Introduction
Low-density lipoprotein receptor-related protein-1
(LRP1) is a multifunctional endocytic receptor that participates in
the metabolism of various extracellular ligands, such as
platelet-derived growth factor and matrix metalloproteinase-9
(MMP-9), by regulating cell signaling pathways, such as the
Wnt/β-catenin pathway, including enzymes involved in tumor invasion
(1). Lipoprotein metabolic
processes are activated in tumors and increased lipid uptake and
storage in a variety of cancers contribute to rapid tumor growth
(2).
Gastrointestinal (GI) cancer is among the most
common malignancies worldwide and comprises esophageal, stomach,
colorectum, liver and pancreatic cancer (3). A total of 4.9 million GI cancer cases
and 3.5 million GI cancer-related deaths were estimated in 2020
(4). GI cancers account for 26% of
global cancer incidence and 35% of all cancer-related deaths
(4). Early stage treatment of GI
cancer is mainly performed by surgical resection (5). Chemotherapy is the primary treatment
for advanced GI cancers; however, the resistance of cancer cells to
chemotherapy drugs leads to chemotherapy failure (6). Clinical diagnosis and treatment need
novel biomarkers of GI cancer to allow for earlier detection. Feng
et al (7) showed that
positive rates of α-fetoprotein and cancer antigen 19-9 (CA19-9)
are relatively low in early-stage stomach cancer, while levels of
carcinoembryonic antigen are an independent risk factor for poor
prognosis of early-stage stomach cancer. Moreover, pancreatic
cancer has a poor survival rate and the high mortality rate is
attributed to the difficulty of making an early-stage diagnosis
(8).
In a previous study, knocking down LRP1 in
pancreatic cancer PANC-1 cells inhibited tumor cell proliferation
(9). Gheysarzadeh et al
(10) showed that upregulation of
LRP1 is associated with poor prognosis and cell invasion in
pancreatic cancer. In pancreatic cancer, lipoprotein metabolic
process results in an increase in the levels of cholesterol and
upregulation of low-density lipoprotein receptor (LDLR) in tumor
cells (11). Pancreatic ductal
adenocarcinoma has no obvious symptoms in the early stage of the
disease, and diagnosis is difficult, which results in later-stage
diagnosis of the disease and poor prognosis. Compared with
low-grade astrocytoma, malignant glioma is characterized by notably
higher levels of LRP1 mRNA and protein (12). Huang et al (13) showed that high expression of LRP1 is
associated with low metastatic potential of hepatocellular
carcinoma.
However, the specific role and molecular mechanism
of LRP1 in GI tumor cells remains unclear. It is hypothesized that
LRP1 is related to cholesterol uptake and the proliferation of
tumor cells and the high expression of LRP1 may promote the
proliferation of tumor cells. To verify this hypothesis,
bioinformatics analysis was used to investigate the expression of
LRP1 in GI tumors and its correlation with prognosis of patients
with GI cancer. Subsequently, in vitro experiments were
performed to evaluate the impact of LRP1 knockdown on GI cancer
cell proliferation, migration and invasion.
Materials and methods
Bioinformatics analysis
RNA sequencing data for normal stomach, pancreas and
liver from the Genotype-Tissue Expression database
(gtexportal.org), and for stomach, pancreas and liver tumors from
The Cancer Genome Atlas database (TCGA; portal.gdc.cancer.gov) were
downloaded from the Xena portal (https://github.com/BD2KGenomics/toil) with data
generated by the Toil pipeline management system (14). The enrichment data of Gene Ontology
(GO; http://geneontology.org/) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
pathway were derived from the KEGG website (www.kegg.jp). The analyses were performed using R
(version 3.6.3; http://www.r-project.org/) with packages ‘ggplot2’
(version 3.3.3), ‘survival’ (version 3.2.10), ‘survminer’ (version
0.4.9), ‘DESeq2’ (version 3.3.3) and ‘pROC’ (version 1.17.0.1).
GSEA
Enrichment analysis was conducted using GSEA
(version 4.2.2) software on data retrieved from the TCGA database
(15,16). The analysis process involved the
following steps: i) Calculating enrichment scores for each gene
set; ii) sorting the gene sets based on the magnitude of their
enrichment scores; iii) considering results as statistically
significant if they met the criteria of P<0.05, false discovery
rate (FDR) <25% and normalized enrichment score >1.0.
Cell lines and culture
HGC-27, HepG2 hepatoblastoma
(cellosaurus.org/CVL_0027), BxPC-3 (authenticated through short
tandem repeat analysis), PANC-1 and CaCo-2 cells, and HUVECs, were
obtained from the Cell Bank of the Chinese Academy of Science
(Shanghai, China). Cells were cultured in Dulbecco's Modified
Eagle's medium or RPMI-1640 (both Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS; HyClone;
Cytiva). All cell lines were cultured at 5% CO2 and
37°C.
Western blotting
The following primary antibodies were used for
western blotting: LRP1 (cat. no. ab92544; 1:10,000; Abcam), ERK1/2
(cat. no. ab184699; 1:5,000, Abcam), phosphorylated ERK1/2
(p-ERK1/2; cat. no. ab201015; 1:5,000; Abcam), AKT (cat. no.
ab38449; 1:10,000; Abcam), p-AKT (cat. no. ab81283; 1:10,000;
Abcam), epidermal growth factor receptor (EGFR; cat. no. ab52849;
1:10,000; Abcam), MMP2 (cat. no. ab92536; 1:5,000; Abcam), MMP9
(cat. no. ab76003; 1:10,000; Abcam) and GAPDH (cat. no. ab8245;
1:5,000; Abcam). Horseradish peroxidase-conjugated anti-mouse (cat.
no. GAM007; 1:5,000; Multi Sciences Biotech Co., Ltd.) or
anti-rabbit IgG antibodies (cat. no. GAR007; 1:5,000; Multi
Sciences Biotech Co., Ltd.) were used as secondary antibodies, and
an enhanced chemiluminescent kit (Multi Sciences Biotech Co., Ltd.)
was used for visualization. The total protein extraction buffer was
prepared by mixing RIPA Lysis Buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology) with phenylmethanesulfonyl fluoride
(cat. no. ST506; Beyotime Institute of Biotechnology) in a 1:1
ratio. The total protein was obtained all aforementioned cell
lines. The protein concentration was determined after total protein
extraction using the BCA Protein Assay Kit (cat. no. P0012;
Beyotime Institute of Biotechnology). Each lane was loaded with 100
µg total protein and separated on a gels using SDS-PAGE (6%
stacking gel, 10% separating gel). Transfer to a PVDF membrane
(cat. no. ISEQ00010; MilliporeSigma) was performed at 200 mA
constant current using the Biorad Powerpac Basic 164–5050 (Bio-Rad
Laboratories Inc.) for 95 min in an ice bath at 4°C. After
transfer, the membranes were blocked with 5% skimmed milk for 2 h
(5% skimmed milk prepared in TBST containing 0.1% Tween 20). The
blocked membranes were then incubated in a primary antibody
solution at 4°C overnight. After the first antibody incubation, the
membrane was washed with TBST three times for 10 min each.
Subsequently, the membrane was incubated with the secondary
antibody at room temperature for 2 h. Finally, protein detection
was performed using an enhanced chemiluminescence kit.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from all the aforementioned cell lines was
extracted with TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.), and cDNA synthesis was performed using a reverse
transcription kit (cat. no. 1708891; Bio-Rad Laboratories, Inc.)
according to the manufacturer's instructions. Next, iTaq™ Universal
SYBR-Green Supermix (Bio-Rad Laboratories, Inc.) was added
according to the qPCR reaction system, the reaction was centrifuged
for a short time (5,000 × g, 4°C, 10 sec) and then placed in an ABI
7500 (Applied Biosystems; Thermo Fisher Scientific, Inc.)
fluorescent instrument for qPCR. The qPCR thermal cycling
conditions consisted of an initial denaturation step at 95C for 30
sec, followed by an annealing step at 95°C for 15 sec and an
extension step at 60°C for 60 sec. The entire cycling process was
repeated for a total of 40 cycles. The relative expression levels
of LRP1 and CD36 mRNA were evaluated using the 2−ΔΔCq
method and normalized to β-actin (17). Primer sequences are listed in
Table I.
 | Table I.Primer sequences for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR.
| Gene | Primer
sequence |
|---|
| Low-density
lipoprotein receptor-related protein 1 | Forward:
5′TAATCCCTCTGCTGTTGCTGC-3′ |
| Reverse:
5′-GGTTTCCAATCTCCACGTTCA-3′ |
| CD36 | Forward: 5′-
GGAACTGTGGGCTCAT-3′ |
|
| Reverse: 5′-
AGAATACCTCCAAACAC −3′ |
| β-actin | Forward:
5′-CATGTACGTTGCTATCCAGGC-3′; |
|
| Reverse:
5′-CTCCTTAATGTCACGCAGGAT-3′ |
Lentiviral transduction
Lentiviral short hairpin RNA LRP1 (LV-shLRP1) and
lentiviral short-hairpin RNA negative control (LV-shNC) were made
by and purchased from Shanghai GeneChem Co., Ltd. LV-shLRP1
transfections were performed to interfere with LRP1 expression in
HGC-27, HepG2 and BxPC-3 cells according to manufacturer's
instructions of the lentivirus. A total of 5,000 cells/well were
seeded into a 6-well plate, with a multiplicity of infection of 10.
