Introduction
XB130, an adaptor protein, plays essential roles in
various biological processes, including cell proliferation,
skeletal remodeling, endocytosis, migration, invasion, and
epithelial-mesenchymal transformation (EMT) (1). XB130 can be phosphorylated by specific
protein tyrosine kinases and subsequently interact with the P85α
subunit of PI3K, thereby activating the PI3K/Akt signaling pathway
(2,3). Dysregulation of XB130 has been
observed in human gastrointestinal cancer, thyroid cancer,
hepatocellular carcinoma, skin basal cell carcinoma, and other
types of cancer, where it acts as either an oncogene or a tumor
suppressor (1,4). Given its significance in cancer,
further understanding of XB130 expression regulation is crucial for
cancer clinical diagnosis, risk prediction, prognosis, and targeted
therapy (1,5).
Previous studies have shown that silencing XB130
inhibits cell proliferation, migration, invasion, and EMT in
non-small cell lung cancer (NSCLC) (6,7).
Upregulated XB130 mRNA expression levels have been associated with
improved survival in patients with NSCLC (1). Similarly, XB130 is poorly expressed in
human gastric cancer and thyroid cancer, but in vitro
experiments have shown that XB130 acts as an oncogene in these
cancer types (1,8–10). To
explain these contradictory findings, it has been suggested that
XB130 is highly expressed in normal tissues, playing a vital role
in maintaining normal cellular physiological activity. However, in
cancer cells, the XB130-mediated signaling pathway may be
manipulated to control cell proliferation, survival, and migration
upon XB130 expression inhibition (1). Thus, investigating the regulatory
mechanism of XB130 expression may provide insights into these
paradoxical observations.
Post-transcriptional regulation is a critical form
of eukaryotic gene expression modulation (11). The 3′-untranslated region (3′-UTR)
of mRNA contains numerous cis-acting elements involved in
post-transcriptional regulation, such as microRNA (miRNA) binding
sites and GU/AU/CU/U-rich elements (G/A/C/UREs), which regulate
mRNA stability, subcellular localization and translation (11–14).
G/A/C/UREs typically interact with specific RNA binding proteins
(RBPs) to modulate gene expression (14–17).
G/A/C/UREs have been found to be closely associated with tumor
occurrence and inhibition (12–14).
For example, the binding of heterogeneous nuclear ribonucleoprotein
F (hnRNP F) to the AREs in the 3′-UTR of snail family
transcriptional repressor 1 (SNAI1) mRNA promotes EMT in bladder
cancer cells by stabilizing SNAI1 mRNA (18). In light of a previous report and
sequence analysis, it was predicted that the XB130 3′-UTR contains
three consensus AREs (sequence: AUUUA; AREI,
AREII, and AREIII), a specific URE
(U19), and several possible G/A/C/UREs (Fig. 1) (19). These elements may play critical
roles in regulating XB130 mRNA stability and translation (19). Therefore, in the present study, the
post-transcriptional regulation of XB130 expression was explored
from this perspective.
In the present study, the entire XB130 3′-UTR (1,218
bp), as well as truncated fragments and mutant fragments with
predicted ARE or URE deletions were cloned into the modified
psiCHECK-2 vector. These recombinant constructs were then
transfected into the PC-9 NSCLC cell line. Luciferase activity
assays and reverse transcription-quantitative PCR (RT-qPCR) were
performed to evaluate the impact of these insertions on reporter
gene expression. Furthermore, candidate proteins that may regulate
XB130 expression by binding to XB130 3′-UTR were screened for using
RNA pull-down assays, followed by mass spectrometry and western
blotting. This study may provide insights for further exploration
of the regulatory mechanisms underlying XB130 expression.
Materials and methods
Cell culture and transfection
PC-9 and A549 NSCLC cell lines were used in the
present study. PC-9 cells were purchased from FuHeng Biology
Company, and A549 cells were obtained from the Type Culture
Collection of the Chinese Academy of Sciences. Cells were
maintained in RPMI 1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) at 37°C in a 5% CO2 humidified atmosphere.
Transfection of plasmids and small interfering RNAs (siRNAs) into
NSCLC cells was performed using Entranster™-D4000 and
Entranster™-R4000 (Engreen Biosystem), respectively, following the
manufacturer's instructions. Unless otherwise noted, cells were
collected for analysis 48 h after transfection. The sequences of
the siRNAs used (Shanghai GenePharma, Co., Ltd.) are provided in
Table I.
 | Table I.Sequences of the small interfering
RNAs. |
Table I.
Sequences of the small interfering
RNAs.
| Target gene | Direction | Sequence |
|---|
| ANXA2 | Sense |
GGAUGCUUUGAACAUUGAATT |
|
| Antisense |
UUCAAUGUUCAAAGCAUCCTT |
| FLNA | Sense |
AGAAGAAGAUGCACCGCAATT |
|
| Antisense |
UUGCGGUGCAUCUUCUUCUTT |
| LMNA | Sense |
CUGGACUUCCAGAAGAACATT |
|
| Antisense |
UGUUCUUCUGGAAGUCCAGTT |
| PLEC | Sense |
CGGAGAUGGAGAAGCAUAATT |
|
| Antisense |
UUAUGCUUCUCCAUCUCCGTT |
| SPTAN1 | Sense |
CCUCAUGUCUUGGAUCAAUTT |
|
| Antisense |
AUUGAUCCAAGACAUGAGGTT |
| TPM1 | Sense |
CACGCUCUCAACGAUAUGATT |
|
| Antisense |
UCAUAUCGUUGAGAGCGUGTT |
| PKM | Sense |
GAACUUCUCUCAUGGAACUTT |
|
| Antisense |
AGUUCCAUGAGAGAAGUUCTT |
| YBX3 | Sense |
GCAGGUGACCUAAAGAAUUTT |
|
| Antisense |
AAUUCUUUAGGUCACCUGCTT |
| MYH9 | Sense |
GGACCUUCCACAUCUUCUATT |
|
| Antisense |
UAGAAGAUGUGGAAGGUCCTT |
| CALU | Sense |
CAGCCAUGAUGGGAAUACUTT |
|
| Antisense |
AGUAUUCCCAUCAUGGCUGTT |
| NC | Sense |
UUCUCCGAACGUGUCACGUTT |
|
| Antisense |
ACGUGACACGUUCGGAGAATT |
Plasmid construction
The dual-luciferase reporter vector psiCHECK-2
(Promega Corporation) was modified for the present study. Briefly,
a chimeric intron was amplified from the psiCHECK-2 vector using
specific primers (intron For and intron Rev) containing MluI and
ApaI recognition sequences, respectively. The chimeric intron was
then inserted into the 5′-UTR of the firefly luciferase gene
(hluc+) using MluI and ApaI cleavage sites. Various
regions of the XB130 3′-UTR (1,218 bp, NM_032550.4) were amplified
from the genomic DNA of PC-9 cells using the primers listed in
Table II. The PCR thermocycling
conditions were as follows: 35 amplification cycles of denaturation
at 98°C for 10 sec, annealing at 60°C for 15 sec and extension at
72°C for 60 sec, followed by final extension at 72°C for 5 min.
Mutant fragments with the deleted seed sequence, ATTTA, or
T19 were obtained through overlap extension PCR
(20). The PCR products were
subsequently cloned into the 3′-UTR of the Renilla luciferase gene
(hRluc) of the modified psiCHECK-2 vector. All plasmid constructs
were confirmed by DNA sequencing.
 | Table II.Sequences of primers. |
Table II.
Sequences of primers.
