Introduction
Soft tissue sarcoma is a type of tumor that
originates from mesenchymal tissues (1). Soft tissue sarcoma accounts for ~0.7%
of adult malignancies in the USA (2) and consists of >100 subtypes
(3). According to the 4th edition
of the World Health Organization Classification of Tumors of Soft
Tissue and Bone, fibrosarcoma belongs to the subclass of
fibroblastic or myofibroblastic tumors, representing ~3.6% of soft
tissue sarcomas (4,5). Fibrosarcoma typically occurs in
middle-aged and elderly patients, with a median age of onset of 50
years (6). It is commonly revealed
that soft tissue sarcoma occurs in the trunk (13%), extremities
(40–50%), and head and neck regions (9%) (7). The prognosis of patients with soft
tissue sarcoma varies and is associated with both histopathological
type and treatment response (7).
Overall, soft tissue sarcoma has a poor prognosis, with a 5-year
survival rate of <50% and a recurrence rate of >50% after
surgery (8,9). Therefore, an in-depth understanding of
the molecular mechanisms underlying fibrosarcoma pathogenesis could
facilitate the development of novel targeted therapies for this
malignant disease.
To date, researchers have identified ~18 members of
the polypeptide N-acetylgalactosaminyltransferase (GALNT) gene
family that serve important roles in tumor progression, lipid
regulation, type 2 diabetes, neurodevelopmental disorders and
various other diseases (10–13).
As a member of the GALNT family, GALNT12 has two isoforms (Q8IXK2-1
and Q8IXK2-2) and is located on chromosome 9 (14). The GALNT12 protein has a molecular
weight of ~66.9 kDa (14). GALNT12
encodes a glycosyltransferase that catalyzes the first step of
mucin-type O-linked protein glycosylation, transferring
N-acetylgalactosamine (GalNAc) from uridine diphosphate-GalNAc to
serine or threonine residues on polypeptide acceptors (15). Abnormal GALNT12 expression has been
reported in various diseases. For example, abnormal GALNT12
expression has been revealed in several autoimmune diseases
including IgA nephropathy, rheumatoid arthritis and autoimmune
inner ear disease (16–18). Previous studies have reported
genetic variations or abnormal GALNT12 expression in tumors such as
colorectal, glioblastoma, endometrial cancer and lymphoma, which is
associated with their malignant phenotypes (19–23).
However, to the best of our knowledge, GALNT12 expression has not
been reported in fibrosarcoma and its expression pattern and impact
on the malignant behavior of fibrosarcoma is currently unknown.
Therefore, the present study aimed to investigate GALNT12
expression in fibrosarcoma and its influence on biological
functions.
Materials and methods
Patients and tissue samples
In the present study, primary fibroblastic or
myofibroblastic tumors from 22 patients that underwent surgical
resection at Xiangya Hospital of Central South University
(Changsha, China) from 01/01/2017-10/31/2022 were sampled. Patients
were aged between 8 and 84 years, and there were 15 male patients
and 7 female patients. After surgical resection, all specimens were
immediately placed in tissue-protective solution (Biosharp), frozen
in liquid nitrogen and stored at −80°C. Tissue samples were used to
prepare sections for immunohistochemistry (IHC). Fibrosarcoma
tissues from 6 of the 22 cases were also used for mRNA and protein
extraction. Clinicopathological characteristics of enrolled
patients were obtained from medical records. Tumors were classified
histologically using guidelines published in the American Joint
Committee on Cancer Limb/Torso Soft Tissue Sarcoma Staging System
(8th edition, 2017) (24). The
present study was approved by the Ethics Committee of Xiangya
Hospital and written informed consent was obtained from all
participants or their guardians (ethics approval no.
201603079).
Public datasets
Fibroblastic or myofibroblastic tumor datasets
(GSE24369 and GSE21124, respectively) were selected from the Gene
Expression Omnibus (GEO) database for gene selection (control
group, n=6; tumor group, n=6; from different patients) and GSE21124
(control group, n=9; tumor group, n=34; from different
patients).
Cell culture
Human fibrosarcoma HT-1080 cells and human skin
fibroblasts (HSF; HSA-S4) were purchased from Abiowell (Changsha
Abiowell Biotechnology Co., Ltd.) with STR certification reports.
Cells were cultured in high-glucose DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Shanghai ExCell Biology, Inc.) and 1% penicillin/streptomycin at
37°C with 5% CO2.
