Introduction
Oncocytic carcinomas (OC) are rare malignant tumors
composed of oncocytes, which are epithelial cells characterized by
a granular eosinophilic cytoplasm containing numerous mitochondria
(1). OC of the breast is uncommon,
accounting for <1% of all types of breast cancer worldwide
(2). Hence, little is known about
its pathological molecular features.
MicroRNAs (miRNAs/miRs) are small, non-coding RNAs
that play an important role in post-transcriptional gene regulation
(3,4). They interact with their target
messenger RNAs (mRNAs) to control translation through mRNA
degradation or translational repression (5,6).
miRNAs are contributors to various biological processes and
previous studies have revealed the relevance of miRNA biology in
cancer (7–9).
Recently, the presence of miRNAs in mitochondria,
known as mitochondrial miRNAs (mito-miRs), have emerged as
modulators that target the mitochondria and regulate mitochondrial
protein expression and function (10,11).
The present study hypothesized that expression of mitochondrial
miRNAs could be altered in OC, given its characteristic features.
The present study reports the case of a 76-year-old woman diagnosed
with oncocytic carcinoma of the breast. To test the aforementioned
hypothesis, the present study assessed the expression levels of the
miRNAs previously reported to be mito-miRs, such as miR-221-3p,
−146a-5p and −16-5p, in tissues from specimens of the patient with
OC compared with that in tissues from specimens of a more typical
type of invasive ductal carcinoma (IDC) of the breast. The present
study also investigated the mRNA expression levels of BCL2, which
plays a pivotal role in apoptosis in mitochondria and is the
corresponding target of the aforementioned miRNAs.
Case report
A 76-year-old woman with a left breast tumor was
referred to JA Hiroshima General Hospital (Hatsukaichi, Japan) in
June 2019. The patient had noticed a gradual enlargement of the
mass over the past 3 years. Physical examination revealed a 65-mm,
hard, non-mobile mass with a focal cystic appearance in the upper
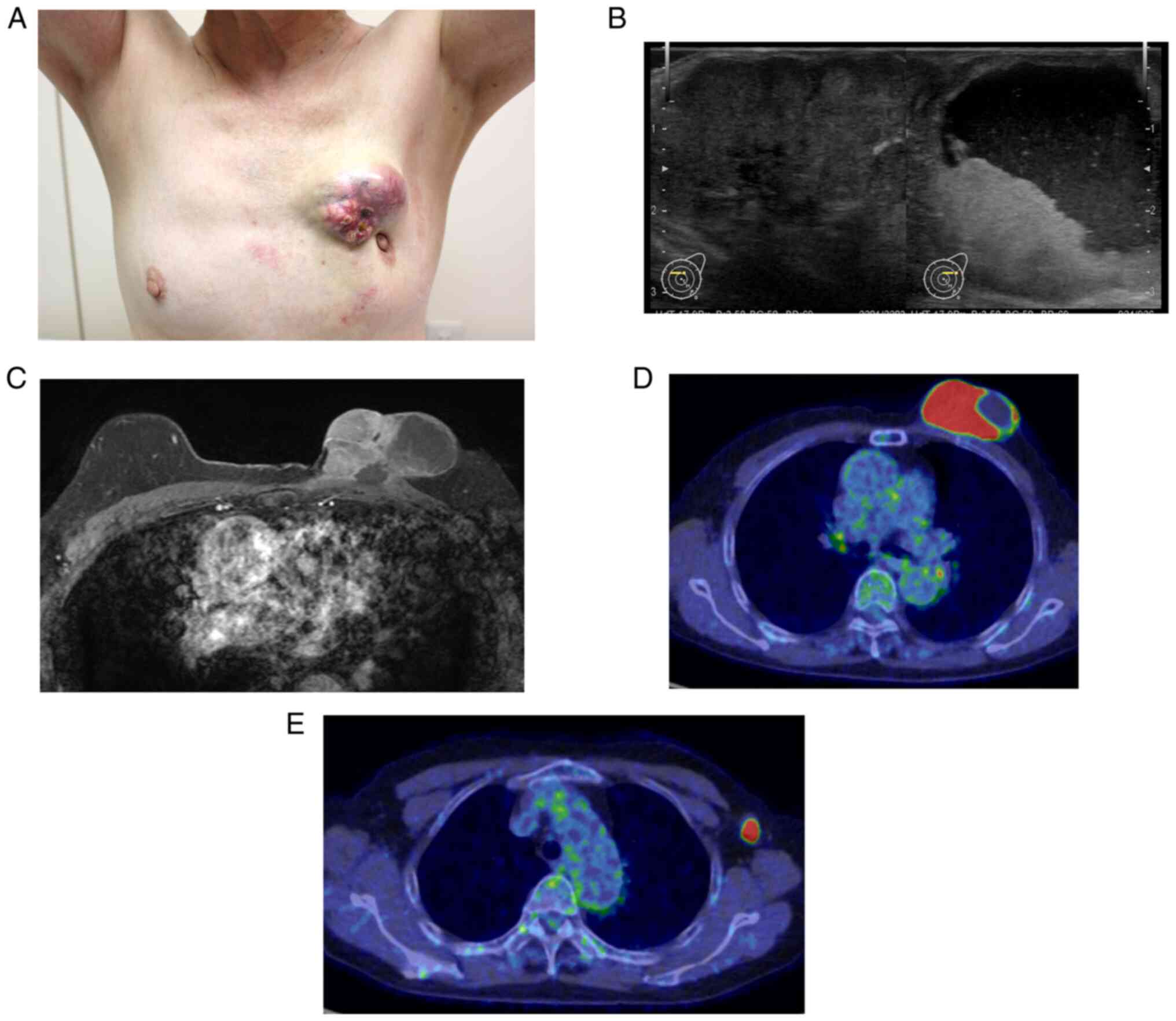
inner quadrant of the left breast (Fig.
1A). Ultrasonography revealed a solid mass with a marked cystic
component, potentially indicating hemorrhage and swelling of the
left axillary lymph nodes (Fig.
1B). Magnetic resonance imaging revealed a solid mass with a
cystic component and apparent involvement of the skin and
pectoralis major muscle (Fig. 1C).
Additionally, positron emission tomography showed abnormal uptake
in the mass and axillary lymph nodes (maximum standardized uptake
values, 10.2 and 8.0, respectively) but no evidence of systemic
metastases (Fig. 1D and E). A core
needle biopsy was performed and the mass was diagnosed as an
