Introduction
Sentinel lymph node (SN) biopsy, commonly known for
diagnosing lymph node metastasis among node-negative patients with
breast cancer clinically (1–3). Rapid
pathological examination and one-step nucleic acid amplification
(OSNA) have been utilized for intraoperatively diagnosing SN
metastasis in practice (4,5). Cytokeratin 19 (CK19) is expressed in
breast cancer cells whereas normal lymph node (LN) cells do not
express it. OSNA measures the amplification of CK19 mRNA in SN
cells to evaluate the presence of SN metastasis with an accuracy
similar to that of histopathological examination (4,5). OSNA
can potentially quantify the total tumor load (TTL) in SNs as the
summation of CK19 mRNA copies, reported to be significant for
forecasting non-SN metastatic state (6–8) and
patient prognosis (9). Though, TTL
determination using OSNA does not sensitively project the total
number of tumor cells in the SN because the copy number of CK19
mRNAs for every tumor cell differs substantially. Actually, a
30-times variance in CK19 mRNA copies amongst tumors of the same
size has been reported (4).
The amount of DNA per tumor cell is considered less
variable than mRNA; hence the identification of total SN tumor
cells from tumor-derived DNA can more accurately ascertain the
extent of LN metastasis. Promoter methylation of Ras association
domain-containing protein 1 (RASSF1A) is an epigenomic change
frequently observed in breast cancer (10,11).
We have recently first developed an assay for detecting
RASSF1A promoter methylation following restriction
enzyme-based digital methylation-specific polymerase chain reaction
(RE-dMSP). RE-dMSP is adequately sensitive to detect ≥3 copies of
methylated RASSF1A per assay. In our previous study, 161 SN
lysates from 71 patients were analyzed using RE-dMSP and showed
high concordance of 95% with the results of OSNA (12). The study also demonstrated that the
variation in methylated RASSF1A copy number determined using
RE-dMSP was remarkably lesser than in CK19 mRNA (2.8 folds vs. 10.5
folds) in 11 breast cancer cell lines (12). Thus, RE-dMSP was indicated to
estimate tumor burden of LN metastasis more precisely than
OSNA.
This previous study suggested that RE-dMSP is a
supplementary method to OSNA in identifying and quantifying
axillary metastatic lymph nodes by quantifying methylated
RASSF1A copy number. However, a further study with more
patients is required for validation. This study was conducted under
the hypothesis that RE-dMSP is a comparable in sensitivity and
specificity to OSNA and is useful in the diagnosis of SN metastases
and designed to perform OSNA and RE-dMSP using SN lysate from
several patients with breast cancer and to re-assess the clinical
usage of RE-dMSP.
Materials and methods
Patients and samples
This study was a diagnostic accuracy study and
included 347 consecutive breast cancer patients who underwent
surgery with sentinel lymph node biopsy (SNB) and whose sentinel
nodes were all assessed using OSNA at Hakuaikai Sagara Hospital
between November 2014 and October 2019. Approval for this research
was issued by the Ethical Review Board of Osaka University Hospital
(approval date/number: January 29, 2019/#18396). All patients
provided opt-out consent for the use of their samples in the
current study. SNB was performed using both dye (patent blue or
indocyanine green) and radiocolloid (technetium-99m tin colloid).
The whole SN tissue was used for OSNA and homogenized to 4-ml
Lynorhag solution (Sysmex Corporation, Kobe, Japan), of which 2-µl
lysate was used for assay. The remaining lysate was kept at −80°C
until RE-dMSP was carried out. The classification of CK19 mRNA copy
number per assay, which is listed below in the present study:
>5,000, (++); >250 and ≤5,000, (+); >0 and ≤250, (−); and
0, (N.D.). As recommended by the manufacturer of OSNA (4), (++) and (+) were considered positive,
and >0 and ≤250 were regarded negative for SN metastasis though
the amplification of CK19mRNA was found. In the analysis, 418 SNs
from 347 patients were included, and 520 lysates were analyzed (the
SN was separated into two lysates in 62 SNs, three lysates in 14
SNs, and four lysates in five SNs because of its large size).