| Name | Sequence |
|---|
| Intron For |
GTGACGCGTGTGGCCTCGAACACCGAGCGACCCTGCAGCGACCCGCTTAAAAGCTTGGCATTCCGGTAAGGTAAGTATCAAGGTTACAAGACAGG |
| Intron Rev | GCAGGGCCCTT CTTAATG
TTCTTAGCATCGGCCATGGTGGCTTTACCAACCTGTGGAGAGAAAGGCAAAG |
| 3′-UTR-1 For |
CCGCTCGAGAAAACAAGCTTCATCTAAAG |
| 3′-UTR-1,218
Rev |
CGGGATCCTTCAAATACAATGAACATTT |
| 3′-UTR-132 Rev |
CGGGATCCCCATTCACAGTACCTCAGTC |
| 3′-UTR-113 For |
CCGCTCGAGGACTGAGGTACTGTGAATGG |
| 3′-UTR-660 Rev |
CGGGATCCCTTGGACTGGAAAGAAGTCT |
| 3′-UTR-661 For |
CCGCTCGAGGCAGGCCTGAATCAACTGTC |
| 3′-UTR-795 Rev |
CGGGATCCTCCTACAGTAAAGAGTCACT |
| 3′-UTR-792 For |
CCGCTCGAGAGGAAAAATATTACTCTGTG |
| 3′-UTR-989 Rev |
CGGGATCCACAGGGGCAGCACAAGGAAC |
| 3′-UTR-970 For |
CCGCTCGAGGTTCCTTGTGCTGCCCCTGT |
| AREI
For |
AACAAGACGTTGTGGGATGTTCTGTGTT |
| AREI
Rev |
CAGAACATCCCACAACGTCTTGTTTACAT |
| AREII
For |
TTAAAATCCTGTCGAACAGTGAAAAGTATC |
| AREII
Rev |
TTTCACTGTTCGACAGGATTTTAAGCA |
| AREIII
For |
AAGGGAAGAGTTATATAAATAGTCTCTGA |
| AREIII
Rev |
GACTATTTATATAACTCTTCCCTTAAAAATG |
| URE For |
TCCTCTGATGGCAAACACAGGTTGAAAC |
| URE Rev |
CAACCTGTGTTTGCCATCAGAGGAAAAGA |
| 3′-UTR-230 Rev |
CGGGATCCACATCCAATAGACTTAGGTC |
| 3′-UTR-231 For |
CCGCTCGAGAAACAAGACGTTGTATTTAG |
| 3′-UTR-342 Rev |
CGGGATCCCACCAAAGTCTAAACAGGAG |
| 3′-UTR-343 For |
CCGCTCGAGGAAGATGAATCAAGGAAGCA |
| 3′-UTR-502 Rev |
CGGGATCCGAGTTGGGCCTGGTAATATT |
| 3′-UTR-503 For |
CCGCTCGAGAGTCTTCAGTGATTTTAAAC |
| 3′-UTR-1,053
Rev | CGGGATCCTAATAGATG
AAATGTTTCAACCTGTGTTAA AAAAAAAAAAAAAAAAATGCC |
| 3′-UTR-1,054
For |
CCGCTCGAGTCTCTGCCTCATTTCTGGA |
| qRL For |
TGGTCGTGAGGCACTGGGCAGGTG |
| qRL Rev |
TGCTCGGGGTCGTACACCTTGGAA |
| qFL For |
AAAGCTTGGCATTCCGGTAAGGTT |
| qFL Rev |
GTGGGCATCGGTGAAGGCAA |
| qANXA2 For |
TGCCTTCGCCTACCAGAGAA |
| qANXA2 Rev |
GCCCAAAATCACCGTCTCC |
| qFLNA For |
GGAGGAGGCAAAAGTGACCG |
| qFLNA Rev |
ACTTATCCACGTACACCTCGAAG |
| qLMNA For |
AATGATCGCTTGGCGGTCTAC |
| qLMNA Rev |
CACCTCTTCAGACTCGGTGAT |
| qPLEC For |
TGTACCGGCAGACCAACCT |
| qPLEC Rev |
GCATGGCGTCATACAGCGA |
| qSPTAN1 For |
GCCAACTCAGGAGCCATTGTT |
| qSPTAN1 Rev |
CGGGTCCGTATGGTTTCAGAT |
| qTPM1 For |
TTGAGAGTCGAGCCCAAAAAG |
| qTPM1 Rev |
CATATTTGCGGTCGGCATCTT |
| qPKM For |
ATGTCGAAGCCCCATAGTGAA |
| qPKM Rev |
TGGGTGGTGAATCAATGTCCA |
| qYBX3 For |
ACCGGCGTCCCTACAATTAC |
| qYBX3 Rev |
GGTTCTCAGTTGGTGCTTCAC |
| qMYH9 For |
CCTCAAGGAGCGTTACTACTCA |
| qMYH9 Rev |
CTGTAGGCGGTGTCTGTGAT |
| qCALU For |
ATGGACCTGCGACAGTTTCTT |
| qCALU Rev |
ACTCTGAGCATCATTGTGAACC |
| qGAPDH For |
GGAGCGAGATCCCTCCAAAAT |
| qGAPDH Rev |
GGCTGTTGTCATACTTCTCATGG |
Dual-luciferase reporter assay
PC-9 cells were seeded into 48-well plates and
transfected with the recombinant psiCHECK-2 constructs. After 48 h
of transfection, Renilla and firefly luciferase activities were
measured using a Luc-PairTM Duo-Luciferase HS Assay Kit
(GeneCopoeia, Inc.) according to the manufacturer's instructions.
Briefly, cells were washed twice with PBS and lysed with 65 µl
1×Luc-Lysis II Buffer for 10 min at room temperature on a rotating
wheel device. The luciferase assays were performed by sequentially
adding 100 µl firefly and Renilla luciferase assay reagents to 20
µl cell lysate. The light output was measured using a BioTek
Synergy2 Multimode Microplate Reader (Biotek Instruments, Inc.)
immediately after adding each luciferase assay reagent. The
hluc+ gene served as the internal control.
RT-qPCR
Total RNA was isolated from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Reverse
transcription was performed using a HyperScript III 1st Strand cDNA
Synthesis Kit with gDNA Remover (NovaBio). The synthesized cDNA was
quantified by qPCR using a 2× SYBR Green qPCR MasterMix (Bimake) on
an Applied Biosystems StepOnePlus Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The primer pairs qRL
For and qRL Rev, and qFL For and qFL Rev (Table II) were used to amplify hRluc and
hluc+ cDNA, respectively. The quantity of hRluc mRNA in
each sample was normalized to the hluc+ content. For the
expression of endogenous genes, GAPDH was used as the internal
reference. The relative quantification of gene mRNA was calculated
using the 2−ΔΔCq method (21).
Reporter gene mRNA decay
To investigate the mRNA decay of a reporter gene,
PC-9 cells were transiently transfected with the recombinant
psiCHECK-2 constructs and incubated for 48 h. The transcription
process was inhibited by adding 10 mg/ml actinomycin D. Cells were
harvested at 0, 2, 4, and 6 h post-treatment, and total RNA was
isolated. The hRluc and hluc+ mRNA contents were
determined by RT-qPCR. The relative hRluc mRNA levels, normalized
to the hluc+ mRNA content, are expressed as a percentage
of the mRNA level at 0 h. Additionally, the levels of the
endogenous gene heat shock protein 90 α family class A member 1
(Hsp90) and GAPDH mRNA were detected by RT-qPCR. GAPDH was used as
the internal reference. The decay of Hsp90 mRNA was calculated
using the aforementioned method.
RNA pull-down assay
Nucleotides 113–230, 503–660, or 970–1,053 of the
XB130 3′-UTR were transcribed in vitro using a T7 RNA
polymerase kit (Roche Diagnostics GmbH) and then labeled with
Biotin using a Pierce RNA 3′End Desthiobiotinylation Kit according
to the manufacturer's protocol (Invitrogen; Thermo Fisher
Scientific, Inc.). The purified biotinylated RNA was incubated with
streptavidin magnetic beads at room temperature for 15–30 min,
followed by incubation with lysates from PC-9 cells at 4°C for
30–60 min. The pulled-down proteins were subjected to mass
spectrometry analysis by Sangon Biotech, Co., Ltd.
In silico co-expression analysis
The in silico co-expression analyses between
RBPs and XB130 were performed using the ENCORI (https://rnasysu.com/encori/panGeneCoExp.php) and
GEPIA2 (http://gepia2.cancer-pku.cn/#correlation)
databases.
Western blotting
Harvested cells were lysed in RIPA Buffer (cat. no.
P0013B; Beyotime Institute of Biotechnology) containing 1 mM PMSF
(Beyotime Institute of Biotechnology) on ice for 30 min. After
centrifugation at 14,000 × g for 15 min at 4°C, the supernatants
were collected. Protein samples were loaded on 12% SDS gels,
resolved using SDS-PAGE, and transferred to PVDF membranes (cat.
no. IPVH00010, MilliporeSigma). Following blocking with 5% BSA for
1 h, the membranes were incubated overnight at 4°C with anti-XB130
(1:2,000, cat. no. 17183-1-AP; ProteinTech Group, Inc.) or
anti-GAPDH (1:5,000, cat. no. 10494-1-AP; ProteinTech Group, Inc.)
antibodies. Subsequently, the membranes were treated with a
horseradish peroxidase-conjugated secondary antibody (1:5,000, cat.
no. SA00001-2; ProteinTech Group, Inc.) for 1 h at 37°C. The
specific protein bands were visualized using an Enhanced
Chemiluminescence Reagent (cat. no. WBKLS0100; MilliporeSigma) and
analyzed using a GeneGnome XRQ Chemiluminescence Imaging System
(Syngene Europe).
Statistical analysis
Data are presented as the mean ± standard deviation
(SD) of three independent experiments. Statistical analysis was
performed using SPSS version 20 (IBM Corp.). Differences between
two groups were determined using a Student's t-test. For
comparisons involving ≥3, a one-way ANOVA followed by a post hoc
Tukey's test was used. In the case of mRNA decay analysis, a
repeated-measures ANOVA followed by a post hoc Dunnett's test was
used. P<0.05 was considered to indicate a statistically
significant difference.
Results
XB130 3′-UTR insertion promotes the
expression of a reporter gene
To investigate the effect of the XB130 3′-UTR on
gene expression, various regions of the XB130 3′-UTR were inserted
into the 3′-UTR of the hRluc reporter gene in the dual-luciferase
reporter vector, psiCHECK-2. hRluc was used as the reporter, while
hluc+ served as the internal control. In all reporter
constructs, hRluc was controlled by identical promoter elements and
followed by different XB130 3′-UTR fragments, thus any changes
observed in luciferase activity would be attributed to alterations
in either mRNA stability or translation rate. Additionally, to
avoid plasmid DNA contamination during mRNA sample measurement, the
psiCHECK-2 vector was modified as described by Tao and Gao
(22). To detect the levels of
hRluc and hluc+ mRNA transcribed from the recombinant
plasmids, two forward primers (qRL For and qFL For) that spanned
the chimeric intron, along with their respective reverse primers,
were designed (Fig. 2A).