invasive carcinoma. Cytological examination of a specimen obtained
with fine-needle aspiration of the lymph node revealed the presence
of malignant cells. Subsequently, ~1 month after the first visit,
the patient underwent mastectomy and axial lymph node
dissection.
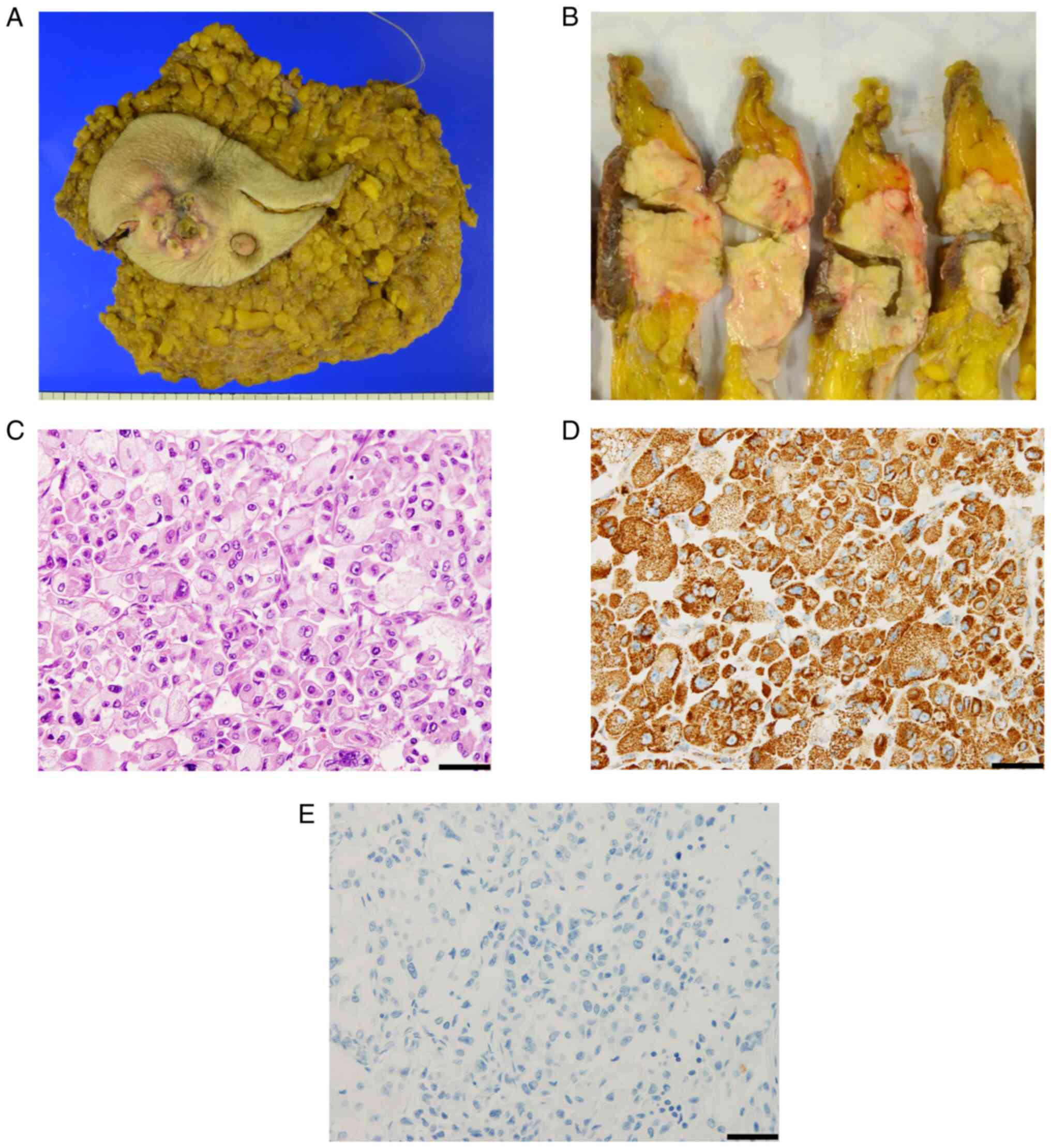
The tumor measured 45×12 mm, and four lymph nodes
were involved (Fig. 2A and B).
Hematoxylin and eosin (HE) staining revealed a granular cytoplasm,
ranging from eosinophilic to clear, in ovoid cancer cells (Fig. 2C). In addition, strongly positive
immunostaining for antimitochondrial antibodies indicated that the
tumor cells were of oncocytic origin (Fig. 2D). Further, immunohistochemical
(IHC) staining showed that the tumor cells were positive for
estrogen and progesterone receptors but negative for human
epidermal growth factor receptor 2 (HER2) and gross cystic disease
fluid protein 15 (GCDFP-15; Fig.
2E). The Ki-67 labeling index was 15%. Based on these results,
the diagnosis of OC of the left breast was made. The patient
underwent postmastectomy radiation therapy 1 month after surgery,
receiving 50 Gy in 25 fractions to the chest wall and
supraclavicular lymph nodes, sequentially followed by adjuvant
endocrine therapy with an aromatase inhibitor. In the 3 years after
surgery, there was no cancer recurrence detected by regular 3-month
follow-up visits.