The lentivirus concentration was 1×107 TU/ml, and 10 µl
lentivirus was added to each well. The transduction was performed
at 37°C in a 5% CO2 incubator for 48 h. After 48 h, the
cells were selected with 2 µl puromycin and maintained at a
concentration of 500 µg/ml for 24 h. The medium was then changed,
and the transfection efficiency was determined by observing green
fluorescence. The efficiency of transfection was verified by
western blotting. shRNA sequences are listed in Table II.
 | Table II.shRNA sequences. |
Table II.
shRNA sequences.
| shRNA | Primer | Sequence |
|---|
| shLRP1 | Sense primer |
5′-GCAGTTTGCCTGCAGAGAT-3′ |
|
| Antisense
primer |
5′-ATCTCTGCAGGCAAACTGC-3′ |
| shNC | Sense primer |
5′-TTCTCCGAACGTGTCACGT-3′ |
|
| Antisense
primer |
5′-ACGTGACACGTTCGGAGAA-3′ |
Cytotoxicity and proliferation
assay
Cell proliferation was quantified using Cell
Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology)
according to manufacturer's instructions. CCK-8 reagent was applied
for 2 h at 37°C. The absorbance was measured at 450 nm with a
microplate reader (Model 680; Bio-Rad Laboratories, Inc.), and
values were calculated as an optical density index to compare
proliferation before and after LRP1 knockdown. EdU staining was
conducted using BeyoClick™ EdU Cell Proliferation kit with Alexa
Fluor 594 (cat. no. C0078S; Beyotime Institute of Biotechnology)
according to the manufacturer's instructions. Cells were observed
under a fluorescence microscope (IX71; Olympus Corporation).
Colony formation assay
Cells infected with LV-shLRP1 and LV-shNC were
transferred to a 6-well plate at a density of 1,000 cells/well.
shNC- and shLRP1-infected cells were seeded in a 6-well plate and
cultured at 37°C for 15 days. Colonies were fixed at room
temperature with glutaraldehyde (6.0% v/v) for 10 min, stained at
room temperature with crystal violet (0.5% w/v) for 15 min before
observation and images captured (Nikon D850 DSLR camera mounted on
a Nikon TS2R-FL fluorescence microscope; Nikon Corporation;
magnification, ×40.).
Transwell assay
A Transwell assay was performed using Transwell
membranes precoated with Matrigel (at 4°C, then at 37°C for 30
min), in accordance with the instructions of the Transwell assay
kit (cat. no. 4322; Corning, Inc.). For cell resuspension (10,000
cells), 200 µl serum-free medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) containing 5% BSA (cat. no. ST023; Beyotime
Institute of Biotechnology) was added to the upper chamber of
Transwell; 500 µl medium containing 20% FBS was added to the lower
chamber, and then cells were incubated at 37°C in a 5%
CO2 cell incubator for 12–48 h. The Transwell plate
culture medium was discarded, and the cells were fixed with 4%
paraformaldehyde for 15 min and stained with 1 ml crystal violet
for 10 min, both at room temperature. DAPI staining was also
performed at room temperature in the dark for 5 min. The cells were
observed under a fluorescence microscope (×200 magnification; Nikon
Corporation).
Wound healing assay
For the wound healing assay, HGC-27 cells, HepG2
cells and BxPC-3 cells (5×104) were cultured in a 6-well
plate until confluent (80%), then scratched with a 200-µl
micropipette tip. The medium was replaced with fresh serum-free
medium containing 10% FBS every 12 h. The wound healing area was
measured at 0 and 48 h at 37°C using a light microscope (×40
magnification; Nikon Corporation). The wound healing rate
(%)=[(average wound area at 0 h/average wound area at 48 h/average
wound area at 0 h] ×100, and the relative wound healing rate (%)
between shLRP1 group and shNC group was calculated with the shNC
group as the control.
Hematoxylin and eosin (H&E)
staining and immunohistochemistry
Paraffin sections (five sections each of gastric,
liver and pancreatic cancer, and adjacent normal tissues were used
as normal controls) were obtained from The Second Affiliated
Hospital of Zhejiang University School of Medicine (Hangzhou,
China). The paraffin section preparation process was as follows:
All tissues (gastric, liver and pancreatic cancer, and
cancer-adjacent tissues located 2 cm away from the lesion) were
fixed in a 4% paraformaldehyde solution at room temperature for 48
h. Subsequently, they were dehydrated in a series of alcohol
solutions with increasing concentrations at room temperature (50,
60, 70, 80 and 90% ethanol for 1 h each, 95% ethanol I for 1 h and
95% ethanol II for 1 h). After that, they were placed in a
xylene-anhydrous ethanol mixture (1:1) for 30 min, xylene I for 10
min and xylene II for 5 min for clearing. The dehydrated and
cleared tissues were then immersed in paraffin for 2 h at 60°C.
Once paraffin embedding was completed using an embedding machine,
the tissue blocks were sectioned into 2-µm slices using a microtome
and baked on a hot plate at 60°C for 24 h. During H&E staining,
the paraffin sections were subjected to deparaffinization and
hydration processes. The sections were sequentially immersed in two
xylene solutions for 10 min each at room temperature, followed by
immersion in 95, 85 and 70% ethanol for 5 min each to achieve full
hydration. After hydration, the sections were rinsed with running
water for 10 min. Subsequently, the sections were stained with
hematoxylin for 10 min at room temperature, followed by 3 min of
staining with 0.5% eosin at room temperature. After staining, the
sections were dehydrated by immersing in 80% ethanol for 5 sec, 95%
ethanol for 2 min and absolute ethanol for 2 min. The dehydrated
tissue sections were then immersed in xylene twice, for 4 min each.
Finally, the sections were air-dried and mounted with neutral
mounting medium. The mounted slides were observed under a light
microscope (×200 magnification; Nikon Corporation).
Immunohistochemical analysis was performed using
Histostain-streptavidin-peroxidase kit (cat. no. SP-0022; BIOSS)
according to the manufacturer's instructions. Immunohistochemistry
staining for the aforementioned paraffin sections was performed as
follows: The fixed, deparaffinized, sectioned and baked sections
were subjected to the same procedures as aforementioned. After
hydration, the sections were placed in 50 ml citrate antigen
retrieval solution (cat. no. P0081; Beyotime Institute of
Biotechnology) and boiled at 100°C for 10 min. After natural
cooling, the sections were washed twice with distilled water for 5
min each. Subsequently, each section was incubated with 200 µl
enhanced endogenous peroxidase blocking buffer (cat. no. P0100B;
Beyotime Institute of Biotechnology) at room temperature for 20
min. Next, 100 µl of the primary antibody was added to cover each
section, and the slides were incubated overnight at 4°C. After
washing three times with PBS for 5 min each, 100 µl HRP-labeled
secondary antibody was added, and the slides were incubated at room
temperature for 2 h. Finally, the sections were stained using the
DAB Horseradish Peroxidase Color Development Kit (cat. no. P0203;
Beyotime Institute of Biotechnology). The following primary
antibodies were used for immunohistochemistry: LRP1 (cat. no.
ab92544; 1:100; Abcam), CD36 (cat. no. ab252922; 1:100; Abcam) and
horseradish peroxidase-conjugated goat-anti-mouse secondary
antibody (cat. no. GAM007; 1:100; Multi Sciences Biotech Co,.
Ltd.). The slides were observed under a light microscope (BX41;
Olympus Corporation) at ×200 magnification.
Oil Red O staining
A total of 5×105 cells/well (HGC-27,
HepG2 and BxPC-3 cells transfected with shNC and shLRP1) were
inoculated in a 6-well plate. After 24 h of culture (at 37°C in a
5% CO2 humidified incubator), the culture medium (HGC-27
cells and BxPC-3 cells were cultured in RPMI-1640 medium, while
HepG2 cells were cultured in DMEM) was discarded. Following
fixation with 4% paraformaldehyde for 15 min, Oil Red O staining
was performed for 15–20 min, both at room temperature.
Decolorization (at room temperature for 5 min) with 60% isopropanol
was performed to remove excess dye. Slides were subsequently
observed under a light microscope (BX41; Olympus Corporation) at
×200 magnification.