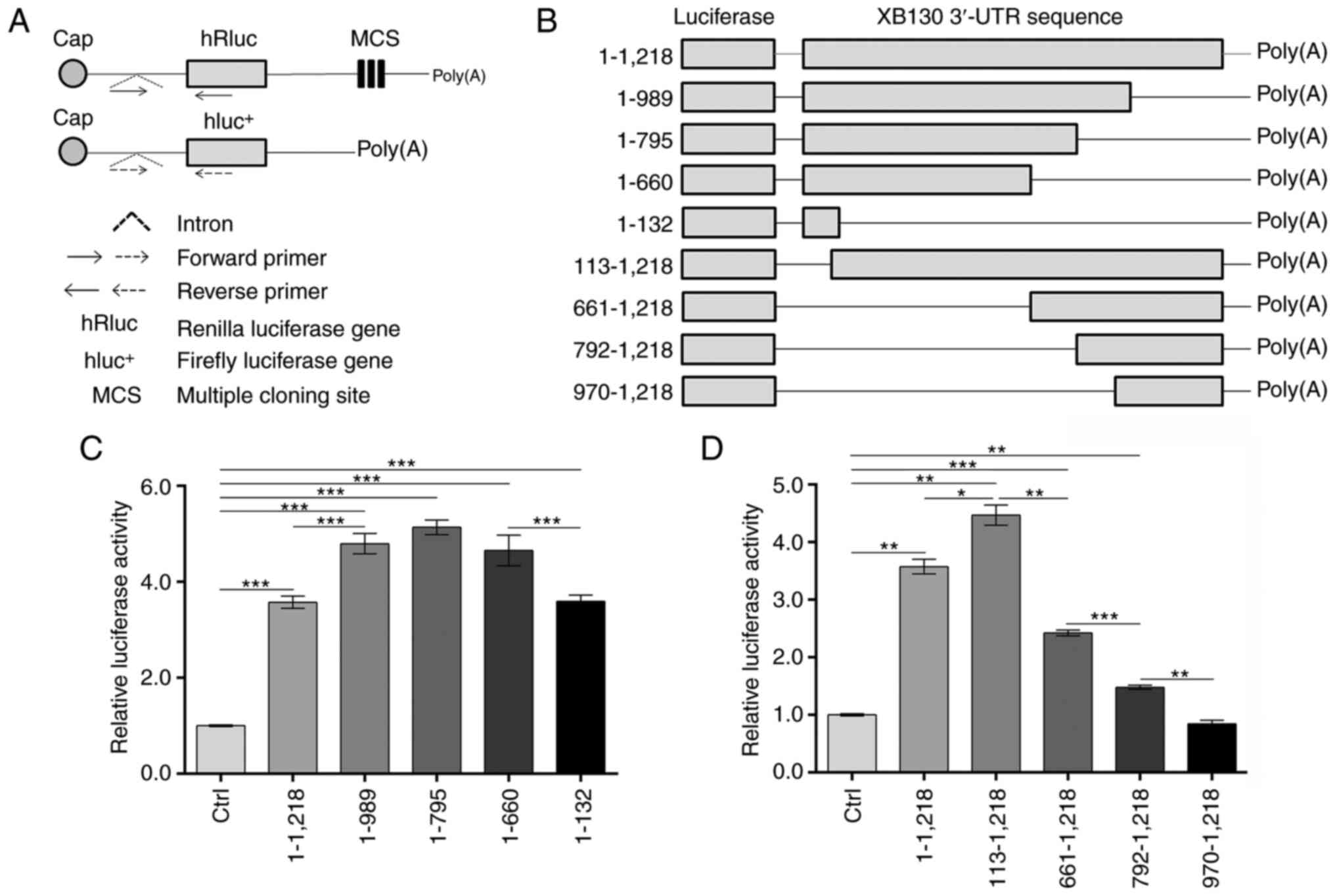 | Figure 2.XB130 3′-UTR insertion enhances
luciferase activity. (A) Schematic of the psiCHECK-2 vector
modification and RT-qPCR primer design. (B) Construction of
recombinant plasmids via insertion of the different regions of the
XB130 3′-UTR into the modified psiCHECK-2 vectors. Numbering starts
from the nucleotide following the stop codon for XB130. (C and D)
PC-9 cells were transfected with the empty modified psiCHECK-2
vector or the recombinants. After 48 h, Renilla and firefly
luciferase activity was measured. The ratio of Renilla/firefly
luciferase activity was calculated, and the relative luciferase
activity is expressed as a multiple of that measured in the Ctrl
group. *P<0.05, **P<0.01, ***P<0.001. UTR, untranslated
region; RT-qPCR, reverse transcription-quantitative PCR; hRluc,
Renilla luciferase gene; MCS, multiple cloning site;
hluc+, firefly luciferase gene; Ctrl, control. |
The recombinant constructs shown in Fig. 2B were transfected into PC-9 cells,
and the luciferase activities were analyzed 48 h after
transfection. As shown in Fig. 2C,
inserting the entire XB130 3′-UTR (nucleotides 1–1,218) into the
hRluc 3′-UTR significantly increased luciferase activity by
3.57-fold compared with the control group (Ctrl). Deletion of
nucleotides 990–1,218 from the entire 3′-UTR led to an increase in
luciferase activity by 1.34-fold compared with the 1–1,218 3′-UTR
group. Truncation of the 3′-UTR from nucleotide 796 or 661 to 1,218
did not have an additional effect on luciferase activity compared
with the 1–989 3′-UTR group. However, further truncation from
nucleotide 660 to 133 resulted in a significant 0.77-fold decrease
in luciferase activity compared with the 1–660 3′-UTR group.
Notably, the 1–132 3′-UTR group still exhibited high luciferase
activity at 3.59-fold higher than the Ctrl group. These findings
suggested the presence of positive regulatory elements in
nucleotides 1–660 and negative regulatory elements in nucleotides
990–1,218 of the XB130 3′-UTR.
Deleting the 5′ regions of the XB130 3′-UTR revealed
different results. As shown in Fig.
2D, the deletion of the first 112 nucleotides increased
luciferase activity by 1.25-fold compared with the 1–1,218 3′-UTR
group, indicating the presence of negative regulatory elements in
nucleotides 1–112. Gradually truncation from the proximal region to
nucleotides 660, 791, and 969 resulted in a decrease in luciferase
activity, suggesting the presence of positive regulatory elements
in nucleotides 113–969. Notably, insertion of the 970–1,218 3′-UTR
into psiCHECK-2 had no significant effect on luciferase activity
compared with the Ctrl group. Overall, these results indicated the
presence of negative regulatory elements in nucleotides 1–112 and
990–1,218, and positive regulatory elements in nucleotides 113–989.
Additionally, the effect of the 660–989 3′-UTR fragment on
luciferase activity appeared to be context-dependent.
A URE in the XB130 3′-UTR inhibits the
expression of a reporter gene
A previous study reported the presence of two
consensus AREs (AREII, nucleotides 742–746 and
AREIII, nucleotides 858–862) in the XB130 3′-UTR
(Fig. 1), which may play a role in
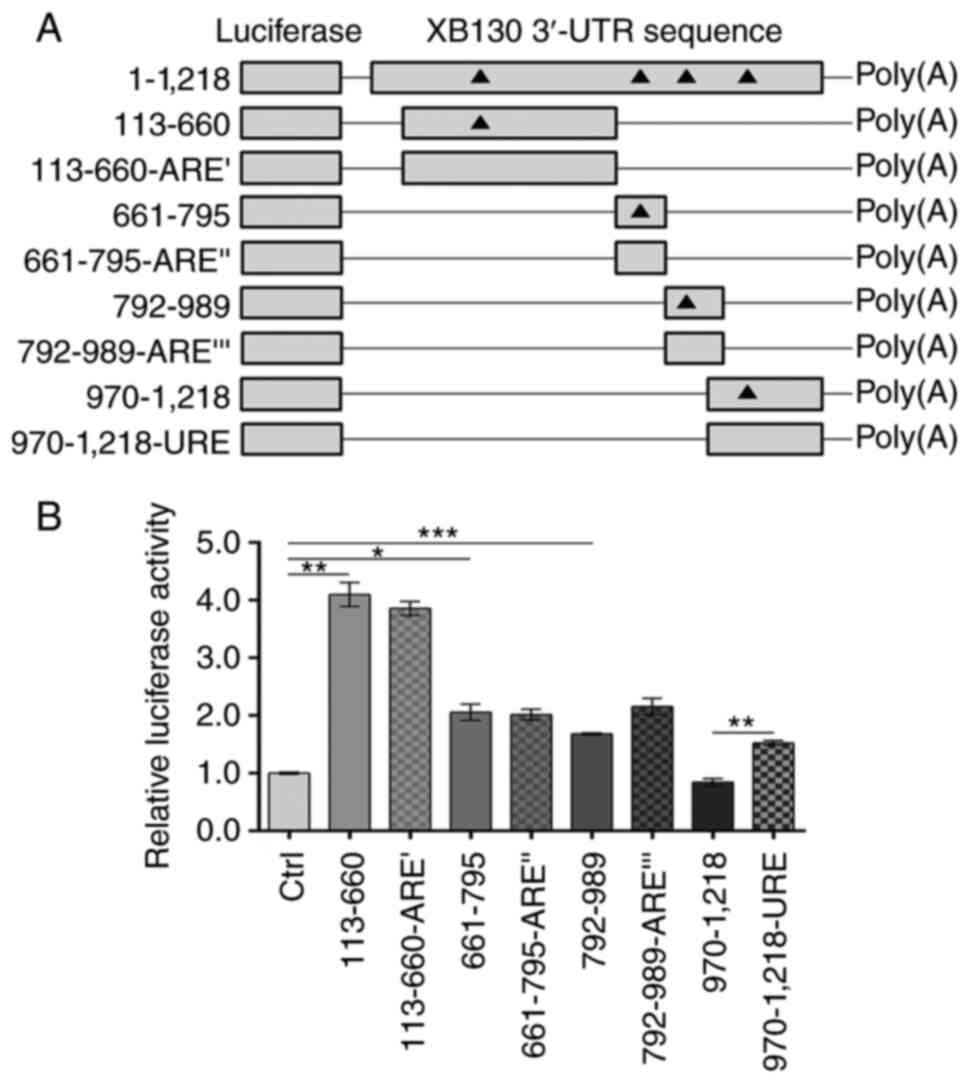
regulating mRNA stability and translation rate (19). Another consensus ARE
(AREI, nucleotides 245–249) and a potential URE
(nucleotides 1,007-1,025) were also identified (Fig. 1). To determine the effects of these
elements on gene expression, recombinant psiCHECK-2 plasmids
containing XB130 3′-UTR fragment 113–660, 661–795, 792–989, or
970–1,218, as well as the corresponding mutant plasmids with ARE or
URE deletions (113-660-AREI, 661–795-AREII,
792–989-AREIII, or 970–1,218-URE), were constructed
(Fig. 3A). As shown in Fig. 3B, the insertion of the 113–660,
661–795, or 792–989 3′-UTR fragments significantly increased
luciferase activity by 4.10-fold, 2.06-fold, and 1.68-fold,
respectively, compared with the Ctrl group. These findings further
supported the presence of positive regulatory elements in the
113–989 region of the 3′-UTR. Notably, deletion of AREI,
AREII, or AREIII did not have a significant
effect on luciferase activity compared with the corresponding
wild-type constructs (Fig. 3B).