Detection of RASSF1A methylation using
RE-dMSP
RASSF1A gene methylation was detected using
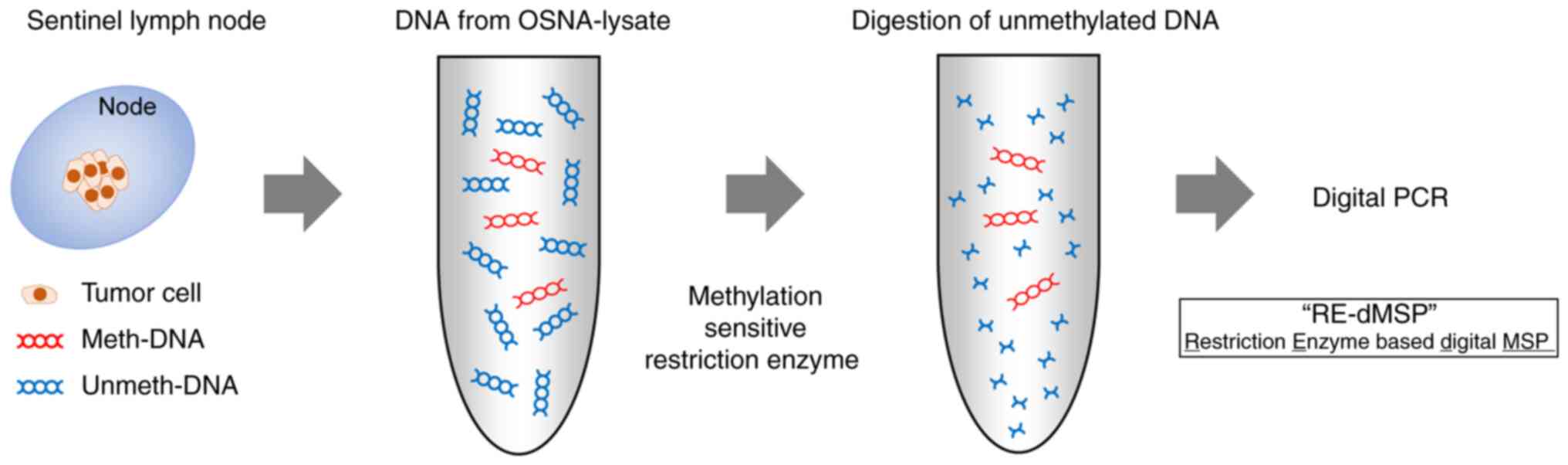
RE-dMSP assay as reported in our previous study (Fig. 1) (12). Briefly, DNA was extracted from
100–150-µl OSNA lysate using the QIAamp Circulating Nucleic Acid
Kit (Qiagen GmbH, Hilden, Germany) and eluted in 50-µl desalted
water. Then, 6.6-µl DNA solution was mixed to 20 µl volume with
following solutions: 1X ddPCR Supermix for probes (Bio-Rad
Laboratories, Inc., Hercules, CA, USA), 900 nM each primer, 250 nM
probe, and to completely digest unmethylated DNA, three
methylation-sensitive restriction enzymes, 10 U HhaI,
HpaII (New England BioLabs, Inc., Ipswich, MA, USA), and
BstUI (Thermo-Fisher Scientific, Inc., Waltham, MA, USA).
The final 20-µl mixture was incubated for 16 h at 37°C. Methylation
analytical process was undertaken using three wells per assay.
2.0-µl DNA solution was also incubated without restriction enzymes
as a control to ensure the presence of DNA (Fig. S1). The sequence of primers and
probes was as follows: forward 3′-AGCTGGCACCCGCTGG-5′, reverse
3′-GTGTGGGGTTGCACGCG-5′, and probe 3′-CTCCAGCC-5′ (Universal Probe
Library #19; Roche #04686926001). Following incubation, droplet
generation oil was added, and subsequently the mixture was
transferred onto a QX100 droplet generator (Bio-Rad Laboratories,
Inc.). Then, 40-µl emulsified mixture was subjected to polymerase
chain reaction (PCR) using a T100 thermal cycler (Bio-Rad
Laboratories, Inc.) at 95°C for 10 min, followed by 40 cycles at
94°C for 30 sec and 60°C for 1 min and 98°C for 10 min. The data
analysis was performed with the QX100 droplet reader and QuantaSoft
software, version 1.7.4 (both Bio-Rad Laboratories, Inc.). The
presence of two or more dots per well was considered positive
result, and the copy numbers of three positive wells were summated.
For cases divided into multiple lysates, the results were
summed.
For analysis of methylation status in primary breast
tumors, DNA was extracted from five 10-µm formalin-fixed
paraffin-embedded (FFPE) tumor sections using the QIAamp DNA FFPE
kit (Qiagen GmbH), and RE-dMSP was carried out. The cutoff for
methylation in primary tumors was set at 4% based on previous
reports to distinguish cancer tissues from noncancer tissues
(13–15).
Analysis of CK19 with
Immunohistochemistry
The protein expression of CK19 in primary tumors was
examined using immunohistochemistry with 4-µm FFPE tissue sections.
As reported in our former study (16), each section was
immunohistochemically stained with mouse monoclonal anti-CK19
primary antibody (clone, RCK 108; 1:100; Dako; Agilent
Technologies, Inc.) and a peroxidase-conjugated secondary antibody
(catalog number: 414131F; Histofine Simple Stain MAX PO (M);
Nichirei, Tokyo, Japan). The sections were visualized subsequently
with 3,3-diaminobenzidine tetrahydrochloride (Wako Pure Chemical
Industries, Ltd., Osaka, Japan) and counterstained with
hematoxylin.