Statistical analysis
Statistical analysis was performed using SPSS
(version 19.0; IBM Corp.), R (version 3.6.3; r-project.org/) and
GraphPad Prism (version 9.3.0; Dotmatics). Groups with multiple
tumor subtypes were compared using Wilcoxon rank-sum test. Survival
analysis was performed using the Kaplan-Meier method, and log-rank
test was used for comparing survival times. Cox proportional
hazards model was employed for univariate and multivariate analysis
to identify prognostic factors. Pearson correlation coefficient was
used to examine the correlation between LRP1 gene and gastric,
liver and pancreatic cancer. Data were normalized using the Z-score
standardization method. Wound healing and invasion assays were
analyzed using a two-tailed unpaired Student's t-test for two-group
comparisons. One-way ANOVA followed by Tukey's post hoc test was
used to assess the differences in mRNA and protein expression among
>2 groups. Data are presented as the mean ± standard deviation
of ≥3 independent experimental repeats. P<0.05 was considered to
indicate a statistically significant difference.
Results
High expression of LRP1 predicts poor
prognosis in GI tumors
The expression of LRP1 in tumor tissue was evaluated
using TCGA database and the GTEx project. LRP1 was expressed at
higher levels in tumor compared with the corresponding normal
tissues (Fig. 1A), including
adrenocortical, bladder urothelial, breast invasive and cervical
squamous cell carcinoma and endocervical adenocarcinoma,
cholangiocarcinoma, colon adenocarcinoma (COAD), lymphoid neoplasm
diffuse large B cell lymphoma, esophageal carcinoma, glioblastoma
multiforme, head and neck squamous cell carcinoma, kidney
chromophobe and renal clear and papillary cell carcinoma, acute
myeloid leukemia, brain lower grade glioma, liver hepatocellular
carcinoma (LIHC), lung adenocarcinoma and squamous cell carcinoma,
ovarian serous cystadenocarcinoma, pancreatic adenocarcinoma
(PAAD), pheochromocytoma and paraganglioma and prostate, rectum and
stomach adenocarcinoma (STAD). Furthermore, in TCGA database,
expression of LRP1 was higher in STAD and PAAD tissues compared
with that in adjacent normal tissue. Differential gene analysis of
the pancreatic cancer data in TCGA database showed that LRP1 was
significantly upregulated in PAAD [P<0.05; log fold-change
(FC)>5; Fig. 1B]. These results
suggested that LRP1 may play a key role in the pathogenesis of GI
tumors. To evaluate the effect of LRP1 expression in predicting the
prognosis of patients with GI cancer, the association between LRP1
expression and overall survival in STAD was analyzed (Fig. 1C). High LRP1 expression was
associated with poor prognosis in STAD (P=0.003). Cox's regression
test was used to analyze prognosis-associated genes in PAAD, STAD,
COAD and LIHC. The expression of the top 13 prognosis-associated
genes in STAD, COAD and LIHC (Fig.
1D) was positively correlated with LRP1 expression in
pancreatic cancer (specifically PAAD) (Fig. 1E).
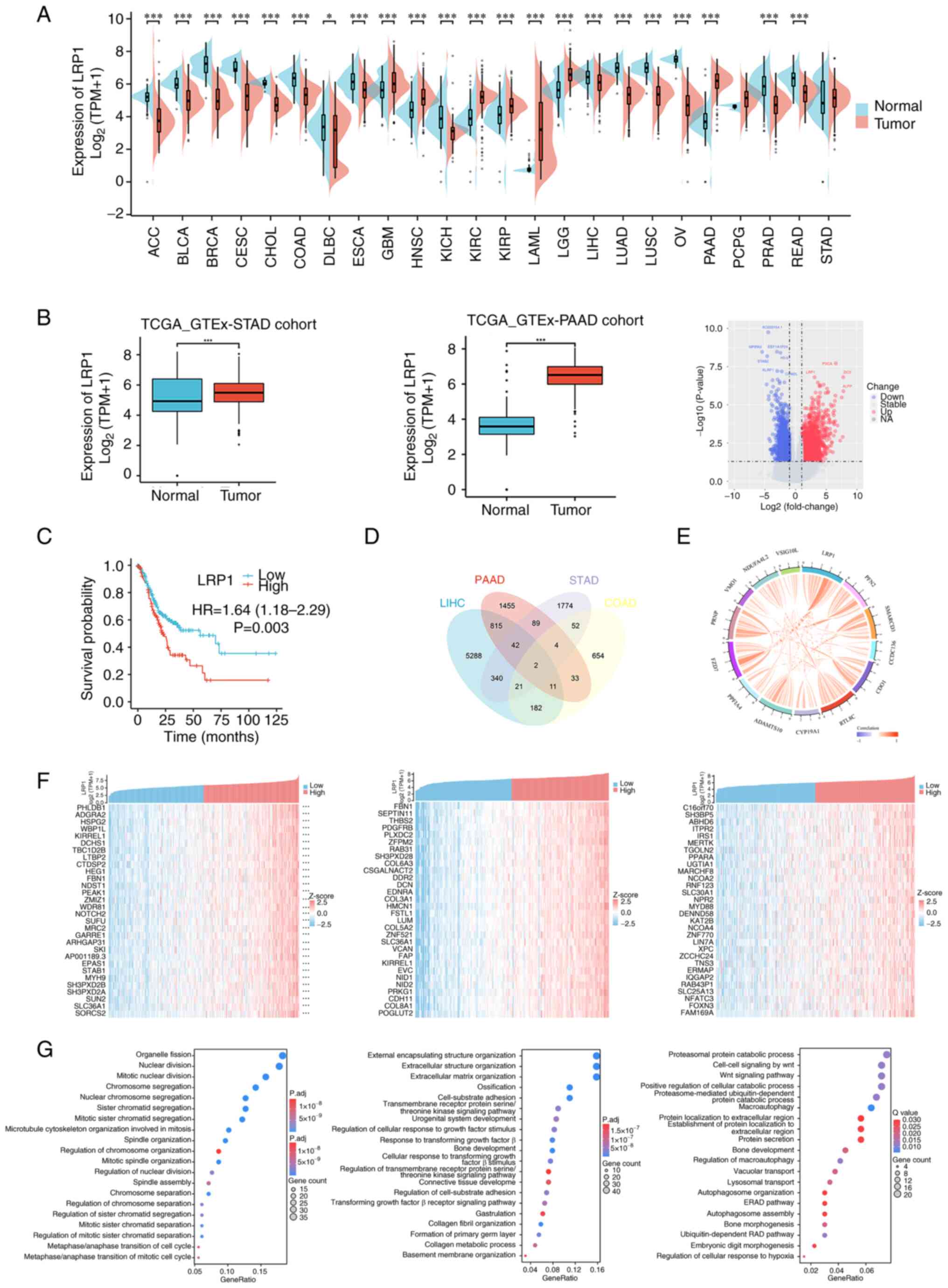 | Figure 1.High expression of LRP1 is associated
with poor prognosis in gastrointestinal tumor. (A) LRP1 expression
profile in various cancers and normal specimens. (B) LRP1
expression in tumor and normal tissues in STAD and PAAD from TCGA
database. (C) Correlation between LRP1 and prognosis of STAD. (D)
Prognosis-associated genes of STAD, PAAD, COAD and LIHC, and (E)
their correlation with LRP1. (F) Top 30 genes positively associated
with LRP1 shown in heatmap. (G) Significant Gene Ontology terms of
the top 300 genes positively associated with LRP1, including cell
proliferation pathway in STAD (left image), extracellular matrix
formation and regulation of transmembrane receptor proteins
pathways in PAAD (middle image), extracellular matrix formation and
regulatory pathways of transmembrane receptor proteins in LIHC
(right image). The statistical method used for GOKEGG enrichment
analysis is Fisher's exact test, with a P-value <0.05 and
FDR=0.05. *P<0.05, ***P<0.001 vs. normal. LRP1, low-density
lipoprotein receptor-related protein 1; STAD, stomach
adenocarcinoma; PAAD, pancreatic adenocarcinoma; TCGA, The Cancer
Genome Atlas; GTEx, Genotype-Tissue Expression; HR, hazard ratio;
TPM, transcripts per million; NA, not available; ACC,
adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA,
breast invasive carcinoma; CESC, cervical and endocervical cancers;
CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse
large B-cell lymphoma; ESCA, esophageal carcinoma; GBM,
glioblastoma multiforme; HNSC, head and neck squamous cell
carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell
carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute
myeloid leukemia; LGG, lower grade glioma; LIHC, liver
hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung
squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma;
PCPG, pheochromocytoma and paraganglioma; PRAD, prostate
adenocarcinoma; READ, rectum adenocarcinoma. |
To explore the function and pathways of LRP1 in GI
tumors, correlation analysis between LRP1 and other mRNAs in STAD,
PAAD and LIHC was performed using TCGA. The top 30 genes that were
positively correlated with LRP1 expression were visualized as a
heatmap (Fig. 1F). R software
package ‘clusterProfiler’ was used to explore the potential
functions and pathways of the top 300 correlated genes. GO
functional enrichment analysis showed that in STAD, LRP1 was
primarily associated with pathways associated with cell
proliferation, including ‘mitotic nuclear division’ and
‘microtubule cytoskeleton organization involved in mitosis’. In
PAAD, LRP1 was primarily associated with pathways such as
‘extracellular matrix organization’ and ‘transmembrane receptor
protein serine/threonine kinase signaling pathway’. In LIHC, LRP1
was primarily associated with pathways associated with protein
metabolism (‘proteasomal protein catabolic process’), ‘Wnt
signaling pathway’ and autophagy (‘autophagosome organization’ and
‘autophagosome assembly’) (Fig.