However, deletion of the URE resulted in 1.81-fold higher
expression of the reporter gene compared with the 970–1,218 3′-UTR
group, suggesting that the URE had a negative effect on gene
expression.
Regions 113–230, 503–660, and
970–1,053 of the XB130 3′-UTR significantly affect the expression
of a reporter gene
Based on the aforementioned results, the 113–660
region and the URE in the XB130 3′-UTR were identified as having
the most pronounced positive and negative effects on the expression
of the reporter gene, respectively. To facilitate subsequent
screening of the combined RBPs, the 113–660 3′-UTR fragment was
further divided into smaller fragments: 113–230, 231–342, 343–502,
and 503–660. Additionally, the 970–1,218 3′-UTR fragment was
truncated into 970–1,053 and 1,054-1,218 fragments, of which the
identified URE was located in the 970–1,053 3′-UTR fragment. The
corresponding recombinant psiCHECK-2 plasmids were constructed
(Fig. 4A) and transfected into PC-9
cells, to measure the luciferase activity and steady-state mRNA
levels. The results demonstrated that the insertion of the 113–230
or 503–660 3′-UTR fragment significantly increased the luciferase
activity (Fig. 4B) and steady-state
mRNA levels (Fig. 4C), compared
with the Ctrl group, indicating that the 113–230 and 503–660
regions of the 3′-UTR promoted reporter gene expression, possibly
through increased mRNA stability. However, the insertion of the
970–1,053 3′-UTR fragment resulted in a significant decrease in
luciferase activity (Fig. 4B), but
an increase in the steady-state mRNA levels of the luciferase gene
(Fig. 4C), compared with the Ctrl
group. This suggested that the 970–1,053 region of the 3′-UTR may
hinder reporter gene expression by reducing the translation rate of
the luciferase mRNA.
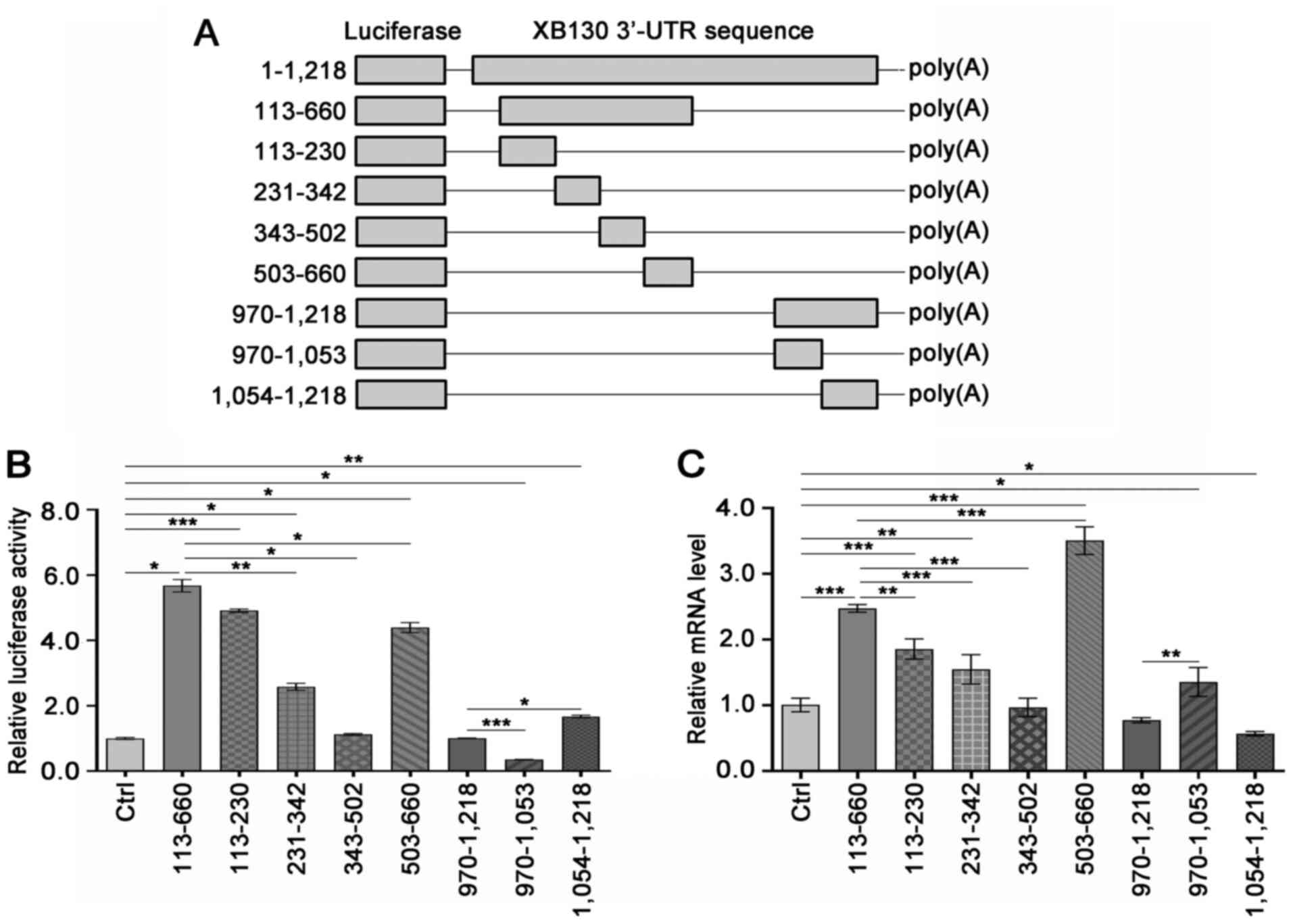 | Figure 4.Regions 113–230, 503–660, and
970–1,053 of the XB130 3′-UTR significantly affect the luciferase
activity and mRNA levels. (A) Overview of the construction of the
recombinant constructs. (B and C) PC-9 cells were transfected with
the empty modified psiCHECK-2 vector or the recombinants. The
luciferase activity and mRNA levels were measured after 48 h. The
hRluc mRNA levels were normalized to the hluc+ mRNA
level, and the relative mRNA level was expressed as a multiple of
that measured in the Ctrl group. *P<0.05, **P<0.01,
***P<0.001. UTR, untranslated region; Ctrl, control;
hluc+, firefly luciferase gene; Ctrl, control. |
XB130 3′-UTR 113–230, 503–660, or
970–1,053 fragment insertion increases the mRNA stability of a
reporter gene
To understand the molecular mechanism underlying the
regulation of reporter gene expression by the aforementioned
fragments, their effects on mRNA stability were investigated. As
shown in Fig. 5, all the tested
fragments increased the mRNA stability of the reporter gene, which
was consistent with the observed steady-state mRNA levels of the
reporter gene in Fig. 4C.
Furthermore, compared to the Ctrl group, no alteration was observed
in the stability of Hsp90 mRNA within the 113–230, 503–660, or
970–1,053 3′-UTR groups (Fig. 5).
These findings suggested that the 113–230 and 503–660 regions of
the 3′-UTR promoted the expression of the reporter gene by
enhancing mRNA stability.