For molecular profiling, the present study collected
fresh tissue samples from the surgical specimens (both tumor and
non-cancerous samples) and stored them at −80°C until use. The
tumor tissues were acquired from the largest cross-section of the
surgical specimen of the breast immediately after resection. At the
same time, the non-cancerous tissue was acquired from another area
≥5 cm distant from the cancerous region of the surgical specimen
that was confirmed grossly as normal breast tissue. The opposite
cross section of this non-cancerous sample was microscopically
confirmed as normal breast tissue. For comparison, fresh tissue
samples of IDC were also collected from a 72-year-old female
patient who underwent a consecutive surgery at the same hospital on
the same day without planned recruitment with a matching phenotype
(positive estrogen and progesterone receptors and negative HER2)
who underwent surgery in JA Hiroshima General Hospital in July
2019. Both tumor and non-cancerous samples were also acquired from
this patient in the same manner for the patient with oncocytic
carcinoma. The present study obtained written permission from this
patient with IDC to utilize surgical specimens for research
purposes.
Reverse transcription-quantitative PCR (RT-qPCR) was
used to evaluate the expression levels of miR-221-3p, −146a-5p and
−16-5p as mito-miRs and BCL2 mRNA. MiRNAs were extracted using a
miRNeasy Mini kit (QIAGEN, Ltd.) according to the manufacturer's
protocol. RT-qPCR was performed using TaqMan MicroRNA Assays
(Applied Biosystems; Thermo Fisher Scientific, Inc.), which
includes TaqMan MGB probes that contains FAM™ dye (Applied
Biosystems; Thermo Fisher Scientific, Inc.) as previously described
(12). All primers were obtained
from Applied Biosystems: MiR-221 (Assay ID: 000524_mat), miR-146a
(000468_mat) and miR-16 (000391_mat). U6 small nuclear RNA and
glyceraldehyde 3-phosphate dehydrogenase were used for
normalization in RT-qPCR with miRNAs and mRNA, respectively. RT-PCR
was performed at 95°C for 10 min, followed by 40 cycles of 95°C for
15 sec and 60°C for 1 min. All experiments were conducted in
triplicate. For RT-qPCR, the results were calculated as the mean ±
standard deviation of triplicate data, and the expression was
illustrated as a fold difference of 1 for non-cancerous samples
(normal).
Additionally, the BCL2 protein level was determined
using IHC staining using 10% neutral buffered formalin-fixed for 24
h at room temperature, paraffin-embedded samples and assessed
H-score, an immunoreactivity semiquantitative scoring system
(13). Paraffin-embedded sections
with a thickness of 4 µm were deparaffinized with xylene and washed
with serially diluted ethanol for rehydration, antigen retrieval
was accomplished using pH 9.0 conditional buffer for 20 min until
the temperature reached 98°C. The endogenous peroxidase activity
was blocked with 3–4% (V/V) hydrogen peroxide solution at room
temperature for 5 min. Sections were incubated with Ready-To-Use
anti-BCL2 antibody (PA0117; Leica Biosystems) at room temperature
for 60 min. Ready-To-Use Post Primary Mouse Linker as secondary
antibody (DS9800; Leica Biosystems) was applied to the slides at
room temperature for 10 min followed by horseradish
peroxidase-labeled anti-rabbit IgG antibody (DS9800; Leica
Biosystems) at room temperature for 10 min. Application of Leica
Bond Polymer Refine Detection at room temperature for 10 min
(DS9800; Leica Biosystems) with 3,3′-diaminobenzidine as the
chromogen was followed by counterstaining with hematoxylin at room
temperature for 5 min. We observed the samples using an light
microscope. The scores were all noted for the staining intensity
(0, no staining; 1, weak staining; 2, moderate staining; and 3,
strong staining). The BCL2 H-score was then calculated using the
following formula: H-score=[1*(% cells 1+) + 2*(% cells 2+) + 3*(%
cells 3+)] (14). Notably, the
expression level of miR-221-3p was increased, while that of
miR-146a-5p and miR-16-5p were decreased in OC compared with that
in IDC (Fig. 3A-C). Moreover, the
mRNA expression of BCL2 was 2.26-fold higher in OC compared with in
IDC (Fig. 3D), which could further
explain the data obtained from IHC staining, which showed a higher
intensity of BCL2 protein in OC (H-score, 98.4) compared with that
in IDC (H-score, 73.6) referred to as non-cancerous sample
(Fig. 3E-G).