1G). These results suggested that high expression of LRP1 in
PAAD, STAD and LIHC may be a result of upregulation of multiple
pathways associated with cancer formation in GI tumors,
particularly those that control cell proliferation and
metastasis.
Differential and Gene Set Enrichment
Analysis (GSEA) of LRP1
Differential expression and correlation analysis was
performed to analyze the potential effects of LRP1 upregulation in
GI tumors. R package ‘DESeq2’ was used to analyze the differential
expression of LRP1 in STAD, PAAD and LIHC, and GSEA was performed
on the top 30 genes with log(FC)>0. Differentially expressed
genes in all the three cancer types were significantly enriched in
pathways that promote cancer occurrence and development, such as
MET and ERK, and other pathways (Fig.
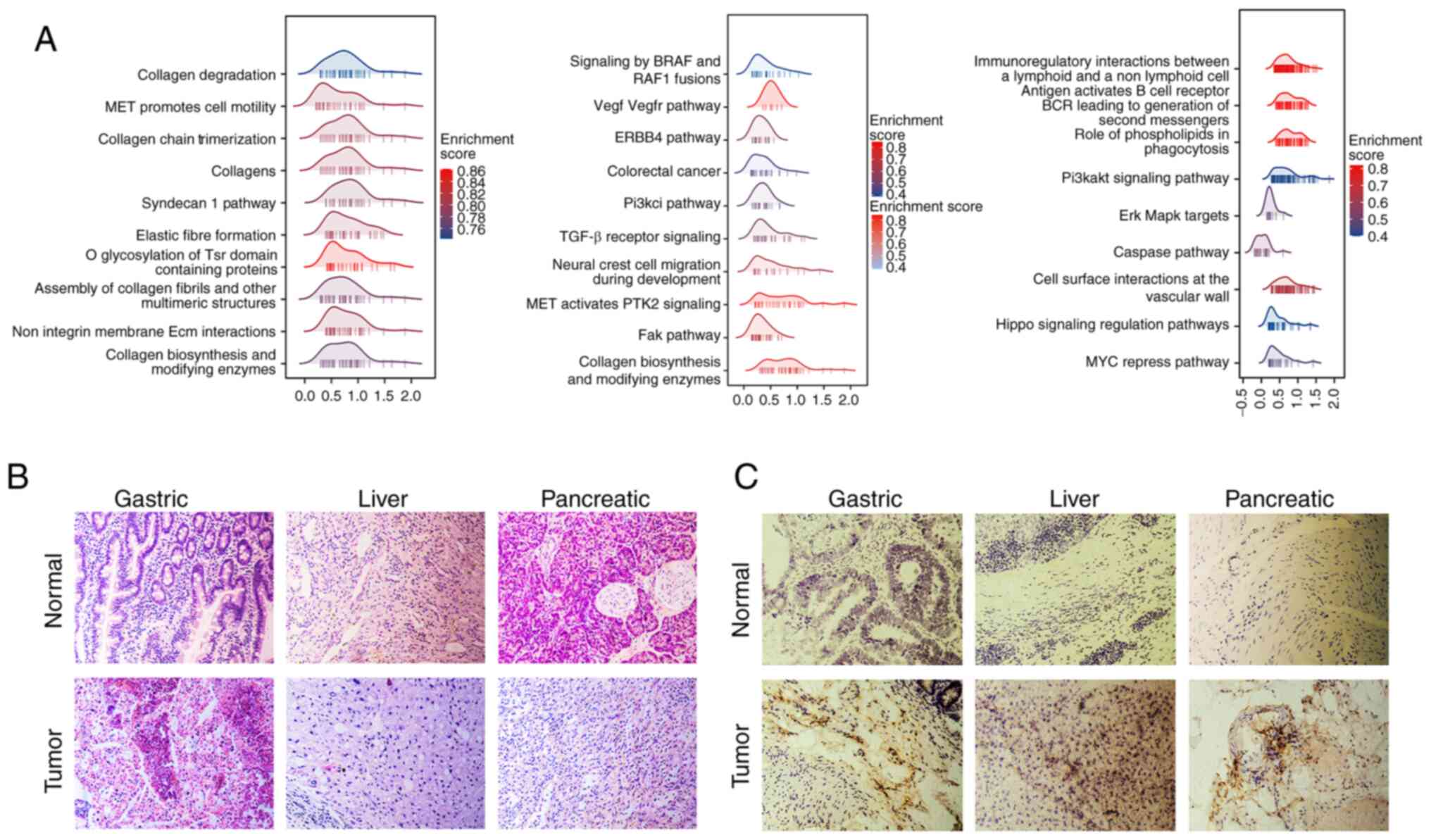
2A; ‘collagen chain trimerization’, ‘assembly of collagen
fibrils and other multimeric structures’ and ‘fak pathway’) that
regulate collagen production and degradation (Fig. 2A).
H&E staining showed the difference between
normal and tumor tissues. Compared with the normal tissues, the
tissue samples from gastric, pancreatic and liver cancer exhibited
structural disorganization, disarrayed cell arrangements, increased
cell density and thickening of the extracellular matrix (Fig. 2B). Immunohistochemical experiments
were conducted to compare the differences in LRP1 expression
between normal and tumor GI tissue. Immunohistochemistry showed
that expression of LRP1 was upregulated in GI cancer compared with
that in normal tissues (Fig. 2B and
C).
LRP1 is expressed at high levels in
HGC-27, HepG2 and BxPC-3 cells
The expression of LRP1 in gastric HGC-27, liver
HepG2 cells and pancreatic BxPC-3 cancer cells was notably
increased compared with that in HUVECs (Fig. 3A). Protein expression of LRP1 was
significantly higher in BxPC-3 compared with in HUVECs (P<0.05).
Protein expression of LRP1 was also significantly higher in HGC-27
and HepG2 cells compared with that in control HUVECs (P<0.01;
Fig. 3B). The mRNA expression of
LRP1 in HGC-27, HepG2 and BxPC-3 cells was significantly higher
than that in HUVECs (P<0.01; Fig.
3C). To investigate the effect of LRP1 on GI cancer cells, LRP1
was knocked down via transfection of HGC-27, HepG2 and BxPC-3 cells
with LV-shLPR1 (Fig. S1B). Western
blotting showed that LRP1 protein expression levels in cells in
which LRP1 was knocked down were decreased compared with those in
cells transfected with shNC (Fig.
3D). Quantitative analysis is shown in Fig. S1A. Transfection efficiency was ≥80%
for shNC and shLRP1 in HGC-27, HepG2 and BxPC-3 cells (Fig. S1B). Based on the findings from the
bioinformatics analysis and experimental results (Fig. 3A), it was observed that LRP1
exhibits low expression levels in both normal tissues and cells.
Consequently, it was hypothesized that attempting to further
suppress LRP1 in cells that already possess a baseline low
expression would not result in significant effects.