Statistical analysis
R, version 4.1.1, was used for statistical
processing. Spearman's rank correlation coefficient was used to
assess the correlation between methylated DNA copy number and CK19
mRNA copy number. P<0.05 was considered significant. Regarding
the sample size in this study, at least 214 cases were needed to
estimate a 95% confidence interval with a specificity of 85% and an
accuracy of 80 to 90% (interval width 10%). In this study, 420 SNs
were provided from Hakuaikai Sagara Hospital as much as
possible.
Results
RE-dMSP using SN lysates for OSNA
The clinicopathological characteristics of the 347
patients in this study are presented in Table I. Overall, 418 SNs were analyzed:
284 patients had one SN, 56 had two SNs, and seven had three or
more SNs. The amount of total DNA in the SN lysates ranged from
4,800 to 5,920,000 copies per 100-µl lysate, confirming successful
DNA extraction from all samples. SN metastases were detected
intraoperatively using OSNA in 284 of the 418 SNs (67.9%), and 266
(63.6%) SNs were metastatic according to RE-dMSP results. The
concordance rate between the OSNA and RE-dMSP results was 83.3%
(Table SIA). In 418 SNs, the
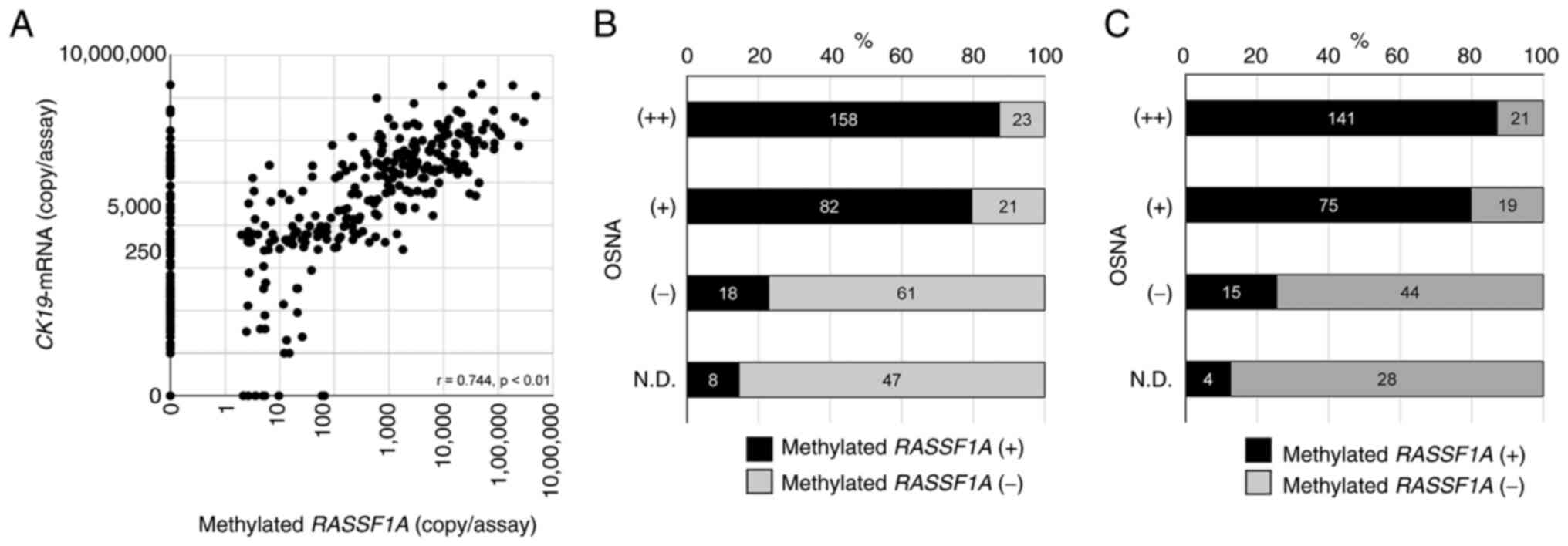
amounts of CK19 mRNA and methylated RASSF1A were
significantly related (r=0.744; P<0.01) (Fig. 2A). Of 134 OSNA-negative [(−) and
N.D.] SNs, 26 (19.4%) were RE-dMSP-positive. Of 284 OSNA-positive
[(++) and (+)] SNs, 44 (15.5%) were RE-dMSP-negative (Fig. 2B). Of 91 patients having
OSNA-negative [(−) and N.D.] SNs, 19 (20.9%) had RE-dMSP-positive
SNs. Of 256 patients having OSNA-positive [(++) and (+)] SNs, 40
(15.6%) had RE-dMSP-negative SNs (Fig.