 | Figure 3.Differential expression of miR-221-3p,
miR-146a-5p, miR-16-5p and BCL2 in OC and IDC. The relative
expression level, examined via RT-qPCR, of (A) miR-221-3p, (B)
miR-146a-5p, (C) miR-16-5p and (D) BCL2 messenger RNA in OC and IDC
compared with non-cancerous tissue (normal). Immunohistochemical
staining of BCL2 in (E) IDC, (F) OC and (G) normal (scale bar, 50
µm). The intensity of staining was stronger in OC compared with in
IDC (magnification, ×400). All RT-qPCR experiments were conducted
in triplicate. Data are represented as mean ± standard deviation,
presented as the fold-change compared with non-cancerous tissue.
GAPDH, glyceraldehyde 3-phosphate dehydrogenase; miRNA, microRNA;
OC, oncocytic carcinoma; IDC, invasive ductal carcinoma; RT-qPCR,
reverse transcription-quantitative PCR. |
Discussion
Although miRNAs have been explored as important
biomarkers in numerous fields, there are few studies on OC of the
breast because of its rarity. The present report presents a case of
OC of the breast, along with expression analysis of three
mito-miRNAs (miR-221-3p, −146a-5p and −16-5p) in tissue samples
from OC compared with those of IDC. Additionally, the present study
assessed the expression level of BCL2 mRNA using the same samples.
The differential expression of these miRNAs and mRNA in this study
may provide valuable insights into the molecular features of OC of
the breast.
The World Health Organization classification defines
OC of the breast as a tumor in which >70% of all cells have
oncocytic features (2). The
distinctive feature of oncocytes is the enlargement of cells
containing abundant eosinophilic granules in their cytoplasm due to
an increase in altered mitochondria (15). OC morphologically resembles apocrine
carcinoma, which also has an abundant eosinophilic granular
cytoplasm (16). Consequently,
distinguishing OC from apocrine carcinoma solely based on HE
staining is challenging. Accurate diagnosis of OC necessitates IHC
staining using mitochondrial markers and GCDFP-15, a marker for
apocrine differentiation, which must be absent for the diagnosis of
OC. Ragazzi et al (17)
reported that oncocytic breast carcinoma shows a trend for shorter
survival based on the analysis of 104 cases, including not
otherwise specified invasive breast carcinomas, using mitochondrial
immunostaining. Therefore, oncocytic breast carcinomas should be
considered in differential diagnosis for cytoplasmic granular
eosinophilia. In the present case, almost all tumor cells showed a
positive staining for antimitochondrial markers and a negative
staining for GCDFP-15 following the diagnosis as OC of the
breast.
Increasing evidence in recent years has indicated
that miRNAs participate in regulating gene expression at the
post-transcriptional level (4).
Consequently, the aberrant expression of miRNAs has garnered
attention as a potential biomarker in disease development,
including cancer (9). A previous
study revealed that miR-221-3p is induced by docosahexaenoic acid
(18). MiR-146a-5p is expressed in
various immune cells, plays a role in regulating inflammatory
response and is a potential biomarker for vascular complications of
diabetes (19,20). Furthermore, miR-16-5p is associated
with diabetes and Alzheimer's disease progression (21). Additionally, differential expression
of mito-miRs has been identified in numerous diseases, such as
heart failure, indicating their specific function in the pathogenic
process (22,23). Despite advancements in understanding
the underlying mechanisms involving mito-miRs, the contribution of
mito-miRs in oncocytic tumorigenesis remains unclear. Using gene
and miRNA expression analyses focusing specifically on
mitochondrial function and its interactions in follicular thyroid
tumors, miRNAs were found to be involved in the development of
oncocytic variants. However, to the best of our knowledge, there
are few reports of oncocytic breast carcinoma and no study has
explored the relationship between mito-miRs and oncocytic variants
(24). Considering mitochondria
accumulation as the hallmark of OC, the present study hypothesized
that the expression of mito-miRs would be altered in OC in contrast
to IDC. A previous study identified the diagnostic utility of
multiple mi-RNAs combination using miR-221, −146-b and −375 for
Hürthle cell (oncocytic) thyroid tumors (25). Additionally, the upregulation of
miR-221 is a characteristic of the OC of the thyroid and kidney
(26). Similarly, the present data
showed that expression of miR-221-3p was significantly elevated in
OC compared with that in IDC. The present study also observed the
differential expression of miR-146a-5p and −16-5p. A previous study
that used cultured breast cancer cells derived from a hormone
receptor negative and HER2 negative phenotype different from the
present study showed that miR-146a is poorly expressed in cancer
cells compared with non-cancer breast cells (27). In addition, Chen et al
(28) reported that miR-146a is
relatively less expressed in lung cancer lesions compared with
normal lung tissues. However, these observations have been poorly
described, and the miRNA expression of OC of the breast has not
been investigated in OC of the breast due to its rarity.