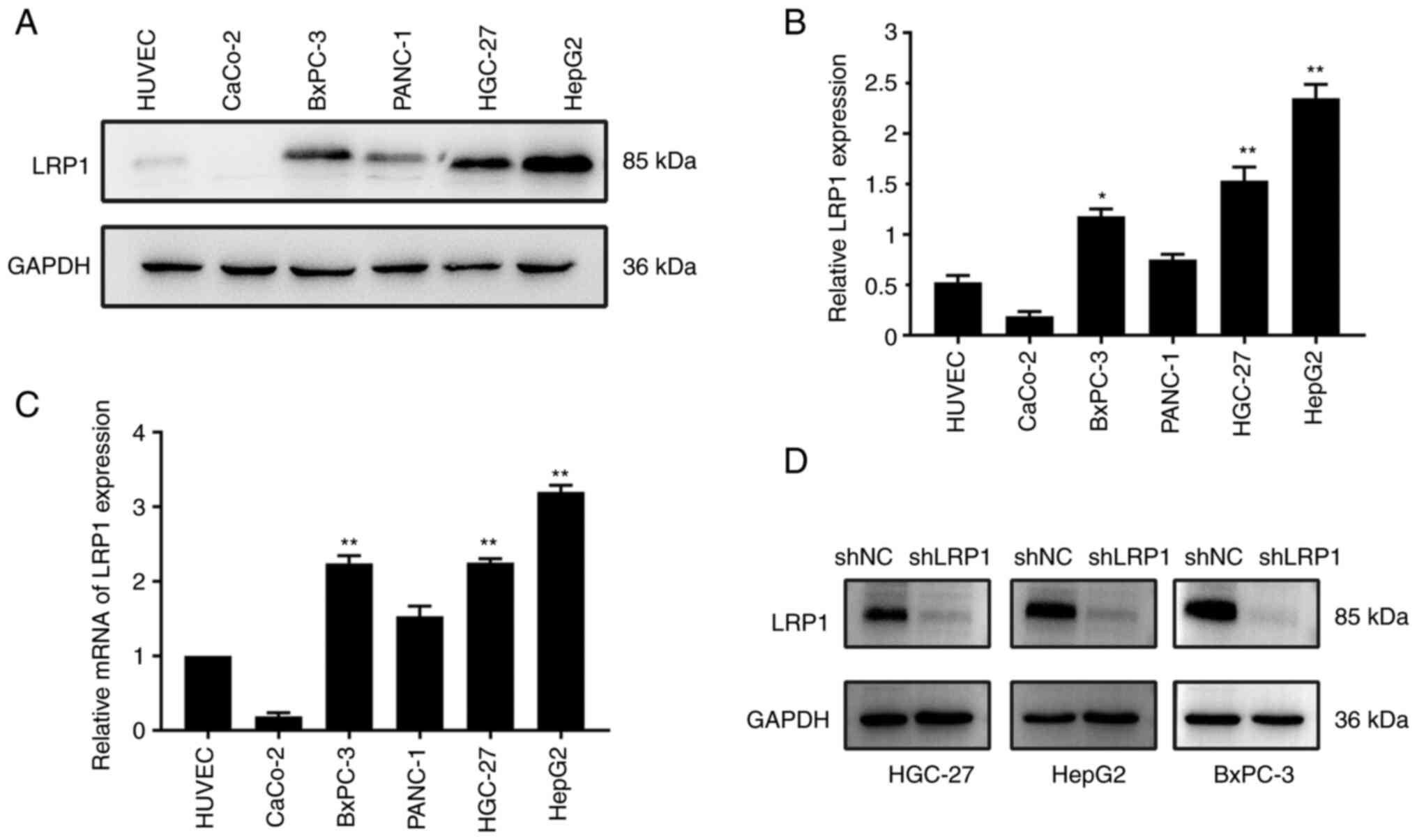 | Figure 3.LRP1 is expressed at high levels in
HGC-27, HepG2 and BxPC-3 cells. (A) Expression of LRP1 and loading
control GAPDH in HUVEC, CaCo-2, BxPC-3, PANC-1, HGC-27 and HepG2
cells was investigated by western blotting. (B) Relative expression
of LRP1 in HUVEC, CaCo-2, BxPC-3, PANC-1, HGC-27 and HepG2 cells.
(C) Expression of LRP1 gene in HUVEC, CaCo-2, BxPC-3, PANC-1,
HGC-27 and HepG2 cells was investigated by quantitative PCR. (D)
Expression of LRP1 and loading control GAPDH in HGC-27, HepG2 and
BxPC-3 cells before and after lentiviral transfection was
investigated by western blotting. *P<0.05, **P<0.01 vs.
HUVEC. LRP1, low-density lipoprotein receptor-related protein 1;
shNC, short hairpin negative control. Unpaired Student's t-test was
used for analysis. |
Decreased LRP1 expression can inhibit
the proliferation of GI tumor cells
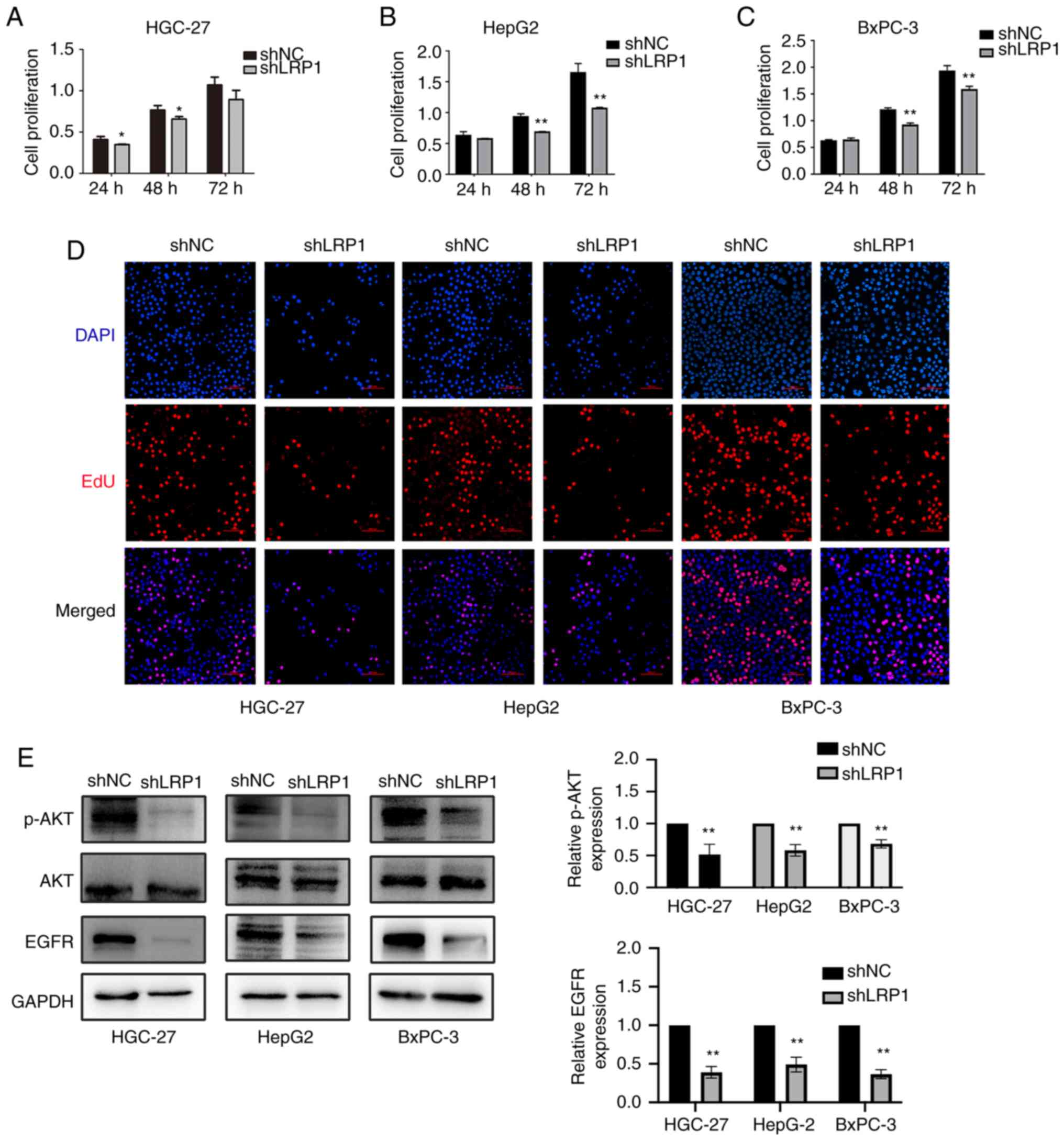
To investigate the effect of LRP1 knockdown on
proliferation of GI cancer cells, CCK-8 and EdU cell proliferation
assays were performed on HGC-27, HepG2 and BxPC-3 cells following
transfection. CCK-8 assay showed that cell proliferation was
decreased after transfection with LV-shLRP1 (Fig. 4A-C). EdU proliferation assay showed
that the proliferation of HGC-27, HepG2 and BxPC-3 cells was
inhibited and the number of dividing cells decreased after LRP1
knockdown (Fig. 4D).
Western blotting showed that expression of p-AKT and
EGFR proteins in HGC-27, HepG2 and BxPC-3 cells was inhibited
following knockdown of LRP1 (Fig.
4E). The aforementioned data showed that the proliferation of
HGC-27, HepG2and BxPC-3 cells was inhibited after LRP1
knockdown.