2C).
 | Table I.Clinicopathological characteristics of
347 patients with breast cancer. |
Table I.
Clinicopathological characteristics of
347 patients with breast cancer.
| Characteristic | No. of
patients |
|---|
| Age, years |
|
|
<50 | 251 |
|
≥50 | 96 |
| Type of
surgery |
|
| Bt | 160 |
| Bp | 187 |
| No. of SLN |
|
| 1 | 284 |
| 2 | 56 |
| ≥3 | 7 |
| ALND |
|
| No | 148 |
|
Yes | 199 |
| Tumor
histology |
|
|
IDC | 287 |
|
ILC | 34 |
|
Othersa | 26 |
| Tumor size |
|
| T1 | 218 |
| T2,
3 | 129 |
| Histological
grade |
|
| 1,
2 | 307 |
| 3 | 40 |
| LVI |
|
|
Positive | 267 |
|
Negative | 80 |
| Subtype |
|
|
HR+b/HER2- | 280 |
|
HER2+ | 55 |
|
TNBC | 5 |
|
Unknown | 7 |
| Recurrence |
|
| No | 343 |
|
Yes | 4 |
CK19 expression in primary tumors with
OSNA-negative/RE-dMSP-positive SNs
In 19 patients whose SNs were
OSNA-negative/RE-dMSP-positive, immunohistochemical staining for
CK19 expression in primary tumors revealed strong homogeneous
expression of CK19 in all tumors (Fig.
S2).
RASSF1A methylation in primary tumors
with OSNA-positive/RE-dMSP-negative SNs
In 40 patients having SNs of
OSNA-positive/RE-dMSP-negative SNs, RASSF1A methylation in
primary tumors were analyzed using RE-dMSP, and of them, 24 (60%)
showed RE-dMSP-negative.
RE-dMSP using SN lysates for OSNA
limited to patients with RASSF1A methylation-positive primary
tumors
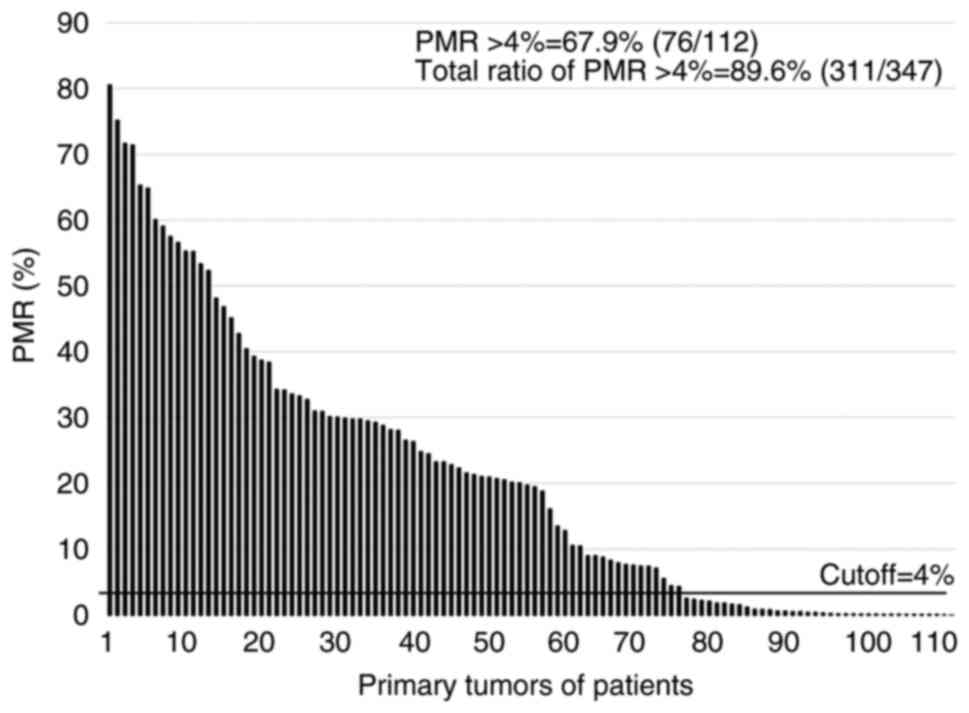
The status of the 112 patients whose SNs were
RE-dMSP-negative regardless of OSNA results was determined, and
RASSF1A methylation (the percent of methylated reference
(PMR) >4%) was positive in 76 (67.9%) of 112 tumors (Fig. 3). The other 235 RE-dMSP-positive
patients could be considered methylation-positive for primary
