of
ABCA3
In the present study, the expression levels of three
EMT-related proteins, E-cadherin, N-cadherin and vimentin, were
explored by western blotting after siRNA transfection. The results
indicated that E-cadherin expression was significantly reduced
after transfection, whereas the expression levels of N-cadherin and
vimentin in ABCA3-KD cells were significantly higher than those in
NC A549 cells, indicating that the EMT process was activated after
the KD of ABCA3 (Fig. 3G and
H).
Discussion
Lung cancer is the most common malignant tumor of
the respiratory system (1). It has
a number of pathological subtypes due to different cell origins,
such as small cell lung cancer, squamous cell carcinoma and
adenocarcinoma. In recent years, smoking cessation has markedly
reduced the incidence of squamous cell carcinoma, while with the
popularization of CT scanning for medical examination, an
increasing number of early lung cancer cases have been found
through health examinations, with most of them being peripheral
lung cancer represented by adenocarcinoma. At the same time, with
the popularization of smoking cessation campaigns, the incidence of
lung squamous cell carcinoma has significantly decreased, and LUAD
has gradually become the most common type of lung cancer (17). On the basis of traditional surgery,
radiotherapy and chemotherapy, gene mutation-oriented targeted
therapy and treatment with immune checkpoint inhibitors provide
individualized treatment strategies for the comprehensive treatment
of patients with LUAD, and notably improve prognosis compared with
past anticancer treatments (18).
At the same time, the issue of acquired drug resistance caused by
these treatments has gradually emerged, and it has become the main
factor affecting the treatment effect and long-term survival of
patients (19). An important way to
solve the issue of drug resistance is to explore the genes and
proteins that could potentially be used as novel therapeutic
targets in the occurrence and development of LUAD. In the present
study, in-depth research was carried out on the survival-related
gene ABCA3 in LUAD to develop novel treatment approaches.
ABCA3 is a gene located on chromosome 16p13.3, and
the protein encoded by this gene belongs to the ABC transporter
superfamily. Schimanski et al (5) demonstrated that ABCA3 expression was
diminished in human breast cancer tissue, and Steinbach et
al (6) hypothesized that ABCA3
may be a possible cause of drug resistance in childhood AML.
Furthermore, de Lima et al (20) reported that ABCA3 4548-91 CC/CA
genotypes were related to a poor complete molecular response in
patients with chronic myeloid leukemia treated with standard-dose
imatinib. However, there have been few studies on ABCA3 in LUAD
(4), and the regulatory effect of
ABCA3 on LUAD cells is unclear.
In the present study, ABCA3 expression was analyzed
using TCGA and CPTAC databases, and a significant decrease in the
mRNA and protein expression levels of ABCA3 was observed in LUAD
cancer tissues compared with in normal tissues. This abnormal
expression may indicate that ABCA3 serves a role in the occurrence
and development of LUAD. In previous studies, the role of ABCA3 in
tumorigenesis and development was not clear. Most studies have been
limited to the difference in ABCA3 expression with regard to
prognosis in tumors (5,8), Previous scholars have studied the
expression of ABCA3 in lung cancer, both in SCLC and in NSCLC, but
have not analyzed lung adenocarcinoma as a separate disease
(8). Previous studies have
demonstrated that LUAD is notably different from other types of
lung cancer in terms of pathogenesis, treatment targets and drug
resistance, all of which should be studied further (18,21).
The follow-up pan-cancer analysis in the present study revealed
that the ABCA3 expression range was different in multiple types of
malignant tumors, with abnormal increases in some tumors and
significant decreases in others. These results indicated that ABCA3
may serve different roles in different tumors. In some tumors, such
as glioblastoma multiforme and LUAD, ABCA3 expression was revealed
to be inhibited. We hypothesized that, in these tumors, ABCA3 has a
negative regulatory effect on tumor cells, with ABCA3 expression
being inhibited during tumor development.
The subsequent survival analysis revealed that
compared with the high ABCA3 expression group, patients with low
ABCA3 expression had significantly worse OS. By contrast, Overbeck
et al (8) reported the
opposite results from an analysis of 89 patients with lung cancer;
patients with high ABCA3 expression had a worse OS time. The
present study examined LUAD cases, while ~70% of cases in the study
by Overbeck et al (8) were
LUSC cases. Through further analysis of the LUSC data in TCGA
(TCGA-LUSC), it was demonstrated that the relationship between
ABCA3 and survival in LUSC is opposite to that in LUAD; in
TCGA-LUSC dataset, the high ABCA3 expression group had a worse OS
than the low ABCA3 expression group.