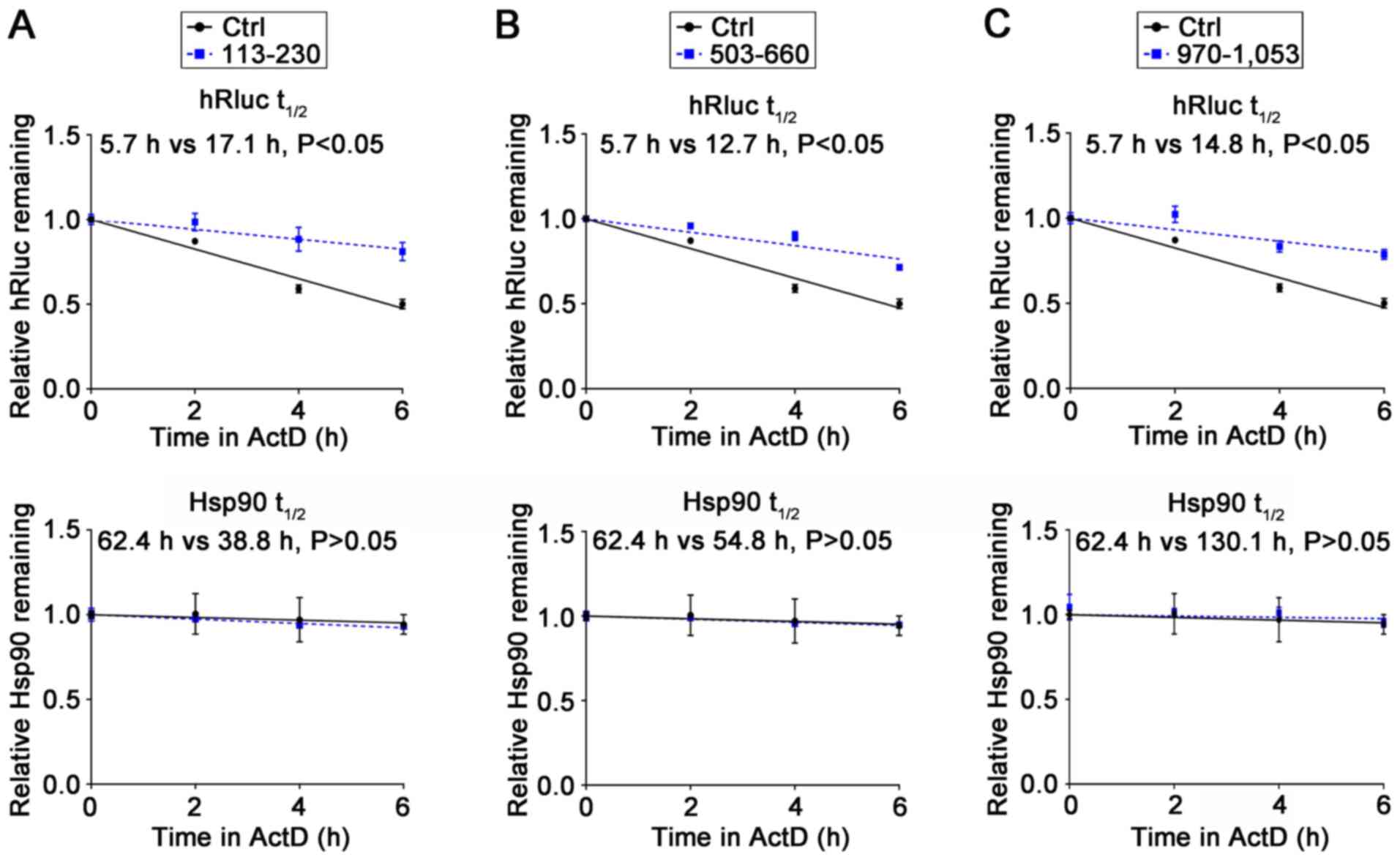 | Figure 5.XB130 3′-UTR 113–230, 503–660, or
970–1,053 fragment insertion increases luciferase mRNA stability.
PC-9 cells were transfected with the empty modified psiCHECK-2
vector or the recombinant containing the (A) 113–230 region, (B)
503–660 region or (C) 970–1,053 region of the XB130 3′-UTR. After
48 h, ActD (10 µg/ml) was added. At 0, 2, 4, and 6 h
post-treatment, cells were collected for RNA extraction, and the
mRNA expression levels of luciferase and Hsp90 were measured by
RT-qPCR. The hRluc mRNA level was normalized to the
hluc+ mRNA levels, and Hsp90 mRNA levels were normalized
to GAPDH mRNA levels. UTR, untranslated region; hluc+,
firefly luciferase gene; Ctrl, control; t1/2, the
half-lives of hRluc and Hsp90 mRNA; ActD, actinomycin D. |
XB130 3′-UTR 113–230, 503–660, or
970–1,053 fragment insertion impairs mRNA translation of a reporter
gene
To assess the relative contribution of mRNA
translation rate to the expression of the reporter gene, the
relative enzymatic activity level (Fig.
4B) to the relative mRNA level (Fig. 4C) of luciferase was calculated for
each group. A ratio of 1 indicated a direct correlation between
protein and mRNA levels, suggesting no contribution of the
translation process to reporter gene expression. Ratios >1
indicated a positive contribution of the translation process to
reporter gene expression, while ratios <1 indicated a negative
contribution. The larger the deviation from 1, the greater the
positive or negative impact of translation.
The results listed in Table III indicated that the 113–230
3′-UTR fragment had a relatively high ratio, the 503–660 fragment
had a ratio slightly >1 and the 970–1,053 fragment had a ratio
significantly <1. These results suggested that the 113–230 and
503–660 regions of the 3′-UTR promoted reporter gene expression
through a combination of positive contributions from mRNA stability
and translation rate. By contrast, the 970–1,053 region of the
3′-UTR inhibited reporter gene expression due to translation
inhibition.
 | Table III.Comparison of the enzymatic activity
and mRNA level of the luciferase. |
Table III.
Comparison of the enzymatic activity
and mRNA level of the luciferase.
| Region of
3′-UTR | Enzymatic activity
level | mRNA level | Ratioa |
|---|
| Control | 1.00±0.03 | 1.00±0.10 | 1.00±0.03 |
| 113-660 | 5.68±0.19 | 2.48±0.06 | 2.29±0.08 |
| 113-230 | 4.91±0.05 | 1.86±0.15 | 2.64±0.03 |
| 231-342 | 2.58±0.11 | 1.55±0.22 | 1.67±0.07 |
| 343-502 | 1.12±0.03 | 0.97±0.14 | 1.16±0.03 |
| 503-660 | 4.40±0.15 | 3.51±0.21 | 1.25±0.04 |
| 970-1,218 | 1.01±0.00 | 0.77±0.04 | 1.30±0.01 |
| 970-1,053 | 0.35±0.01 | 1.35±0.22 | 0.26±0.01 |
| 1,054-1,218 | 1.67±0.04 | 0.57±0.03 | 2.94±0.07 |
Seven RBPs potentially regulate XB130
expression by binding to the 113–230, 503–660, or 970–1,053 region
of the XB130 3′-UTR
Typically, the mRNA 3′-UTR influences mRNA stability
or translation rate by binding to specific RBPs. Given the
significant influence of the 113–230, 503–660, or 970–1,053 XB130
3′-UTR fragment insertion on reporter gene expression, further
screening for RBPs that bind to these regions may shed light on the
regulatory mechanism of XB130 expression. RNA pull-down assays and
mass spectrometry analysis were conducted, resulting in the
identification of 29, 31, or 12 candidate RBPs that bound to the
113–230, 503–660, or 970–1,053 XB130 3′-UTR fragment, respectively
(Tables IV, SI and SII). To validate the potential
interactions between candidate RBPs and XB130, in silico
co-expression analyses were performed using the ENCORI and GEPIA2
databases. Considering that the insertion of the 113–230, 503–660,
or 970–1,053 XB130 3′-UTR fragment increased reporter gene mRNA
stability, the mRNA expression levels of candidate RBPs were
expected to be positively correlated with XB130 mRNA expression
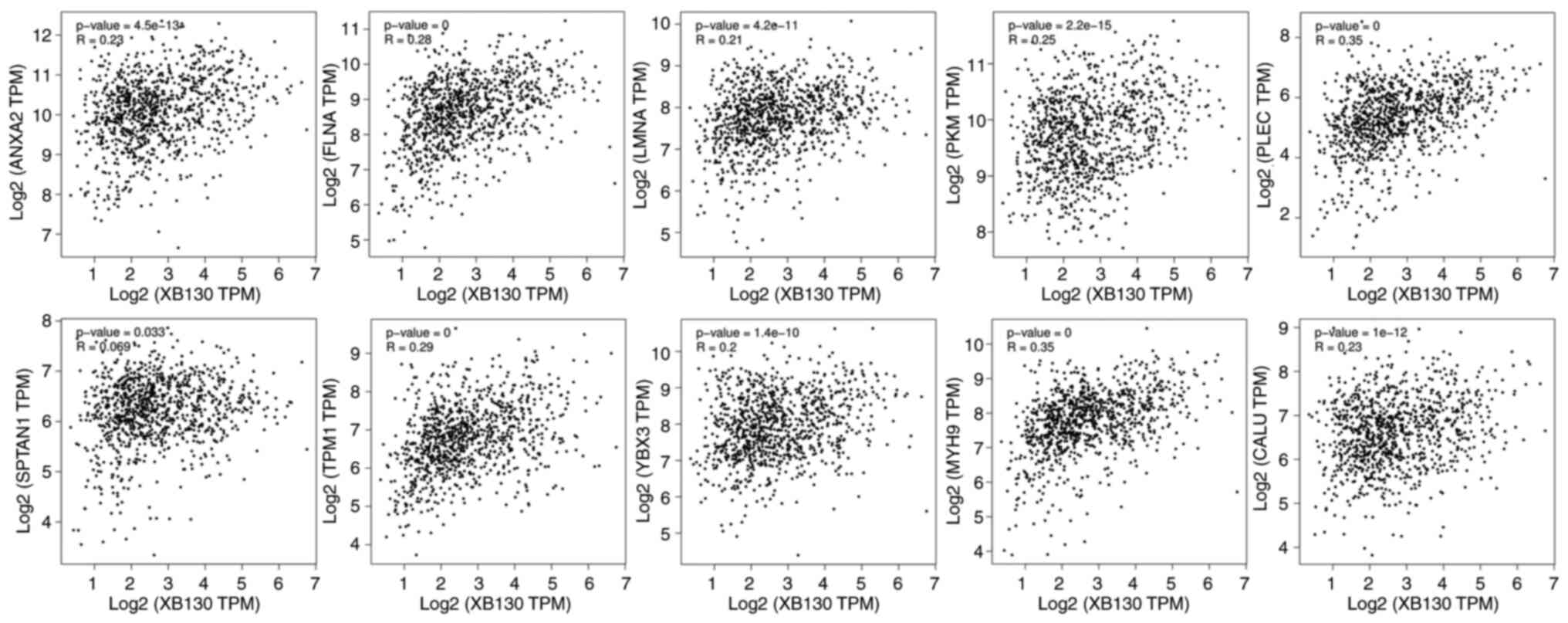
level. As shown in Fig. 6, 10
candidate RBPs were screened for co-expression with XB130 in both
databases. Among them, Annexin A2 (ANXA2), Filamin-A (FLNA), Lamin
A/C (LMNA), Plectin (PLEC), Spectrin α, non-erythrocytic 1
(SPTAN1), and Tropomyosin 1 (TPM1) were identified as candidate
RBPs for the 113–230 region of the 3′-UTR, while ANXA2, FLNA,
Pyruvate kinase M1/2 (PKM), and Y-box binding protein 3 (YBX3) were
identified as candidate RBPs for the 503–660 region of the 3′-UTR.