Decreased LRP1 expression can inhibit
the invasion and migration of GI cancer cells
To investigate whether LRP1 is related to the
invasion and migration of GI cancer cells, Transwell and wound
healing assays were performed; invasion and migration ability of
HGC-27, HepG2 and BxPC-3 cells decreased following LRP1 knockdown.
Compared with cells transfected with shNC, invasion of HGC-27,
HepG2 and BxPC-3 cells was reduced after LRP1 knockdown (Fig. 5A). Wound healing assay confirmed
that the migration of GI tumor cells was decreased following LRP1
knockdown (Fig. 5B). Based on
preliminary investigations, it was determined that 48 h
post-transfection gave the optimal efficiency of lentiviral
transduction. Compared with cells transfected with shNC, colony
formation assay demonstrated that LRP1 knockdown effectively
suppressed cell proliferation (Fig.
5C). Western blotting showed that protein expression of p-ERK
and MMP-9 in HGC-27, HepG2 and BxPC-3 cells was significantly
downregulated following LRP1 knockdown. Expression of ERK and MMP-2
proteins in BxPC-3 cells decreased significantly (Fig. 5D).
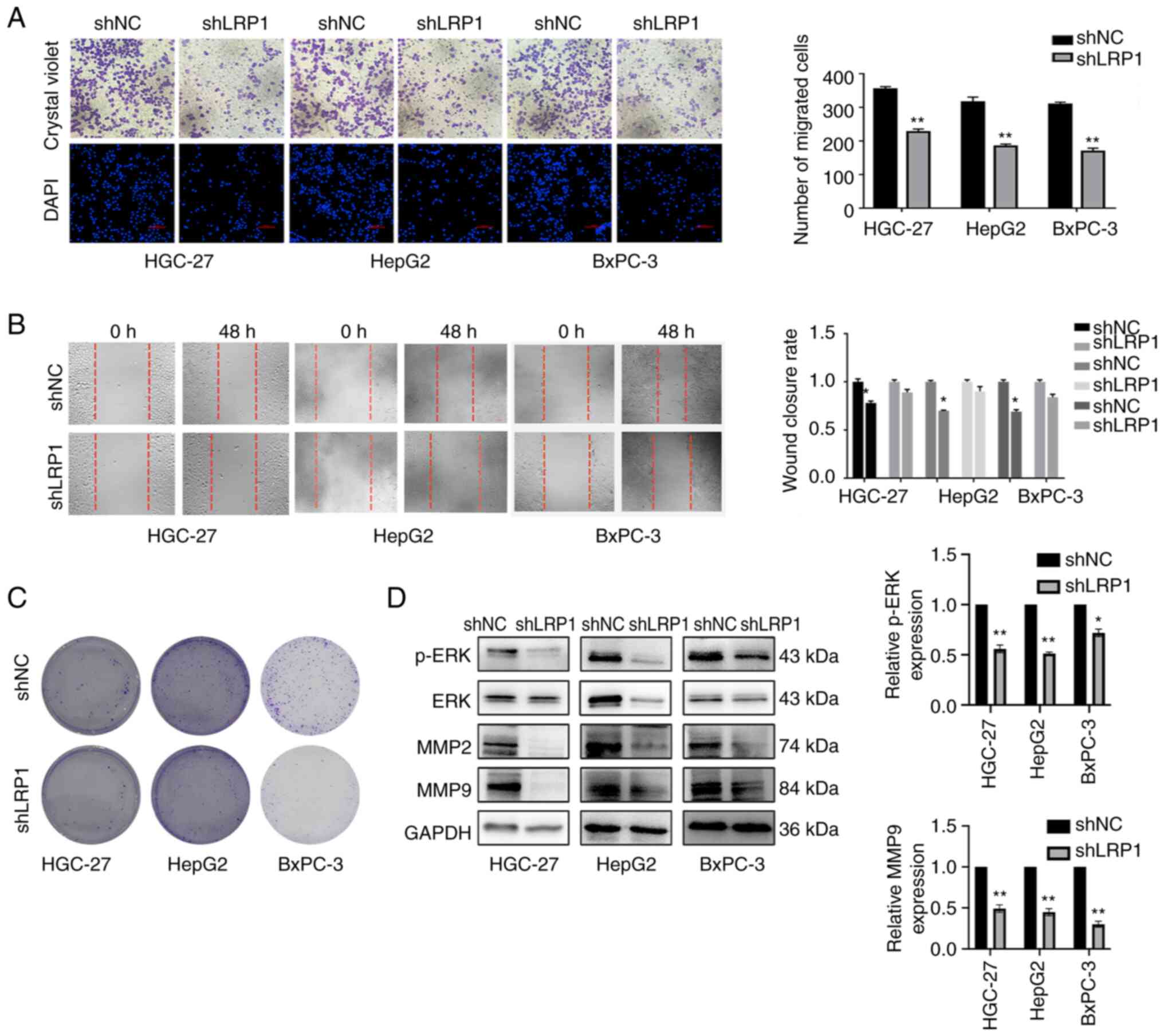 | Figure 5.Decreased LRP1 expression inhibits
invasion and migration of gastrointestinal tumor cells. (A)
Transwell assay of HGC-27 cells, HepG2 cells and BxPC-3 cells
before and after lentivirus transfection. Magnification, ×200. (B)
Wound healing assay of HGC-27, HepG2 and BxPC-3 cells before and
after lentivirus transfection. Magnification, ×100. (C) Colony
formation before and after lentivirus transfection of HGC-27 cells,
HepG2 cells and BxPC-3 cells. Magnification, ×40. (D) Expression of
p-ERK, ERK, MMP-2, MMP-9 and loading control GAPDH in HGC-27, HepG2
and BxPC-3 cells before and after lentivirus transfection was
investigated by western blotting. *P<0.05 and **P<0.01 vs.
shNC. p, phosphorylated; LRP1, low-density lipoprotein
receptor-related protein 1; shNC, short hairpin negative control.
Unpaired Student's t-test was used for analysis. |
Decreased LRP1 expression can inhibit
lipid absorption
Lipid metabolism is mainly accomplished via the
digestive system (18). To explore
the impact of LRP1 knockdown on lipid metabolism in GI tumor cells,
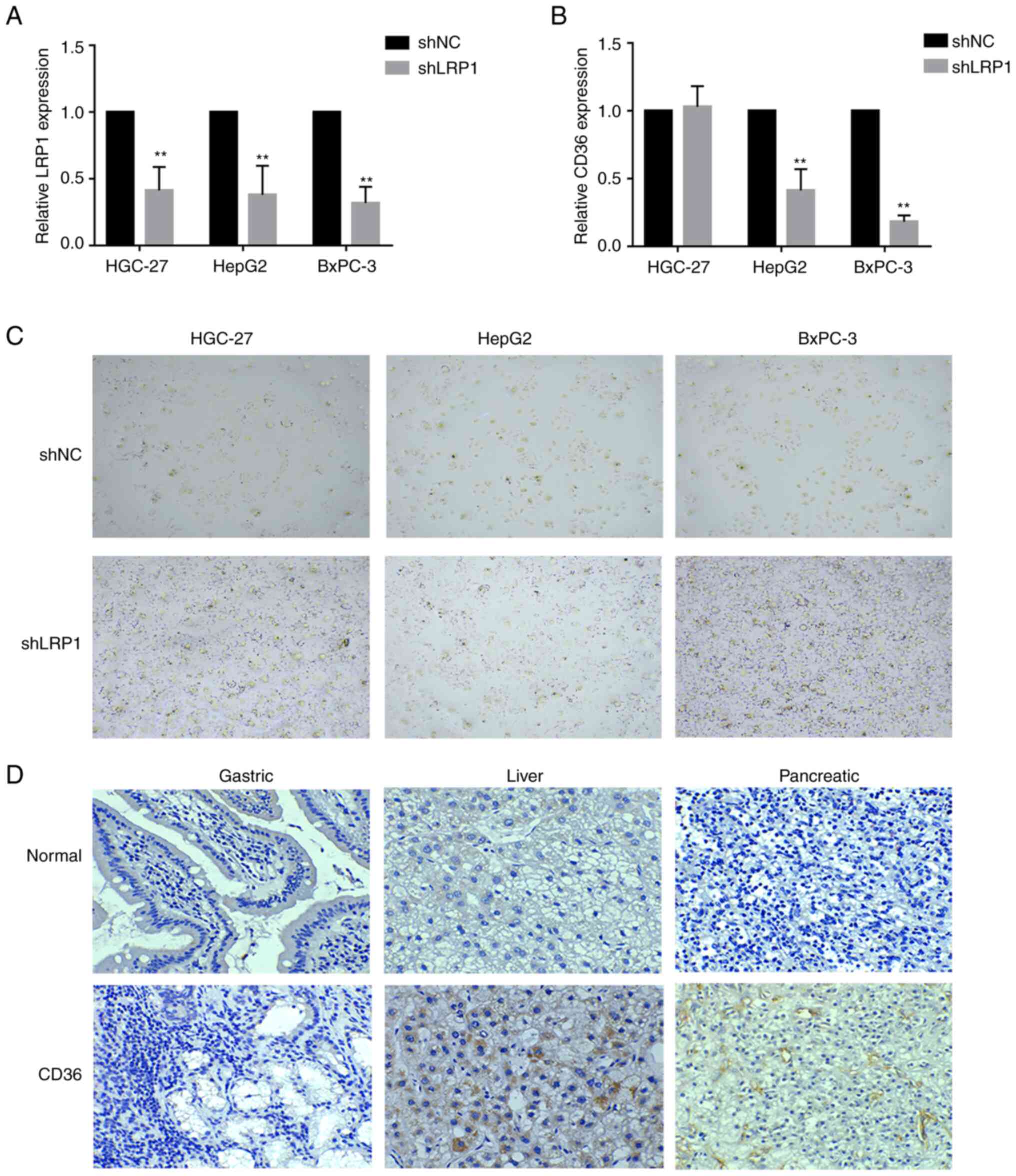
the expression of related genes (LRP1 and CD36) was detected via
qPCR. LRP1 gene expression was significantly downregulated in
HGC-27, HepG2 and BxPC-3 cells following LRP1 knockdown compared
with that in cells transfected with shNC (Fig. 6A). CD36 gene expression in HepG2 and
BxPC-3 cells was significantly downregulated after LRP1 knockdown
compared with that in cells transfected with shNC (P<0.01;
Fig. 6B). Oil Red O staining showed
that lipids accumulated in GI tumor cells after LRP1 knockdown
(Fig. 6C). CD36 is a scavenger
receptor responsible for the uptake/transport of lipids and
oxidized LDL (19). CD36 was
upregulated in liver and pancreatic cancer tissues compared with
that in normal GI tissue (Fig. 6D).