The present study performed molecular profiling
focused on BCL2 as the potential target gene of miR-221-3p,
−146a-5p and −16-5p. Although it has not been proven that
miR-221-3p directly regulates BCL2, miR-221-3p has been reported to
target PUMA (p53 upregulated modulator of apoptosis), a BCL2 family
member also called BCL2 binding component 3, showing that
downregulation of BCL2 is observed after miR-221 reduction
(29). Current research has
suggested that miR-146a-5p functions as a tumor suppressor that
regulates apoptosis via the mitochondrial pathway by targeting
anti-apoptotic proteins in aging-related diseases, including cancer
(30). Apoptosis is a specific
mechanism for programmed cell death, and its dysregulation can
induce carcinogenesis. BCL2 is one of the fundamental
anti-apoptotic factors involved in maintaining homeostasis between
cell formation and death and controlling mitochondrial function
(31–33). Although the underlying mechanisms
have not been elucidated, a recent study demonstrated that
mito-miRs appear to have a close relationship with mitochondrial
protein expression (34). The
higher expression of BCL2 and lower expression of miR-146a-5p in OC
of the breast when compared with that in IDC in the present study
suggests that miR-146a-5p may be involved in the altered expression
of BCL2 through mitochondrial metabolism. Direct interaction
between miR-16 and BCL2 has been reported in chronic lymphocytic
leukemia, and loss of miR-16 is involved in overexpression of BCL2
(35). Although its interaction in
breast cancer has not been shown, the overexpression of miR-16 has
been reported to inhibit the activation of BMI1, a member of the
BCL2 family, resulting in apoptosis. The present findings are
consistent with these reports, as a lower expression of miR-16-5p
and higher expression of BCL2 was observed in OC compared with in
the more common IDC (36,37).
The present study has several limitations. First,
only one sample of OC was investigated; multiple OC samples could
not be obtained because of its rarity. We acknowledge that our
results would be insufficient to generalize to all cases of OC;
however, providing descriptions of molecular profiling in a fresh
sample is important and beneficial for patients with rare tumors,
from both clinical and translational standpoints (38). In addition, the current analysis was
conducted using tissue samples; hence, there is no proof that the
current study observed expression differences directly related to
miRNAs, specifically inside the mitochondrial compartment. Further,
the present study was unable to identify the mechanisms underlying
the association between the three miRNAs and BCL2 in OC.
Nevertheless, the present findings may help improve
the understanding of this rare tumor, given the current lack of
knowledge regarding the expression and function of miRNA in OC of
the breast.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
RT collected the clinical data. YK wrote the
manuscript. KK, YK, and MO acquired and interpreted clinical data.
YD performed the pathological examination. YY, SF, and HT performed
and analyzed the experimental examination. MO revised the
manuscript. YK and MO confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethical Committee for
Human Genome Research of Hiroshima University (approval no.
IRINHIM129). Written informed consent was obtained from the patient
before enrollment.
Patient consent for publication
Written informed consent for the publication of the
clinical data, including photos and images, was obtained from the
patient.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chang A and Harawi SJ: Oncocytes,
oncocytosis, and oncocytic tumors. Pathol Annu. 27(Pt 1): 263–304.