breast cancer, and therefore, of the 347 patients, 311 (89.6%) were
positive for RASSF1A methylation in primary tumors (Fig. 3). SN metastases were detected
intraoperatively using OSNA in 258 of the 374 SNs (68.9%). Then,
266 (71.1%) SNs were positive according to RE-dMSP results
(Table SIB). Only in cases where
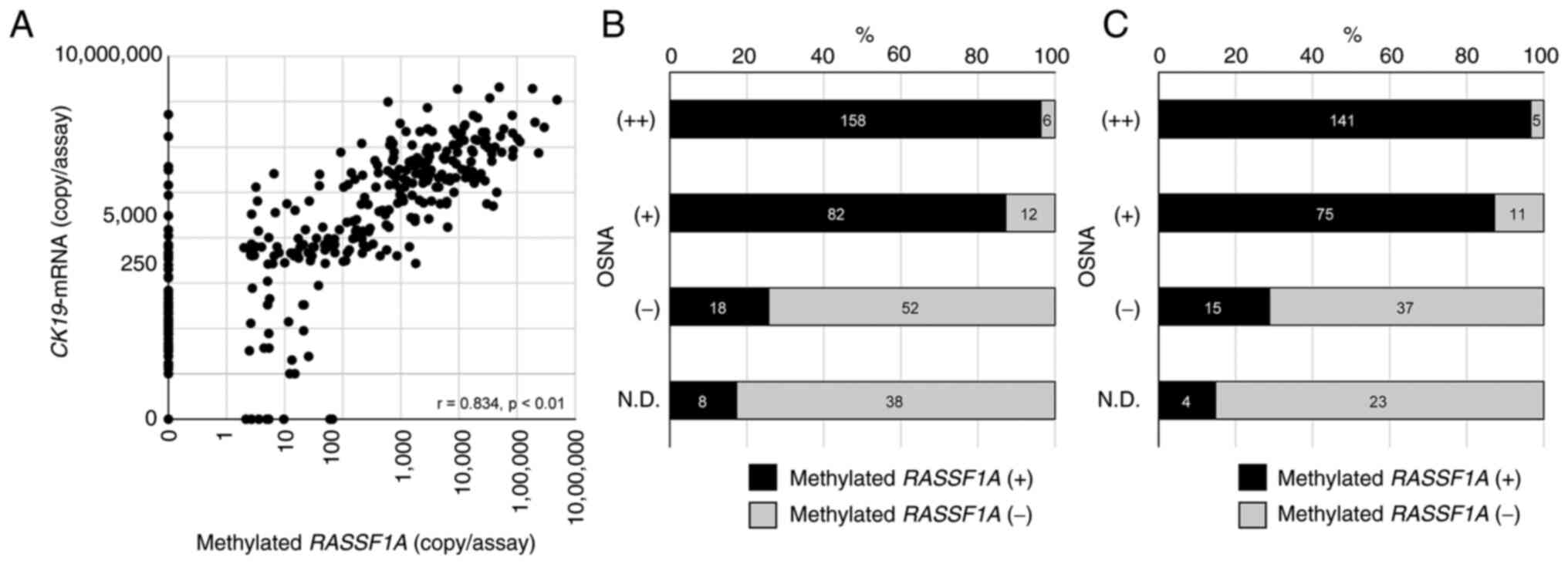
the primary tumors were positive for methylation (n=311), the copy
number of CK19 mRNA by OSNA and RASSF1A methylation by
RE-dMSP assay showed a better correlation than that in all cases
(r=0.834; P<0.01) (Fig. 4A). The
concordance rate between the OSNA and RE-dMSP results was 88.2%
(Table SIB). Of 116 OSNA-negative
[(−) and N.D.] SNs, 26 (22.4%) were RE-dMSP-positive. Of 258
OSNA-positive [(++) and (+)] SNs, 18 (6.9%) were RE-dMSP-negative
(Fig. 4B). Of 79 patients having
OSNA-negative [(N.D.) or (−)] SNs, 19 (24.1%) had RE-dMSP-positive
SNs. Of 232 patients having OSNA-positive [(++) or (+)] SNs, 16
(6.9%) had RE-dMSP-negative SNs (Fig.
4C).
Correlation PMR of the primary tumors
with tumor size in OSNA-positive/RE-dMSP-negative SNs
In 16 patients whose SNs were
OSNA-positive/RE-dMSP-negative, the PMR of the primary tumors of
them ranged from 4.25 to 75.1% (median=22.4%) and showed a positive
correlation with tumor size (r=0.405) (Fig. S3).
Discussion
We have previously reported the use of RE-dMSP in
detecting tumor-derived DNA in SNs from 71 patients. This study was
conducted to validate this finding with more SNs. In this study,
418 SNs from 347 patients with breast cancer were analyzed, and we
found a high correlation between the results of OSNA and RE-dMSP.
RE-dMSP could be a supplementary tool to OSNA in diagnosing SN
metastasis of breast cancer.