To verify the hypothesis that ABCA3 may serve a
protective role in LUAD as a tumor suppressor that is downregulated
in tumor tissue, cell behavior experiments were performed. The A549
cell line, as the most commonly used LUAD cell line, was used in
the current study. siRNA was transfected into A549 cells to knock
down ABCA3 expression. The results demonstrated that, in the A549
cell line, ABCA3 expression was negatively associated with
malignant behavior. When ABCA3 expression was knocked down, the
proliferation, migration and invasion of A549 cells was enhanced.
This result indicated that ABCA3 may inhibit the development of
tumors, which confirmed that ABCA3 has an inhibitory effect on the
occurrence and development of LUAD, and this may provide novel
potential ideas for the treatment of LUAD.
In further experiments, the pathological pathways
that ABCA3 and co-expressed genes were enriched in were identified
through GO analysis; a number of these pathways were related to the
regulation of EMT, epithelial cells and endothelial cells, which
indicated that the EMT process may serve an important role in ABCA3
inhibition of tumor cells. EMT is an important process in the
occurrence and development of malignant tumors (22). The EMT process can regulate the
proliferation, migration and invasion of tumor cells, and affect
the growth and metastasis of tumors in patients, which has an
important impact on their prognosis (23). When the EMT process occurs, the
polarity of cells begins to disappear, the epithelial
characteristics are weakened and the mesenchymal characteristics
are notably enhanced (24).
Therefore, in subsequent experiments, the expression of the
following EMT-related proteins was investigated: The epithelial
marker E-cadherin (19), and the
mesenchymal markers N-cadherin (25) and vimentin (26). The results demonstrated that when
ABCA3 expression was knocked down, the expression levels of
E-cadherin were markedly decreased, whereas those of N-cadherin and
vimentin were significantly increased, indicating that the EMT
process was activated.
In addition, GO pathway analysis showed that ABCA3
was related to the RNA regulation function of some substances in
the cell and before protein synthesis. Kaminski et al
(27) proposed that ABCA3 may
function as a lung surfactant lipid transporter, and ABCA-subfamily
transporters serve critical functions in human physiology, which,
when they are defective, cause disease. Matsumura et al
(7) reported that ABCA3 mediated
ATP-dependent choline-phospholipid uptake into intracellular
lysosomal associated membrane protein 3-positive vesicles in A549
cells. Fish et al (28)
demonstrated that silencing of ABCA3 could markedly increase lung
cancer growth in a study of the regulatory process mediated by
transactivation-responsive RNA-binding protein 2 (TARBP2), and a
notable increase in the mature mRNA levels and a decrease in the
relative pre-mRNA levels of ABCA3 upon TARBP2 KD were observed in
H1299 LUAD cells. These results supported the previous conclusions
of the bioinformatics analysis that ABCA3 may have a negative
regulatory role in the development of tumor cells, and ABCA3 could
act as a protective factor against tumor development.
In conclusion, ABCA3 may have an inhibitory role in
the development of lung adenocarcinoma, as observed in the present
cytological assays. The underlying pathway mechanisms require
further exploration in subsequent research. The present findings
indicated that ABCA3 may be a promising target for the treatment of
lung adenocarcinoma, and inhibitors targeting ABCA3 may have the
potential to provide survival benefits to patients with lung
adenocarcinoma.
Acknowledgements
The authors would like to acknowledge the assistance
of Hebei Medical University, from which the cell line in the
present study was obtained.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in The Cancer Genome Atlas (https://www.cancer.gov/tcga), NIH CPTAC database
(https://proteomics.cancer.gov//),
cBioportal (http://www.cbioportal.org/), University of Alabama at
Birmingham Cancer Data Analysis Portal (http://ualcan.path.uab.edu/analysis.html), UCSC XENA
(https://xenabrowser.net/) and Search Tool for the
Retrieval of Interacting Genes/Proteins (https://cn.string-db.org/) repositories. The other
datasets used and/or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
MS designed the study, conducted the experiments and
drafted the manuscript. LG and JZ designed the study and analyzed
the data. XX designed the study, revised the manuscript and given
final approval of the version to be published. LG and JZ confirm
the authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethical approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maomao C, He L, Dianqin S, Siyi H, Xinxin
Y, Fan Y, Shaoli Z, Changfa X, Lin L, Ji P and Wanqing C: Current
cancer burden in China: Epidemiology, etiology, and prevention.