In addition, myosin heavy chain 9 (MYH9) and calumenin (CALU) were
candidate RBPs for the 970–1,053 region of the 3′-UTR.
 | Table IV.Candidate RBPs that bind to the
113–230, 503–660, or 970–1,053 region of the XB130 3′-UTR
identified by RNA pull-down and mass spectrometry analyses. |
Table IV.
Candidate RBPs that bind to the
113–230, 503–660, or 970–1,053 region of the XB130 3′-UTR
identified by RNA pull-down and mass spectrometry analyses.
| XB130 3′-UTR
region | Gene | Protein name | mW, kDa |
|---|
| 113-230 | TUBB3 | Tubulin β 3 class
III | 50.433 |
|
| ACTC1 | Actin α cardiac
muscle 1 | 42.019 |
|
| TUBA1C | Tubulin α1c | 49.895 |
|
| TPM1 | Tropomyosin 1 | 31.753 |
|
| HIST1H2BK | H2B clustered
histone 12 | 13.89 |
|
| HIST1H2AB | H2A clustered
histone 4 | 14.135 |
|
| HNRNPR | Heterogeneous
nuclear ribonucleoprotein R | 70.943 |
|
| LDHA | Lactate
dehydrogenase A | 36.689 |
|
| YWHAB | Tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein
β | 28.082 |
|
| LMNB2 | Lamin-B2 | 69.948 |
|
| SYNCRIP | Heterogeneous
nuclear ribonucleoprotein Q | 69.603 |
|
| NDUFS3 | NADH: ubiquinone
oxidoreductase core subunit S3 | 30.242 |
|
| HSP90B1 | Heat shock protein
90 β family member 1 | 92.469 |
|
| IMMT | Inner membrane
mitochondrial protein | 83.678 |
|
| PPL | Periplakin | 204.747 |
|
| PHB | Prohibitin 1 | 29.804 |
|
| SPTAN1 | Spectrin α,
non-erythrocytic 1 | 284.539 |
|
| FLNA | Filamin-A | 280.739 |
|
| RNH1 |
Ribonuclease/angiogenin inhibitor 1 | 49.973 |
|
| INA | Internexin neuronal
intermediate filament protein α | 55.391 |
|
| NCL | Nucleolin | 76.614 |
|
| GAPDH |
Glyceraldehyde-3-phosphate
dehydrogenase | 36.053 |
|
| ANXA2 | Annexin A2 | 38.604 |
|
| RAE1 | Ribonucleic acid
export 1 | 40.968 |
|
| PLEC | Plectin | 531.791 |
|
| LMNB1 | Lamin-B1 | 66.408 |
|
| ATP5PD | ATP synthase
peripheral stalk subunit d | 18.491 |
|
| LMNA | Lamin A/C | 74.139 |
|
| HSPA8 | Heat shock protein
family A (Hsp70) member 8 | 70.898 |
| 503-660 | TUBB3 | Tubulin β3 class
III | 50.433 |
|
| HIST1H2BK | H2B clustered
histone 12 | 13.89 |
|
| HIST1H2AB | H2A clustered
histone 4 | 14.135 |
|
| HNRNPR | Heterogeneous
nuclear ribonucleoprotein R | 70.943 |
|
| LDHA | Lactate
dehydrogenase A | 36.689 |
|
| HNRNPK | Heterogeneous
nuclear ribonucleoprotein K | 50.976 |
|
| H2AFZ | H2A.Z variant
histone 1 | 13.553 |
|
| PKM | Pyruvate kinase
M1/2 | 57.937 |
|
| YWHAB | Tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein
β | 28.082 |
|
| SYNCRIP | Heterogeneous
nuclear ribonucleoprotein Q | 69.603 |
|
| NDUFS3 | NADH: ubiquinone
oxidoreductase core subunit S3 | 30.242 |
|
| YBX3 | Y-box binding
protein 3 | 40.09 |
|
| YWHAE | Tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein
ε | 29.174 |
|
| HNRNPD | Heterogeneous
nuclear ribonucleoprotein D | 38.434 |
|
| IMMT | Inner membrane
mitochondrial protein | 83.678 |
|
| ALDOA | Aldolase,
fructose-bisphosphate A | 39.42 |
|
| HSP90B1 | Heat shock protein
90 β family member 1 | 92.469 |
|
| RPL4 | Ribosomal protein
L4 | 47.697 |
|
| GAPDH |
Glyceraldehyde-3-phosphate
dehydrogenase | 36.053 |
|
| EIF2AK2 | Eukaryotic
translation initiation factor 2 α kinase 2 | 62.094 |
|
| FLNA | Filamin-A | 280.739 |
|
| RBMX | RNA binding motif
protein X-linked | 42.332 |
|
| RPS10 | Ribosomal protein
S10 | 18.898 |
|
| RPS14 | Ribosomal protein
S14 | 16.273 |
|
| HIST1H2BJ | H2B clustered
histone 11 | 13.904 |
|
| ANXA2 | Annexin A2 | 38.604 |
|
| RPL12 | Ribosomal protein
L12 | 17.819 |
|
| RPS19 | Ribosomal protein
S19 | 16.061 |
|
| LRRC59 | Leucine rich repeat
containing 59 | 34.93 |
|
| HSPD1 | Heat shock protein
family D (Hsp60) member 1 | 61.055 |
|
| HNRNPAB | Heterogeneous
nuclear ribonucleoprotein A/B | 30.303 |
| 970-1,053 | HNRNPD | Heterogeneous
nuclear ribonucleoprotein D | 38.434 |
|
| H2AFZ | H2A.Z variant
histone 1 | 13.553 |
|
| PTBP1 | Polypyrimidine
tract binding protein 1 | 57.221 |
|
| CD2BP2 | CD2 cytoplasmic
tail binding protein 2 | 37.646 |
|
| PFN1 | Profilin-1 | 15.054 |
|
| HNRNPC | Heterogeneous
nuclear ribonucleoproteins C | 33.67 |
|
| ESRP1 | Epithelial splicing
regulatory protein 1 | 75.585 |
|
| HNRNPAB | Heterogeneous
nuclear ribonucleoprotein A/B | 30.303 |
|
| PGRMC1 | Progesterone
receptor membrane component 1 | 21.671 |
|
| MYH9 | Myosin heavy chain
9 | 226.532 |
|
| HNRNPDL | Heterogeneous
nuclear ribonucleoprotein D-like | 46.438 |
|
| CALU | Calumenin | 37.107 |
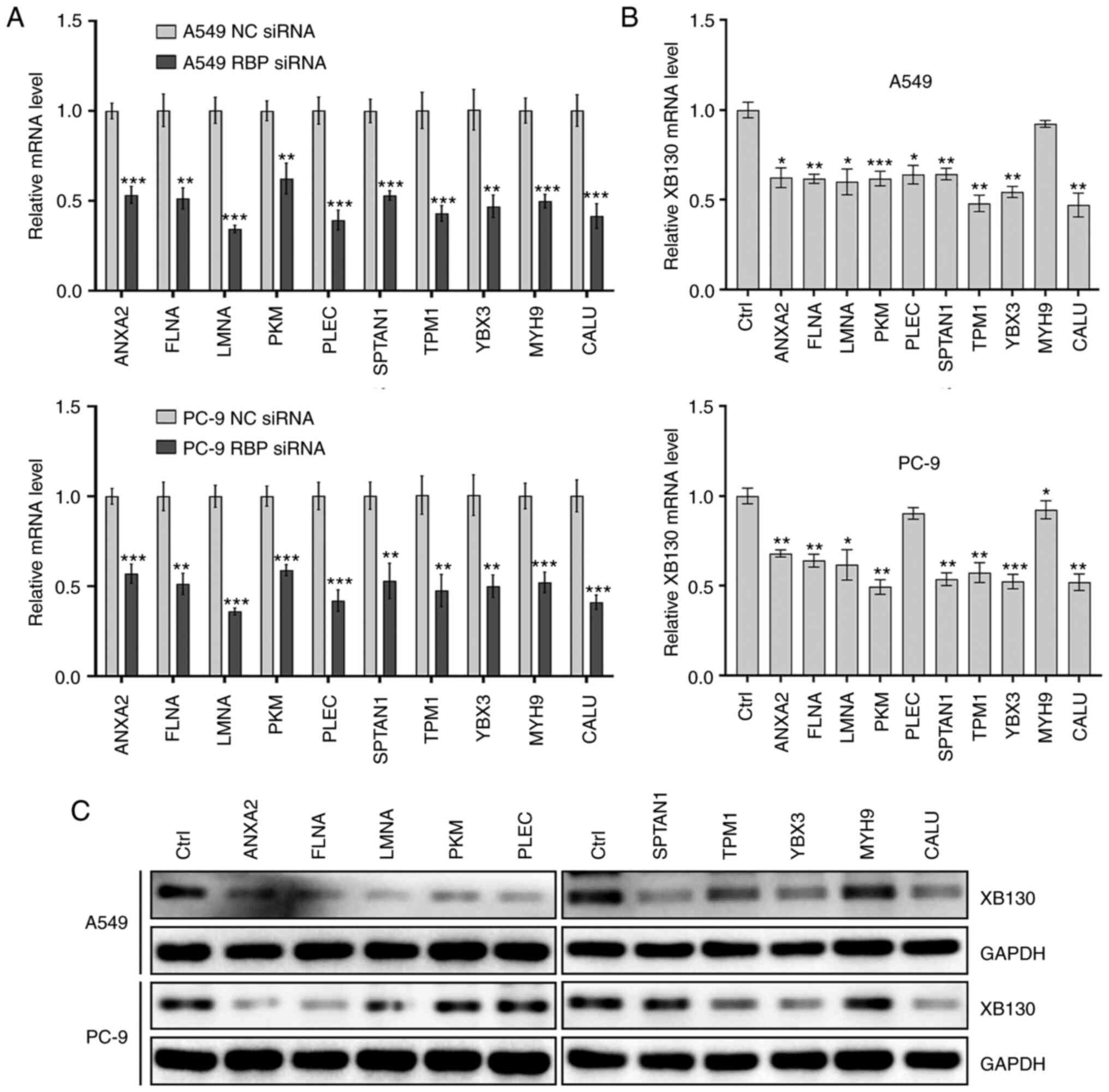
Following this analysis, the expression of each RBP
candidate was knocked down in two NSCLC cell lines, and the XB130
mRNA and protein levels were measured. The results demonstrated
that knockdown of ANXA2, FLNA, LMNA, SPTAN1, TPM1, YBX3, or CALU
downregulated XB130 mRNA and protein expression in both NSCLC cell
lines (Fig. 7). This indicated that
these seven RBPs may regulate XB130 expression by binding to the
113–230, 503–660, or 970–1,053 region of the XB130 3′-UTR.