These results suggested that LRP1 knockdown inhibited lipid
absorption of GI tumors.
Discussion
In recent years, the incidence of GI cancers has
increased worldwide. The stage at diagnosis of GI cancer is closely
associated with the survival rate; early stage detection could
reduce the mortality rate. However, there are no specific symptoms
characterizing early stage of GI cancers; due to its insidious
onset and deep anatomical location, diagnosis of early-stage GI
cancer is challenging (20).
Although biomarkers such as CA19-9 (21) and CA-125 (22) are available, the association between
these indicators and tumor metastasis, invasion or prognosis
remains unclear. Therefore, it is important to find an effective
therapeutic target and novel biomarkers. Based on bioinformatics
analysis, high expression of LRP1 was associated with poor
prognosis in GI tumor in the present study. It was found that LRP1
was expressed at high levels in GI cancer cell lines HGC-27, HepG2
and BxPC-3, and the knockdown of LRP1 changed the biological
characteristics (cell proliferation, invasion, migration and
viability) of the GI tumor cells. The knockdown of LRP1 could
inhibit the proliferation of GI tumor cells. EdU cell proliferation
experiments showed that compared with the control (shNC) group,
proliferation of gastric (HGC-27), liver (HepG2) and pancreatic
(BxPC-3) cancer cells was suppressed following LRP1 knockdown. In
addition, cell population dependence increased after LRP1
knockdown, which inhibited the formation of cell clones. To
elucidate the mechanism by which LRP1 knockdown could inhibit the
proliferation of GI tumor cells, AKT and EGFR protein expression
was measured using western blotting. LRP1 knockdown inhibited
phosphorylation of AKT and the expression of EGFR protein (Fig. 3E). AKT is a key survival signal
transduction protein and it is a downstream target of EGFR
(23). MMP2 and MMP9 are proteases
associated with tumor invasion and migration (24). LRP1-mediated regulation of MMP
expression promotes cancer cell migration and invasion (25). LRP1 serves as an endocytic receptor
for MMP2 and MMP9, thus regulating tumor invasion and migration
(25). A previous study (1) demonstrated that LRP1 induces protein
expression of MMP2 and MMP9, thereby promoting migration and
invasion of human glioblastoma U87 cells. In the present study,
expression of MMP2 and MMP9 was selectively decreased in cells
following LRP1 knockdown. p-ERK in LRP1-knockdown cells was also
significantly decreased, which suggested that LRP1 regulates MMP2
and MMP9 via the ERK signaling pathway (26). In the present study, the expression
of p-ERK, MMP2 and MMP9 was reduced. Transwell and wound healing
assay showed that the invasion and migration of HGC-27, HepG2 and
BxPC-3 cells were decreased compared with those in the control
group. The ERK signaling pathway transmits extracellular stimuli to
the nucleus and regulates tumorigenesis, proliferation, apoptosis
and drug resistance (27,28). Decreased LRP1 expression inhibited
the proliferation, invasion and migration of GI tumor cells. Tumor
cells have a strong metabolism to grow rapidly, especially the
digestive tract cells which are involved in digestion and
absorption of nutrients. Compared with normal cells, tumor cells
possess enhanced metabolic capacity and exhibit accelerated growth.
Therefore, we hypothesize that following the transformation of
digestive tract cells involved in nutrient digestion and absorption
to provide nutrients to cells, the metabolic and growth
capabilities become more prominent. Compared with normal cells,
cancer cells obtain more energy via lipid metabolism to promote
cell proliferation, invasion and migration, which accounts for high
expression of LRP1 in GI tumor cells (29). Moreover, inactivation of LRP1 in
adipocytes could lead to delayed lipid clearance after a meal,
change in adipocyte tissue metabolism, glucose tolerance and
obesity resistance induced by a high-fat diet (30). In mouse intestinal polyps, the
expression of LRP1 is ~3-fold higher than that in normal tissues
(31). LRP1 is expressed at high
levels in both human lung adenocarcinoma A549 and colorectal cancer
cells (32). Another study found
that lipid-associated metabolic pathways are activated at high
levels in pancreatic cancer; LRP1 expression in pancreatic cancer
is double that in normal pancreatic tissue (33). Blocking LRP1-mediated cholesterol
endocytosis via LRP1 gene knockdown disrupts the homeostasis of
cholesterol inside and outside the cell, affecting the
proliferation and tumorigenicity of pancreatic cancer cells, and
inhibits ERK-dependent survival pathways (34). Therefore, the expression of LRP1 may
be associated with mediating the uptake of cholesterol in
pancreatic cancer cells, consequently exerting further influence on
the growth of tumor cells. Rohlmann et al (35) showed that LDLR partially compensates
for the loss of LRP1 in hepatocytes and increased expression of
LDLR indicates that LRP1 plays an important role in elimination of
residual lipoproteins in the liver. In another study, plasma
triglycerides in LRP1-deficient mice receiving normal diet
increases by 2-fold. Under a high-fat diet, morphological
examination and Oil Red O staining of liver sections show notable
accumulation of lipid droplets in LRP1-deficient mice (36). In the present study, BxPC-3 cells
had the most significant decrease in CD36 gene expression following
LRP1 knockdown and Oil Red O staining showed that decreased LRP1
expression increased lipid accumulation in BxPC-3 cells. A previous
study showed that relative expression levels of CD36 are important
for lipid absorption in mammals (37). Thus, it was hypothesized that LRP1
affects cholesterol absorption in tumor cells by regulating
expression of CD36. Here, LRP1 was expressed at high levels in
BxPC-3, HGC-27 and HepG2 cells and high expression of LRP1 was
associated with poor prognosis in GI tumors and increased
proliferation of GI tumor cells. Knocking down expression of LRP1
may interfere with tumor cell lipid metabolism, resulting decreased
EGFR, p-AKT, p-ERK proteins and membrane molecule CD36, MMP2 and
MMP9 expression, thereby inhibiting metastasis and invasion of GI
tumors. The present study demonstrated the regulatory role of LRP1
in invasion and prognosis of GI cancer and also suggested that LRP1
may be a novel target for the treatment of GI cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science
Foundation of Zhejiang Province (grant nos. LY19H060001,
LQ19H160044 and LGF21H160016).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MZ, HS, BW conceived and designed the study. HS and
BW analyzed the data. MZ drafted the manuscript. YH, JC, JR, ZZ and
XJ participated in the statistical analysis and had input in the
experimental design. YH, JC, JR, ZZ and XJ confirm the authenticity
of all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by Institutional
Animal Care and Use Committee (approval no. IACUC-20190429-09) and
Medical Ethics Committee of Zhejiang Chinese Medical University
(approval no. 20221011-5).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GI
|
gastrointestinal
|
|
LRP1
|
low density lipoprotein
receptor-related protein 1
|
|
TCGA
|
The Cancer Genome Atlas
|
|
CCK-8
|
Cell Counting Kit-8
|
|
CA19-9
|
carbohydrate antigen 19-9
|
|
LDLR
|
low-density lipoprotein receptor
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
GO
|
Gene Ontology
|
|
COAD
|
colon adenocarcinoma
|
|
LIHC
|
liver hepatocellular carcinoma
|
|
PAAD
|
pancreatic adenocarcinoma
|
|
STAD
|
stomach adenocarcinoma
|
|
LV
|
lentiviral
|
|
shNC
|
short hairpin negative control
|
|
p
|
phosphorylated
|
|
EGFR
|
epidermal growth factor receptor
|
|
MMPs
|
matrix metalloproteinase
|
References
|
1
|
Song H, Li Y, Lee J, Schwartz AL and Bu G:
Low-density lipoprotein receptor-related protein 1 promotes cancer
cell migration and invasion by inducing the expression of matrix
metalloproteinases 2 and 9. Cancer Res. 69:879–886. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cheng C, Geng F, Cheng X and Guo D: Lipid
metabolism reprogramming and its potential targets in cancer.