Cancer Biol Med. 19:1121–1138. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Passaro A, Jänne PA, Mok T and Peters S:
Overcoming therapy resistance in EGFR-mutant lung cancer. Nat
Cancer. 2:377–391. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schimanski S, Wild PJ, Treeck O, Horn F,
Sigruener A, Rudolph C, Blaszyk H, Klinkhammer-Schalke M, Ortmann
O, Hartmann A and Schmitz G: Expression of the lipid transporters
ABCA3 and ABCA1 is diminished in human breast cancer tissue. Horm
Metab Res. 42:102–109. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Steinbach D, Gillet JP, Sauerbrey A, Gruhn
B, Dawczynski K, Bertholet V, de Longueville F, Zintl F, Remacle J
and Efferth T: ABCA3 as a possible cause of drug resistance in
childhood acute myeloid leukemia. Clin Cancer Res. 12:4357–4363.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Matsumura Y, Sakai H, Sasaki M, Ban N and
Inagaki N: ABCA3-mediated choline-phospholipids uptake into
intracellular vesicles in A549 cells. FEBS Let. 581:3139–3144.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Overbeck TR, Arnemann J,
Waldmann-Beushausen R, Trümper L, Schöndube FA, Reuter-Jessen K and
Danner BC: ABCA3 phenotype in non-small cell lung cancer indicates
poor outcome. Oncology. 93:270–278. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47(D1): D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Detterbeck FC, Boffa DJ, Kim AW and Tanoue
LT: The eighth edition lung cancer stage classification. Chest.
151:193–203. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hutchinson BD, Shroff GS, Truong MT and Ko
JP: Spectrum of lung adenocarcinoma. Semin Ultrasound CT MR.
40:255–264. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ruiz-Cordero R and Devine WP: Targeted
therapy and checkpoint immunotherapy in lung cancer. Surg Pathol
Clin. 13:17–33. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Leonetti A, Sharma S, Minari R, Perego P,
Giovannetti E and Tiseo M: Resistance mechanisms to osimertinib in
EGFR-mutated non-small cell lung cancer. Br J Cancer. 121:725–737.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
de Lima LT, Bueno CT, Vivona D, Hirata RD,
Hirata MH, Hungria VT, Chiattone CS, Zanichelli MA, Chauffaille Mde
L and Guerra-Shinohara EM: Relationship between SLCO1B3 and ABCA3
polymorphisms and imatinib response in chronic myeloid leukemia
patients. Hematology. 20:137–142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu J, Ren L, Li S, Li W, Zheng X, Yang Y,
Fu W, Yi J, Wang J and Du G: The biology, function, and
applications of exosomes in cancer. Acta Pharm Sin B. 11:2783–2797.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Singh M, Yelle N, Venugopal C and Singh
SK: EMT: Mechanisms and therapeutic implications. Pharmacol Ther.
182:80–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Loh CY, Chai JY, Tang TF, Wong WF, Sethi
G, Shanmugam MK, Chong PP and Looi CY: The E-cadherin and
N-cadherin switch in epithelial-to-mesenchymal transition:
Signaling, therapeutic implications, and challenges. Cells.
8:11182019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Satelli A and Li S: Vimentin in cancer and
its potential as a molecular target for cancer therapy. Cell Mol
Life Sci. 68:3033–3046. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kaminski WE, Piehler A and Wenzel JJ: ABC
A-subfamily transporters: Structure, function and disease. Biochim
Biophys Acta. 1762:510–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fish L, Navickas A, Culbertson B, Xu Y,
Nguyen HCB, Zhang S, Hochman M, Okimoto R, Dill BD, Molina H, et
al: Nuclear TARBP2 drives oncogenic dysregulation of RNA splicing
and decay. Mol Cell. 75:967–981.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|