Discussion
To date, the function of XB130 protein has been well
elucidated, but there has been limited research on the regulation
of its expression (1–6,10,19,23–27).
Previous studies have demonstrated that overexpression of
miR-30a/b/c/d/e, 203, 219, or 4782-3p inhibited XB130 expression,
impacting the proliferation, migration, invasion, and EMT of NSCLC
cells (6,7). The present study demonstrated that,
apart from miRNA binding sites, other positive and negative
regulatory elements are present in the XB130 3′-UTR, of which
negative regulatory elements were primarily located in the 1–112
and 990–1,218 regions of the 3′-UTR, and positive regulatory
elements in the 113–989 region.
The present study indicated that the insertion of
the 661–795 or 792–989 3′-UTR fragments into the psiCHECK-2 vector
increased the luciferase activity of the reporter gene, compared
with the Ctrl group. Furthermore, compared with the 970–1,218
3′-UTR group, the addition of 792–969 or 661–969 3′-UTR fragment to
the 5′-end of the 970–1,218 3′-UTR fragment further enhanced
luciferase activity, suggesting the presence of positive regulatory
elements in the 661–989 region of the 3′-UTR. However, compared
with the 1–660 3′-UTR group, the insertion of the 661–795 or
661–989 3′-UTR fragments at the 3′-end of the 1–660 3′-UTR fragment
did not further promote reporter gene expression. This observation
indicated that the presence of the 1–660 region of the 3′-UTR
inhibited the activity of positive regulatory elements in the
661–989 region of the 3′-UTR. Similarly, Cok and Morrison (28) reported that the 373–792 region of
the Cyclooxygenase-2 (COX-2) 3′-UTR alone exerted an inhibitory
effect on gene expression, but exhibited a positive regulatory
property when combined with the 1–373 region of the COX-2
3′-UTR.
Based on the A/URE classification principles, XB130
3′-UTR contains three typical IIE AREs (AREI,
AREII, and AREIII) and one potential URE
(15,29,30).
In the present study, these elements were investigated through
deletion mutations. The results demonstrated that the deletion of
the three AREs did not affect luciferase activity, while the
deletion of the URE increased luciferase activity, indicating that
the URE acts as a negative regulation element for XB130 expression.
Additionally, the insertion of the 970–1,053 3′-UTR fragment, where
the URE is located, increased luciferase mRNA stability but reduced
luciferase activity. Based on these results, it is hypothesized
that the predicted URE may possess functions that promote mRNA
stability while inhibiting mRNA translation rate. Similar functions
of G/A/C/UREs have been previously reported. For example, CUGBP
Elav-like family member 2 (CUGBP2) increased the stability of COX-2
mRNA and inhibited mRNA translation by binding to two AREs in the
COX-2 3′-UTR, resulting in decreased COX-2 expression (31). In addition, human antigen R (HuR) or
AU-rich binding factor 1 (AUF1) reduced the expression of tumor
necrosis factor-α or granulocyte-macrophage colony-stimulating
factor, respectively, through a similar mechanism (32,33).
Among the G/A/C/UREs, AREs and GREs have been more
extensively studied. AREs are present in 10–15% of human mRNAs and
they regulate cell signaling, RNA metabolism, growth, and
development by interacting with specific RBPs, such as
Tristetraprolin, HuR, AUF1, and KH-type splicing regulatory protein
(16,34,35).
Comparatively, research on GRE is relatively scarce (16). GRE is found in at least 5% of human
mRNA, its core sequence is GUUUG, and it is primarily involved in
cell transcription, cell cycle, cell metabolism, and cell-cell
communication (14,16). Several GRE-binding proteins have
been identified, including CUGBP Elav-like family member 1, CUGBP2,
and HuR (14,17). Regarding CRE, it has been reported
that the core sequence is
(C/U)CCANxCCC(U/A)(C/U)yUC(C/U)CC, and hnRNP
K or E2/E1 can stabilize mRNA by binding to the CRE (36,37).
In the present study, it was observed that the insertion of the
113–230 or 503–660 XB130 3′-UTR fragment significantly increased
luciferase activity. However, the core sequences of ARE, GRE, and
CRE were not identified in the 113–230 and 503–660 regions of the
3′-UTR. Therefore, it is speculated that atypical G/A/C/URE
sequences may exist in these regions, such as AGUUU, which requires
further investigation through truncation or mutation
experiments.
In the present study, seven candidate RBPs were
identified that regulated XB130 mRNA and protein expression in two
cell lines. Among them, LMNA is a major component of the mammalian
nuclear lamina and is involved in regulating cellular signaling and
gene transcription (38). CALU is a
Ca2+-binding protein located within the endoplasmic
reticulum (39). SPTAN1 and TPM1
are cytoskeletal proteins associated with cell adhesion, DNA
repair, and cell migration (40,41).
Currently, their participation in post-transcriptional regulation
of gene expression remains unclear. In the present study, knockdown
of CALU, a candidate RBP of the 970–1,053 region of the XB130
3′-UTR, decreased XB130 mRNA and protein expression levels,
suggesting that CALU primarily regulated XB130 mRNA stability. The
970–1,053 region of the 3′-UTR may suppress XB130 translation by
binding to other unidentified RBPs. ANXA2 has been reported to
post-transcriptionally regulate the expression of c-Myc and prolyl
4-hydroxylase subunit α 1 genes by binding to the AA(C/G)(A/U)G
consensus sequence in their 3′-UTRs (42). Furthermore, Cheng and Tong (43) reported that FLNA and ANXA2
interacted and cooperatively mediated gefitinib resistance in
NSCLC. Given the presence of the AA(C/G)AG sequence in the 113–230
and 503–660 regions of the XB130 3′-UTR, it is plausible that the
interaction between FLNA and ANXA2 promotes ANXA2 binding to these
regions of the 3′-UTR, subsequently enhancing XB130 mRNA stability
or translation rate. YBX3 has been shown to bind to the 3′-UTR of
solute carrier family 7 member 5 and stabilize its transcript for
translation (44). Therefore, it is
suggested that YBX3 may bind to the 503–660 region of the XB130
3′-UTR, thereby enhancing the stability of XB130 mRNA.
In conclusion, the present study highlighted the
significant role of the XB130 3′-UTR as a crucial regulator of
XB130 expression by modulating mRNA stability and translational
rate. Specifically, the 113–230, 503–660, and 970–1,053 regions of
the 3′-UTR may play a critical role by interacting with specific
RBPs, including ANXA2, FLNA, LMNA, SPTAN1, TPM1, YBX3 and CALU. It
is important to note that further research is needed to verify the
binding between these RBPs and the mentioned 3′-UTR fragments, as
well as their impact on XB130 mRNA stability and translation.
Despite these limitations, the findings of the present study
provide a strong basis for future investigations into the
mechanisms governing XB130 expression.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science
Foundation of China (grant nos. 81660474 and 81960655), the Guizhou
Provincial Basic Research Program (Natural Science) (grant nos.