The concordance rate between the OSNA and RE-dMSP
results for SNs in this study was lesser compared to our former
study, although there was a predominant correlation. OSNA is an
assay that targets CK19 mRNA, whereas RE-dMSP targets methylated
RASSF1A. Considering the differences between the OSNA and
RE-dMSP results in SNs, the expression of CK19 and RASSF1A
methylation in the primary tumor are important.
It has been reported that 3.0-20.5% of patients with
breast cancer show low expression of CK19 (17–22).
In this study, OSNA-negative/RE-dMSP-positives SNs were observed in
19 patients, and CK19 expression was strongly positive in all these
primary tumors, indicating that this concordance was unlikely to be
attributable to the lack of CK19 expression. Four of these 19
patients had N.D. SNs according to the OSNA results. These
OSNA-negative/RE-dMSP-positive SNs may be false negatives when
assessed using OSNA. Furthermore, RE-dMSP may have identified true
metastases that could not be identified using OSNA. RE-dMSP can
identify as few as three copies of methylated RASSF1A per
assay, which corresponds to 150 tumor cells per node, which is much
smaller than micro-metastasis (>200 µm in diameter) (12). Therefore,
OSNA-negative/RE-dMSP-positive is probably because of the high
sensitivity of RE-dMSP. We examined 374 SNs from 311 patients with
RASSF1A methylation in the primary tumor and found an even
higher correlation than all 347 patients.
The prevalence of low RASSF1A methylation in
the primary tumors has been reported to be 14.8-24% (13,20).
In this study, low RASSF1A methylation of primary tumors was
found in 10.4% (36/347), which was almost consistent with those
reported in previous studies. Of the 232 patients with
OSNA-positive SNs, 16 (6.9%) were RE-dMSP-negative (Fig. 4C). The total copy number of CK19
mRNA in the SNs of the 16 patients ranged from 340 to 521,700
copies, including 11 OSNA (+) and 5 OSNA (++). The PMR of the
primary tumors of these 16 patients showed a positive correlation
with tumor size (r=0.405). This indicated that the discordance of
OSNA-positive/RE-dMSP-negative tended to be observed in small
tumors with low methylation or highly methylated but large tumors,
as previously reported (23). The
existence of regional and spatial heterogeneity of methylation
within the same tumor has been reported (24). A study revealed heterogeneity within
a tumor by showing differences in the rate of methylation between
blocks of the same tumor and between regions of a block within the
same tumor. In this study, the PMR in each case was assessed in the
representative portion of the tumor. Therefore, based on the
correlation between tumor diameter and the PMR, it is likely that
larger tumors contain unmethylated tumor cells in other areas not
used for PMR evaluation.
Therefore, OSNA-positive/RE-dMSP-negative SNs can be
attributable to metastasis of unmethylated tumor cells from the
partially or heterogeneously methylated primary tumors. Considering
that four of them had non-SN metastases after axillary dissection,
OSNA-positive/RE-dMSP-negative is probably a false-negative result
of RE-dMSP. Additionally, as discussed in a previous section, 16
patients were ineligible because the methylation in the primary
tumor was partial or heterogeneous. This means that almost 15%
(51/347) of the patients cannot undergo RE-dMSP assay because of
unfavorable methylation status. In contrast, the loss of CK19 mRNA
expression was reported to be much less frequent than that of
RASSF1A methylation. Even though RE-dMSP can provide more
accurate TTL, it tends to yield false-negative results compared
with OSNA. Using RE-dMSP alone to diagnose SN metastases is
difficult. Additionally, RE-dMSP is not suggested for
intraoperative diagnosis. The use of this assay should be
investigated for its potential contribution to prognosis prediction
and treatment strategy development.
This study has some limitations. We cannot examine
the OSNA and RE-dMSP false-positive results. Furthermore, the other
limitation of this assay may be due to its ability to target
RASSF1A methylation only. A previous report by Abe et
al (12) has analyzed
PIK3CA mutation and RASSF1A methylation in the SN
lysates of patients with PIK3CA mutation-positive tumors and
reported the completed agreement between mutation and methylation
status. Moreover, whole-genome/exon sequencing can identify at
least one mutation in breast tumors (25), suggesting that mutation can be a
more appropriate target for DNA-based SN diagnosis than
RASSF1A methylation, where it can cover all patients.
In conclusion, RE-dMSP can diagnose SN metastasis
with high sensitivity and accuracy and can be a supplementary tool
to OSNA. However, it was revealed that false-negative results
because of heterogeneous methylation and RE-dMSP's inability to
target all patients with breast cancer had a nonnegligible effect
on the results. Therefore, it is not a perfect complement to OSNA.
Targeting genomic mutations will be a solution to these problems,
and studies on this solution are required.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Ms. Megumi Take and
the staff of clinical and pathological laboratories at Hakuaikai
Sagara Hospital (Kagoshima, Japan) for helping prepare the
samples.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to information that
could compromise the privacy of the research participants but are
available from the corresponding author on reasonable request.