1992.PubMed/NCBI
|
|
2
|
Tan PH, Ellis I, Allison K, Brogi E, Fox
SB, Lakhani S, Lazar AJ, Morris EA, Sahin A, Salgado R, et al: The
2019 WHO classification of tumours of the breast. Histopathology.
77:181–185. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zaratiegui M, Irvine DV and Martienssen
RA: Noncoding RNAs and gene silencing. Cell. 128:763–776. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Weiland M, Gao XH, Zhou L and Mi QS: Small
RNAs have a large impact: Circulating microRNAs as biomarkers for
human diseases. RNA Biol. 9:850–859. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Giuliani A, Prattichizzo F, Micolucci L,
Ceriello A, Procopio AD and Rippo MR: Mitochondrial (Dys) function
in inflammaging: Do mitomiRs influence the energetic, oxidative,
and inflammatory status of senescent cells? Mediators Inflamm.
2017:23090342017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jaquenod De Giusti C, Roman B and Das S:
The influence of microRNAs on mitochondrial calcium. Front Physiol.
9:12912018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu D, Takeshita F, Hino Y, Fukunaga S,
Kudo Y, Tamaki A, Matsunaga J, Takahashi RU, Takata T, Shimamoto A,
et al: miR-22 represses cancer progression by inducing cellular
senescence. J Cell Biol. 193:409–424. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu D, Li L, Zhang XX, Wan DY, Xi BX, Hu
Z, Ding WC, Zhu D, Wang XL, Wang W, et al: IX1 promotes tumor
lymphangiogenesis by coordinating TGFβ signals that increase
expression of VEGF-C. Cancer Res. 74:5597–5607. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goulding H, Pinder S, Cannon P, Pearson D,
Nicholson R, Snead D, Bell J, Elston CW, Robertson JF, Blamey RW,
et al: A new immunohistochemical antibody for the assessment of
estrogen receptor status on routine formalin-fixed tissue samples.
Hum Pathol. 26:291–294. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Asa SL: My approach to oncocytic tumours
of the thyroid. J Clin Pathol. 57:225–232. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Durham JR and Fechner RE: The histologic
spectrum of apocrine lesions of the breast. Am J Clin Pathol.
113((suppl_1)): S3–S18. 2000.PubMed/NCBI
|
|
17
|
Ragazzi M, de Biase D, Betts CM, Farnedi
A, Ramadan SS, Tallini G, Reis-Filho JS and Eusebi V: Oncocytic
carcinoma of the breast: Frequency, morphology and follow-up. Hum
Pathol. 42:166–175. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gil-Zamorano J, Martin R, Daimiel L,
Richardson K, Giordano E, Nicod N, García-Carrasco B, Soares SM,
Iglesias-Gutiérrez E, Lasunción MA, et al: Docosahexaenoic acid
modulates the enterocyte Caco-2 cell expression of microRNAs
involved in lipid metabolism. J Nutr. 144:575–585. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Frørup C, Mirza AH, Yarani R, Nielsen LB,
Mathiesen ER, Damm P, Svare J, Engelbrekt C, Størling J, Johannesen
J, et al: Plasma exosome-enriched extracellular vesicles from
lactating mothers with type 1 diabetes contain aberrant levels of
miRNAs during the postpartum period. Front Immunol. 12:7445092021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barutta F, Corbetta B, Bellini S, Guarrera
S, Matullo G, Scandella M, Schalkwijk C, Stehouwer CD, Chaturvedi
N, Soedamah-Muthu SS, et al: MicroRNA 146a is associated with
diabetic complications in type 1 diabetic patients from the
EURODIAB PCS. J Transl Med. 19:4752021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Alamro H, Bajic V, Macvanin MT, Isenovic
ER, Gojobori T, Essack M and Gao X: Type 2 diabetes mellitus and
its comorbidity, Alzheimer's disease: Identifying critical microRNA
using machine learning. Front Endocrinol (Lausanne).
13:10846562023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bandiera S, Matégot R, Girard M, Demongeot
J and Henrion-Caude A: MitomiRs delineating the intracellular
localization of microRNAs at mitochondria. Free Radic Biol Med.