Cancer Commun (Lond). 38:272018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arnold M, Abnet CC, Neale RE, Vignat J,
Giovannucci EL, McGlynn KA and Bray F: Global burden of 5 major
types of gastrointestinal cancer. Gastroenterology. 159:335–349.
e152020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Malvicini M, Aquino JB and Mazzolini G:
Combined therapy for gastrointestinal carcinomas: Exploiting
synergies between gene therapy and classical chemo-radiotherapy.
Curr Gene Ther. 15:151–160. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu Q, Zong L, Chen X, Jiang Z, Nan L, Li
J, Duan W, Lei J, Zhang L, Ma J, et al: Resveratrol in the
treatment of pancreatic cancer. Ann N Y Acad Sci. 1348:10–19. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feng F, Tian Y, Xu G, Liu Z, Liu S, Zheng
G, Guo M, Lian X, Fan D and Zhang H: Diagnostic and prognostic
value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC
Cancer. 17:7372017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang L, Sanagapalli S and Stoita A:
Challenges in diagnosis of pancreatic cancer. World J
Gastroenterol. 24:2047–2060. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang B, Shen C, Li Y, Zhang T, Huang H,
Ren J, Hu Z, Xu J and Xu B: Oridonin overcomes the gemcitabine
resistant PANC-1/Gem cells by regulating GST pi and LRP/1 ERK/JNK
signalling. Onco Targets Ther. 12:5751–5765. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gheysarzadeh A, Ansari A, Emami MH, Razavi
AE and Mofid MR: Over-expression of low-density lipoprotein
receptor-related Protein-1 is associated with poor prognosis and
invasion in pancreatic ductal adenocarcinoma. Pancreatology.
19:429–435. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guillaumond F, Bidaut G, Ouaissi M,
Servais S, Gouirand V, Olivares O, Lac S, Borge L, Roques J, Gayet
O, et al: Cholesterol uptake disruption, in association with
chemotherapy, is a promising combined metabolic therapy for
pancreatic adenocarcinoma. Proc Natl Acad Sci USA. 112:2473–2478.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gopal U, Bohonowych JE, Lema-Tome C, Liu
A, Garrett-Mayer E, Wang B and Isaacs JS: A novel extracellular
Hsp90 mediated co-receptor function for LRP1 regulates EphA2
dependent glioblastoma cell invasion. PLoS One. 6:e176492011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang XY, Shi GM, Devbhandari RP, Ke AW,
Wang Y, Wang XY, Wang Z, Shi YH, Xiao YS, Ding ZB, et al: Low level
of low-density lipoprotein receptor-related protein 1 predicts an
unfavorable prognosis of hepatocellular carcinoma after curative
resection. PLoS One. 7:e327752012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vivian J, Rao AA, Nothaft FA, Ketchum C,
Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD,
Musselman-Brown A, et al: Toil enables reproducible, open source,
big biomedical data analyses. Nat Biotechnol. 35:314–316. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Subramanian A, Tanayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mootha VK, Lindgren CM, Erlksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Font-Burgada J, Sun B and Karin M: Obesity
and cancer: The oil that feeds the flame. Cell Metab. 23:48–62.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J and Li Y: CD36 tango in cancer:
Signaling pathways and functions. Theranostics. 9:4893–4908. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Necula L, Matei L, Dragu D, Neagu AI,
Mambet C, Nedeianu S, Bleotu C, Diaconu CC and Chivu-Economescu M:
Recent advances in gastric cancer early diagnosis. World J
Gastroenterol. 25:2029–2044. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei X, Li YB, Li Y, Lin BC, Shen XM, Cui
RL, Gu YJ, Gao M, Li YG and Zhang S: Prediction of lymph node
metastases in gastric cancer by serum APE1 expression. J Cancer.
8:1492–1497. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zang R, Li Y, Jin R, Wang X, Lei Y, Che Y,
Lu Z, Mao S, Huang J, Liu C, et al: Enhancement of diagnostic
performance in lung cancers by combining CEA and CA125 with
autoantibodies detection. Oncoimmunology. 8:e16256892019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tiemin P, Fanzheng M, Peng X, Jihua H,
Ruipeng S, Yaliang L, Yan W, Junlin X, Qingfu L, Zhefeng H, et al:
MUC13 promotes intrahepatic cholangiocarcinoma progression via
EGFR/PI3K/AKT pathways. J Hepatol. 72:761–773. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Komatsu K, Nakanishi Y, Nemoto N, Hori T,
Sawada T and Kobayashi M: Expression and quantitative analysis of
matrix metalloproteinase-2 and −9 in human gliomas. Brain Tumor
Pathol. 21:105–112. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xing P, Liao Z, Ren Z, Zhao J, Song F,
Wang G, Chen K and Yang J: Roles of low-density lipoprotein
receptor-related protein 1 in tumors. Chin J Cancer. 35:62016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu K, Yang J, Tanaka S, Gonias SL, Mars WM
and Liu Y: Tissue-type plasminogen activator acts as a cytokine
that triggers intracellular signal transduction and induces matrix
metalloproteinase-9 gene expression. J Biol Chem. 281:2120–2127.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shen H, Xu W, Luo W, Zhou L, Yong W, Chen
F, Wu C, Chen Q and Han X: Upregulation of mdr1 gene is related to
activation of the MAPK/ERK signal transduction pathway and YB-1
nuclear translocation in B-cell lymphoma. Exp Hematol. 39:558–569.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng X and Zhang S: MAPK cascades in plant
disease resistance signaling. Annu Rev Phytopathol. 51:245–266.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bian X, Liu R, Meng Y, Xing D, Xu D and Lu
Z: Lipid metabolism and cancer. J Exp Med. 218:e202016062021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yancey PG, Blakemore J, Ding L, Fan D,
Overton CD, Zhang Y, Linton MF and Fazio S: Macrophage LRP-1
controls plaque cellularity by regulating efferocytosis and Akt
activation. Arterioscler Thromb Vasc Biol. 30:787–795. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mutoh M, Komiya M, Teraoka N, Ueno T,
Takahashi M, Kitahashi T, Sugimura T and Wakabayashi K:
Overexpression of low-density lipoprotein receptor and lipid
accumulation in intestinal polyps in Min mice. Int J Cancer.
125:2505–2510. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gueddari N, Favre G, Hachem H, Marek E,
Gaillard FL and Soula G: Evidence for up-regulated low density
lipoprotein receptor in human lung adenocarcinoma cell line A549.
Biochimie. 75:811–819. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vasseur S and Guillaumond F: LDL Receptor:
An open route to feed pancreatic tumor cells. Mol Cell Oncol.
3:e10335862016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miyabayashi K, Ijichi H, Mohri D, Tada M,
Yamamoto K, Asaoka Y, Ikenoue T, Tateishi K, Nakai Y, Isayama H, et
al: Erlotinib prolongs survival in pancreatic cancer by blocking
gemcitabine-induced MAPK signals. Cancer Res. 73:2221–2234. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rohlmann A, Gotthardt M, Hammer RE and
Herz J: Inducible inactivation of hepatic LRP gene by cre-mediated
recombination confirms role of LRP in clearance of chylomicron
remnants. J Clin Invest. 101:689–695. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ding Y, Xian X, Holland WL, Tsai S and
Herz J: Low-density lipoprotein receptor-related protein-1 protects
against hepatic insulin resistance and hepatic steatosis.
EBioMedicine. 7:135–145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Petersen C, Bell R, Klag KA, Lee SH, Soto
R, Ghazaryan A, Buhrke K, Ekiz HA, Ost KS, Boudina S, et al: T
cell-mediated regulation of the microbiota protects against
obesity. Science. 365:eaat93512019. View Article : Google Scholar : PubMed/NCBI
|