Authors' contributions
SAP participated in data analysis and
interpretation, and wrote the manuscript. NK was involved in
designing the experiments and drafted the manuscript. NM, YO, NG,
KA, TY, YoS, TM, TT, MS, and YaS were responsible for providing the
resources, analyzing and interpretating the data, and revising the
discussion. KS conceptualized and supervised the study. SAP and NM
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
This study involving human samples was approved by
the Ethics Review Board of the Osaka University Hospital and was
conducted according to The Declaration of Helsinki. All patients
provided opt-out consent for participation and the use of their
samples in the current study. This form indicated that their
personal data could be utilized for academic or paper
presentations, with the assurance of maintaining absolute
anonymity.
Patient consent for publication
Not applicable.
Competing interests
NK received a research grant from AstraZeneca. KS
received a research grant from AstraZeneca and ROHTO Pharmaceutical
Co., Ltd. KS has received honoraria from Sysmex and AstraZeneca.
KA, TY, and TM have received honoraria from AstraZeneca. The other
authors declare that they have no competing interests.
Glossary
Abbreviations
Abbreviations:
|
SN
|
sentinel lymph node
|
|
OSNA
|
One-Step Nucleic acid
Amplification
|
|
CK19
|
cytokeratin 19
|
|
LN
|
lymph node
|
|
TTL
|
total tumor load
|
|
RASSF1A
|
Ras association domain-containing
protein 1
|
|
SNB
|
sentinel lymph node biopsy
|
|
dPCR
|
digital polymerase chain reaction
|
|
RE-dMSP
|
restriction enzyme-based digital
methylation-specific polymerase chain reaction
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
PMR
|
percent of methylated reference
|
References
|
1
|
Lyman GH, Giuliano AE, Somerfield MR,
Benson AB III, Bodurka DC, Burstein HJ, Cochran AJ, Cody HS III,
Edge SB, Galper S, et al: American society of clinical oncology
guideline recommendations for sentinel lymph node biopsy in
early-stage breast cancer. J Clin Oncol. 23:7703–7720. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Veronesi U, Paganelli G, Viale G, Luini A,
Zurrida S, Galimberti V, Intra M, Veronesi P, Maisonneuve P, Gatti
G, et al: Sentinel-lymph-node biopsy as a staging procedure in
breast cancer: Update of a randomised controlled study. Lancet
Oncol. 7:983–990. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lyman GH, Temin S, Edge SB, Newman LA,
Turner RR, Weaver DL, Benson AB III, Bosserman LD, Burstein HJ,
Cody H III, et al: Sentinel lymph node biopsy for patients with
early-stage breast cancer: American society of clinical oncology
clinical practice guideline update. J Clin Oncol. 32:1365–1383.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsujimoto M, Nakabayashi K, Yoshidome K,
Kaneko T, Iwase T, Akiyama F, Kato Y, Tsuda H, Ueda S, Sato K, et
al: One-step nucleic acid amplification for intraoperative
detection of lymph node metastasis in breast cancer patients. Clin
Cancer Res. 13:4807–4816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tamaki Y: One-step nucleic acid
amplification assay (OSNA) for sentinel lymph node biopsy. Breast
Cancer. 22:230–234. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rubio IT, Espinosa-Bravo M, Rodrigo M,
Amparo Viguri Diaz M, Hardisson D, Sagasta A, Dueñas B and Peg V:
Nomogram including the total tumoral load in the sentinel nodes
assessed by one-step nucleic acid amplification as a new factor for
predicting nonsentinel lymph node metastasis in breast cancer
patients. Breast Cancer Res Treat. 147:371–380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Teramoto A, Shimazu K, Naoi Y, Shimomura
A, Shimoda M, Kagara N, Maruyama N, Kim SJ, Yoshidome K, Tsujimoto
M, et al: One-step nucleic acid amplification assay for
intraoperative prediction of non-sentinel lymph node metastasis in
breast cancer patients with sentinel lymph node metastasis. Breast.