ZK[2022]041 and ZK[2022]372), the Academic Seedling Project of
Guizhou Medical University (grant no. 21NSFCP04), and the Natural
Science Foundation of Guizhou Provincial Health Commission (grant
no. gzwkj2023-254).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
QW, YL, and KS designed the study. QW and LL
performed the experiments, prepared the figures, and wrote the
manuscript. XG and TZ performed the statistical analysis. YZ, YX,
and JZ interpreted the data. QW, YL, and KS confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bai XH, Cho HR, Moodley S and Liu M:
XB130-A novel adaptor protein: Gene, function, and roles in
tumorigenesis. Scientifica (Cairo). 2014:9030142014.PubMed/NCBI
|
|
2
|
Lodyga M, De Falco V, Bai XH, Kapus A,
Melillo RM, Santoro M and Liu M: XB130, a tissue-specific adaptor
protein that couples the RET/PTC oncogenic kinase to PI 3-kinase
pathway. Oncogene. 28:937–949. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shiozaki A, Shen-Tu G, Bai X, Iitaka D, De
Falco V, Santoro M, Keshavjee S and Liu M: XB130 mediates cancer
cell proliferation and survival through multiple signaling events
downstream of Akt. PLoS One. 7:e436462012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cho HR, Wang Y, Bai X, Xiang YY, Lu C,
Post A, Al Habeeb A and Liu M: XB130 deficiency enhances
carcinogen-induced skin tumorigenesis. Carcinogenesis.
40:1363–1375. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shiozaki A and Liu M: Roles of XB130, a
novel adaptor protein, in cancer. J Clin Bioinforma. 1:102011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Q, Yang G, Jiang Y, Luo M, Li C, Zhao
Y, Xie Y, Song K and Zhou J: XB130, regulated by miR-203, miR-219,
and miR-4782-3p, mediates the proliferation and metastasis of
non-small-cell lung cancer cells. Mol Carcinog. 59:557–568. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song K, Jiang Y, Zhao Y, Xie Y, Zhou J, Yu
W and Wang Q: Members of the miR-30 family inhibit the
epithelial-to-mesenchymal transition of non-small-cell lung cancer
cells by suppressing XB130 expression levels. Oncol Lett.
20:682020.PubMed/NCBI
|
|
8
|
Shi M, Zheng D, Sun L, Wang L, Lin L, Wu
Y, Zhou M and Liao W, Liao Y, Zuo Q and Liao W: XB130 promotes
proliferation and invasion of gastric cancer cells. J Transl Med.
12:12014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi M, Huang W, Lin L, Zheng D, Zuo Q,
Wang L, Wang N, Wu Y, Liao Y and Liao W: Silencing of XB130 is
associated with both the prognosis and chemosensitivity of gastric
cancer. PLoS One. 7:e416602012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shiozaki A, Lodyga M, Bai XH, Nadesalingam
J, Oyaizu T, Winer D, Asa SL, Keshavjee S and Liu M: XB130, a novel
adaptor protein, promotes thyroid tumor growth. Am J Pathol.
178:391–401. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mayr C: Evolution and Biological Roles of
Alternative 3′UTRs. Trends Cell Biol. 26:227–237. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Khabar KS: Hallmarks of cancer and AU-rich
elements. Wiley Interdiscip Rev RNA. 8:e13682017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kovarik P, Ebner F and Sedlyarov V:
Posttranscriptional regulation of cytokine expression. Cytokine.
89:21–26. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vlasova-St Louis I and Bohjanen PR:
Feedback regulation of kinase signaling pathways by AREs and GREs.
Cells. 5:42016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Barreau C, Paillard L and Osborne HB:
AU-rich elements and associated factors: Are there unifying
principles? Nucleic Acids Res. 33:7138–7150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Halees AS, Hitti E, Al-Saif M, Mahmoud L,
Vlasova-St Louis IA, Beisang DJ, Bohjanen PR and Khabar K: Global
assessment of GU-rich regulatory content and function in the human
transcriptome. RNA Biol. 8:681–691. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vlasova IA, Tahoe NM, Fan D, Larsson O,
Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS,
Bitterman PB and Bohjanen PR: Conserved GU-rich elements mediate
mRNA decay by binding to CUG-binding protein 1. Mol Cell.
29:263–270. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li F, Zhao H, Su M, Xie W, Fang Y, Du Y,
Yu Z, Hou L and Tan W: HnRNP-F regulates EMT in bladder cancer by
mediating the stabilization of Snail1 mRNA by binding to its 3′
UTR. EBioMedicine. 45:208–219. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang R, Zhang J, Wu Q, Meng F and Liu C:
XB130: A novel adaptor protein in cancer signal transduction.
Biomed Rep. 4:300–306. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hussain H and Chong NF: Combined overlap
extension PCR method for improved site directed mutagenesis. Biomed
Res Int. 2016:80415322016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tao X and Gao G: Tristetraprolin recruits
eukaryotic initiation factor 4E2 To repress translation of AU-Rich
element-containing mRNAs. Mol Cell Biol. 35:3921–3932. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lodyga M, Bai XH, Kapus A and Liu M:
Adaptor protein XB130 is a Rac-controlled component of lamellipodia
that regulates cell motility and invasion. J Cell Sci. 123((Pt
23)): 4156–4169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moodley S, Hui Bai X, Kapus A, Yang B and
Liu M: XB130/Tks5 scaffold protein interaction regulates
Src-mediated cell proliferation and survival. Mol Biol Cell.
26:4492–4502. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen J, Jin C, Liu Y, Rao H, Liu J and Li
J: XB130 enhances invasion and migration of human colorectal cancer
cells by promoting epithelial-mesenchymal transition. Mol Med Rep.
16:5592–5598. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu Q, Nadesalingam J, Moodley S, Bai X and
Liu M: XB130 translocation to microfilamentous structures mediates
NNK-induced migration of human bronchial epithelial cells.
Oncotarget. 6:180501–180565. 2015.
|
|
27
|
Yamanaka D, Akama T, Chida K, Minami S,
Ito K, Hakuno F and Takahashi S: Phosphatidylinositol
3-Kinase-Associated Protein (PI3KAP)/XB130 crosslinks actin
filaments through its actin binding and multimerization properties
in vitro and enhances endocytosis in HEK293 cells. Front Endocrinol
(Lausanne). 7:892016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cok SJ and Morrison AR: The
3′-untranslated region of murine cyclooxygenase-2 contains multiple
regulatory elements that alter message stability and translational
efficiency. J Biol Chem. 276:23179–23185. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baou M, Jewell A and Murphy JJ: TIS11
family proteins and their roles in posttranscriptional gene
regulation. J Biomed Biotechnol. 2009:6345202009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fallmann J, Sedlyarov V, Tanzer A, Kovarik
P and Hofacker IL: AREsite2: An enhanced database for the
comprehensive investigation of AU/GU/U-rich elements. Nucleic Acids
Res. 44((D1)): D90–D95. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mukhopadhyay D, Houchen CW, Kennedy S,
Dieckgraefe BK and Anant S: Coupled mRNA stabilization and
translational silencing of cyclooxygenase-2 by a novel RNA binding
protein, CUGBP2. Mol Cell. 11:113–126. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dean JL, Wait R, Mahtani KR, Sully G,
Clark AR and Saklatvala J: The 3′ untranslated region of tumor
necrosis factor alpha mRNA is a target of the mRNA-stabilizing
factor HuR. Mol Cell Biol. 21:721–730. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Raineri I, Wegmueller D, Gross B, Certa U
and Moroni C: Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent
mRNA turnover studied by RNA interference. Nucleic Acids Res.
32:1279–1288. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bakheet T, Williams BR and Khabar KS: ARED
3.0: The large and diverse AU-rich transcriptome. Nucleic Acids
Res. 34((Database Issue)): D111–D114. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Halees AS, El-Badrawi R and Khabar KS:
ARED Organism: Expansion of ARED reveals AU-rich element cluster
variations between human and mouse. Nucleic Acids Res. 36((Database
Issue)): D137–D140. 2008.PubMed/NCBI
|
|
36
|
Ostareck DH, Ostareck-Lederer A, Wilm M,
Thiele BJ, Mann M and Hentze MW: mRNA silencing in erythroid
differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase
translation from the 3′ end. Cell. 89:597–606. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang S, Zhang J, Theel S, Barb JJ, Munson
PJ and Danner RL: Nitric oxide activation of Erk1/2 regulates the
stability and translation of mRNA transcripts containing CU-rich
elements. Nucleic Acids Res. 34:3044–3056. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Alcorta-Sevillano N, Macías I, Rodríguez
CI and Infante A: Crucial role of Lamin A/C in the migration and
differentiation of MSCs in bone. Cells. 9:13302020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Y, Sun Y, Fu Y, Guo X, Long J, Xuan
LY, Wei CX and Zhao M: Calumenin relieves cardiac injury by
inhibiting ERS-initiated apoptosis during viral myocarditis. Int J
Clin Exp Pathol. 10:7277–7284. 2017.PubMed/NCBI
|
|
40
|
Ackermann A and Brieger A: The role of
nonerythroid spectrin αII in cancer. J Oncol. 2019:70796042019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Du HQ, Wang Y, Jiang Y, Wang CH, Zhou T,
Liu HY and Xiao H: Silencing of the TPM1 gene induces
radioresistance of glioma U251 cells. Oncol Rep. 33:2807–2814.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vedeler A, Hollås H, Grindheim AK and
Raddum AM: Multiple roles of annexin A2 in post-transcriptional
regulation of gene expression. Curr Protein Pept Sci. 13:401–412.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cheng L and Tong Q: Interaction of FLNA
and ANXA2 promotes gefitinib resistance by activating the Wnt
pathway in non-small-cell lung cancer. Mol Cell Biochem.
476:3563–3575. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cooke A, Schwarzl T, Huppertz I, Kramer G,
Mantas P, Alleaume AM, Huber W, Krijgsveld J and Hentze MW: The
RNA-Binding Protein YBX3 controls amino acid levels by regulating
SLC mRNA abundance. Cell Rep. 27:3097–3106.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|