64:12–19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jagannathan R, Thapa D, Nichols CE,
Shepherd DL, Stricker JC, Croston TL, Baseler WA, Lewis SE,
Martinez I and Hollander JM: Translational regulation of the
mitochondrial genome following redistribution of mitochondrial
microRNA in the diabetic heart. Circ Cardiovasc Genet. 8:785–802.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jacques C, Guillotin D, Fontaine JF, Franc
B, Mirebeau-Prunier D, Fleury A, Malthiery Y and Savagner F: DNA
microarray and miRNA analyses reinforce the classification of
follicular thyroid tumors. J Clin Endocrinol Metab. 98:E981–E989.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Titov SE, Poloz TL, Veryaskina YA and
Anishchenko VV: Cytological and molecular diagnosis of Hürthle cell
thyroid tumors: Analysis of three cases. Mol Clin Oncol.
15:1492021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Di Meo A, Saleeb R, Wala SJ, Khella HW,
Ding Q, Zhai H, Krishan K, Krizova A, Gabril M, Evans A, et al: A
miRNA-based classification of renal cell carcinoma subtypes by PCR
and in situ hybridization. Oncotarget. 9:2092–2104. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Si C, Yu Q and Yao Y: Effect of
miR-146a-5p on proliferation and metastasis of triple-negative
breast cancer via regulation of SOX5. Exp Ther Med. 15:4515–4521.
2018.PubMed/NCBI
|
|
28
|
Chen G, Umelo IA, Lv S, Teugels E, Fostier
K, Kronenberger P, Dewaele A, Sadones J, Geers C and De Grève J:
miR-146a inhibits cell growth, cell migration and induces apoptosis
in non-small cell lung cancer cells. PLoS One. 8:e603172013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang C, Zhang J, Zhang A, Wang Y, Han L,
You Y, Pu P and Kang C: PUMA is a novel target of miR-221/222 in
human epithelial cancers. Int J Oncol. 37:1621–1626.
2010.PubMed/NCBI
|
|
30
|
Rippo MR, Olivieri F, Monsurrò V,
Prattichizzo F, Albertini MC and Procopio AD: MitomiRs in human
inflamm-aging: A hypothesis involving miR-181a, miR-34a and
miR-146a. Exp Gerontol. 56:154–163. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hockenbery D, Nuñez G, Milliman C,
Schreiber RD and Korsmeyer SJ: Bcl-2 is an inner mitochondrial
membrane protein that blocks programmed cell death. Nature.
348:334–336. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bender CE, Fitzgerald P, Tait SW, Llambi
F, McStay GP, Tupper DO, Pellettieri J, Sánchez Alvarado A,
Salvesen GS and Green DR: Mitochondrial pathway of apoptosis is
ancestral in metazoans. Proc Natl Acad Sci USA. 109:4904–4909.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oltvai ZN, Milliman CL and Korsmeyer SJ:
Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that
accelerates programmed cell death. Cell. 74:609–619. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Giuliani A, Cirilli I, Prattichizzo F,
Mensà E, Fulgenzi G, Sabbatinelli J, Graciotti L, Olivieri F,
Procopio AD, Tiano L and Rippo MR: The mitomiR/Bcl-2 axis affects
mitochondrial function and autophagic vacuole formation in
senescent endothelial cells. Aging (Albany NY). 10:2855–2873. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Patel N, Garikapati KR, Ramaiah MJ,
Polavarapu KK, Bhadra U and Bhadra MP: miR-15a/miR-16 induces
mitochondrial dependent apoptosis in breast cancer cells by
suppressing oncogene BMI1. Life Sci. 164:60–70. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun
S, Hong L, Liu J and Fan D: miR-15b and miR-16 modulate multidrug
resistance by targeting BCL2 in human gastric cancer cells. Int J
Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nissen T and Wynn R: The clinical case
report: A review of its merits and limitations. BMC Res Notes.
7:2642014. View Article : Google Scholar : PubMed/NCBI
|