23:579–585. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shimazu K, Sato N, Ogiya A, Sota Y,
Yotsumoto D, Ishikawa T, Nakamura S, Kinoshita T, Tsuda H, Ohi Y,
et al: Intraoperative nomograms, based on one-step nucleic acid
amplification, for prediction of non-sentinel node metastasis and
four or more axillary node metastases in breast cancer patients
with sentinel node metastasis. Ann Surg Oncol. 25:2603–2611. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peg V, Espinosa-Bravo M, Vieites B,
Vilardell F, Antúnez JR, de Salas MS, Delgado-Sánchez JJ, Pinto W,
Gozalbo F, Petit A, et al: Intraoperative molecular analysis of
total tumor load in sentinel lymph node: A new predictor of
axillary status in early breast cancer patients. Breast Cancer Res
Treat. 139:87–93. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park SY, Kwon HJ, Choi Y, Lee HE, Kim SW,
Kim JH, Kim IA, Jung N, Cho NY and Kang GH: Distinct patterns of
promoter CpG island methylation of breast cancer subtypes are
associated with stem cell phenotypes. Mod Pathol. 25:185–196. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamamoto N, Nakayama T, Kajita M, Miyake
T, Iwamoto T, Kim SJ, Sakai A, Ishihara H, Tamaki Y and Noguchi S:
Detection of aberrant promoter methylation of GSTP1, RASSF1A, and
RARβ2 in serum DNA of patients with breast cancer by a newly
established one-step methylation-specific PCR assay. Breast Cancer
Res Treat. 132:165–173. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abe M, Kagara N, Miyake T, Tanei T, Naoi
Y, Shimoda M, Shimazu K, Kim SJ and Noguchi S: Highly sensitive
detection of sentinel lymph node metastasis of breast cancer by
digital PCR for RASSF1A methylation. Oncol Rep. 42:2382–2389.
2019.PubMed/NCBI
|
|
13
|
Eads CA, Lord RV, Kurumboor SK,
Wickramasinghe K, Skinner ML, Long TI, Peters JH, DeMeester TR,
Danenberg KD, Danenberg PV, et al: Fields of aberrant CpG island
hypermethylation in Barrett's esophagus and associated
adenocarcinoma.pdf. cancer Res. 60:5021–5026. 2000.PubMed/NCBI
|
|
14
|
Ogino S, Kawasaki T, Brahmandam M, Cantor
M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ,
Laird PW, Loda M and Fuchs CS: Precision and performance
characteristics of bisulfite conversion and real-time PCR
(MethyLight) for quantitative DNA methylation analysis. J Mol
Diagn. 8:209–217. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park SY, Kwon HJ, Lee HE, Ryu HS, Kim SW,
Kim JH, Kim IA, Jung N, Cho NY and Kang GH: Promoter CpG island
hypermethylation during breast cancer progression. Virchows Arch.
458:73–84. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Morimoto K, Kim SJ, Tanei T, Shimazu K,
Tanji Y, Taguchi T, Tamaki Y, Terada N and Noguchi S: Stem cell
marker aldehyde dehydrogenase 1-positive breast cancers are
characterized by negative estrogen receptor, positive human
epidermal growth factor receptor type 2, and high Ki67 expression.
Cancer Sci. 100:1062–1068. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vilardell F, Novell A, Martin J, Santacana
M, Velasco A, Díez-Castro MJ, Cuevas D, Panadés MJ, González S,
Llombart A, et al: Importance of assessing CK19 immunostaining in
core biopsies in patients subjected to sentinel node study by OSNA.
Virchows Arch. 460:569–575. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alvarenga CA, Paravidino PI, Alvarenga M,
Dufloth R, Gomes M, Zeferino LC and Schmitt F: Expression of CK19
in invasive breast carcinomas of special histological types:
Implications for the use of one-step nucleic acid amplification. J
Clin Pathol. 64:493–497. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Parikh RR, Yang Q, Higgins SA and Haffty
BG: Outcomes in young women with breast cancer of triple-negative
phenotype: the prognostic significance of CK19 expression. Int J
Radiat Oncol Biol Phys. 70:35–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Willipinski-Stapelfeldt B, Riethdorf S,
Assmann V, Woelfle U, Rau T, Sauter G, Heukeshoven J and Pantel K:
Changes in cytoskeletal protein composition indicative of an
epithelial-mesenchymal transition in human micrometastatic and
primary breast carcinoma cells. Clin Cancer Res. 11:8006–8014.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Abd El-Rehim DM, Pinder SE, Paish CE, Bell
J, Blamey RW, Robertson JF, Nicholson RI and Ellis IO: Expression
of luminal and basal cytokeratins in human breast carcinoma. J
Pathol. 203:661–671. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chu PG and Weiss LM: Keratin expression in
human tissues and neoplasms. Histopathology. 40:403–439. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gupta V, Agarwal P and Deshpande P: Impact
of RASSF1A gene methylation on clinico-pathological features of
tumor and non-tumor tissue of breast cancer. Ann Diagn Pathol.
52:1517222021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moelans CB, de Groot JS, Pan X, van der
Wall E and van Diest PJ: Clonal intratumor heterogeneity of
promoter hypermethylation in breast cancer by MS-MLPA. Mod Pathol.
27:869–874. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nik-Zainal S, Davies H, Staaf J,
Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB,
Martin S, Wedge DC, et al: Landscape of somatic mutations in 560
breast cancer whole-genome sequences. Nature. 534:47–54. 2016.
View Article : Google Scholar : PubMed/NCBI
|