IHC
IHC was performed as previously described (25). Staining intensity and percentage of
positivity were analyzed by ImageJ/Fiji (2.15.0; Github; http://imagej.net/software/fiji/) (26). Staining was evaluated and graded by
two independent investigators. The staining intensity scoring was 0
(negative), 1 (weak), 2 (moderate) and 3 (strong). The degree of
staining was stratified as 0 (0%), 1 (1–25%), 2 (26–50%), 3
(51–75%) and 4 (76–100%), and was defined as the percentage of
positive staining area in the total tumor invasion area. The final
score of GALNT12 expression was determined by adding the intensity
of staining to the degree of staining; its value was 0–7. The
samples were divided into two groups: Low GALNT12 expression (0–3
points) and high GALNT12 expression (4–7 points). Sections were
classified into high and low expression groups. The primary
antibody used was: Anti-GALNT12 (1:100; cat. no. K108365P; Beijing
Solarbio Science & Technology Co., Ltd.). A HRP-conjugated
antibody (1:200; cat. no. GB23303; Wuhan Servicebio Technology Co.,
Ltd. 1:200 for IHC) was used as a secondary antibody.
Immunocytochemistry
Immunocytochemistry was performed as previously
described (25). The primary
antibody used was: Anti-GALNT12 (1:50; cat. no. K108365P; Beijing
Solarbio Science & Technology Co., Ltd.).
CoraLite594-conjugated Goat Anti-Rabbit IgG (H+L) (1:200; cat. no.
SA00013-4; Proteintech Group, Inc.) was used as a secondary
antibody.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA of fibrosarcoma tissues or HT-1080 cells
was extracted using TransZol Up (TransGen Biotech Co., Ltd.)
according to the manufacturer's instructions. The quality and
concentration of the isolated RNA were analyzed using a NanoDdrop
2000 (Thermo Fisher Scientific, Inc.) and reverse transcribed into
cDNA using the Evo M-MLV RT kit (Accurate Biology) according to the
manufacturer's instructions. RT-qPCR was performed on the Vii7
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using
ribosomal protein S18 (RPS18) as an internal normalization control.
Relative mRNA expression was calculated using the 2−ΔΔCq
method (27). Primer sequences are
listed in Table SI. For qPCR, 2X
SYBR Green qPCR Master Mix (Low ROX) (cat. no. G3321-15; Wuhan
Servicebio Technology Co., Ltd.) was used. The recommended qPCR
procedure was as follows: Stage 1, 95°C for 30 sec for one cycle;
stage 2, 95°C for 15 sec and 60°C for 30 sec for 40 cycles; stage
3, 95°C for 15 sec, 60°C for 60 sec and 95°C for 15 sec for one
cycle.
Western blotting
HSF cells, HT-1080 cells and fibrosarcoma tissues
were lysed in RIPA buffer (Beyotime Institute of Biotechnology)
with protease inhibitors (Wuhan Servicebio Technology Co., Ltd.)
(RIPA buffer:protease inhibitors, 100:1) for 15 min on ice to
extract total proteins. The BCA method was used to determinate
protein concentrations. Subsequently, proteins were boiled with 5X
loading buffer (Wuhan Servicebio Technology Co., Ltd.) for 10 min.
Nuclear proteins were isolated using a commercial kit (cat. no.
R0050; Beijing Solarbio Science & Technology Co., Ltd.)
following the manufacturer's protocol. Proteins (50 µg/lane) were
separated using a 10% gel with SDS-PAGE and transferred to PVDF
membranes. Membranes were blocked with 5% BSA (BioFroxx; neoFroxx
GmbH) or 5% milk (BD Biosciences) in TBS for 1 h at room
temperature before incubating with primary antibodies (overnight at
4°C) and secondary antibodies (1 h at room temperature). 5% BSA was
used to block membranes to assess p-YAP1; 5% milk was used to block
membranes to assess other molecules (GALNT12, GAPDH, YAP1 and Lamin
B1). The primary antibodies used were: GALNT12 (cat. no. K108365P;
Beijing Solarbio Science & Technology Co., Ltd.), GAPDH (cat.
no. GB15004; Wuhan Servicebio Technology Co., Ltd.), yes1
associated transcriptional regulator (YAP1; cat. no. ET1608-30;
HUABIO), phosphorylated (p)-YAP1 (cat. no. ET1611-69; HUABIO) and
Lamin B1 (cat. no. ET1606-27; HUABIO). The HRP-conjugated secondary
antibodies (cat. no. GB23303; Wuhan Servicebio Technology Co.,
Ltd.) were used. Protein bands were visualized using ECL reagent
(Wuhan Servicebio Technology Co., Ltd.) in ChemiDoc™ (BioRad
Laboratories, Inc.). ImageJ/Fiji (2.15.0; Github) was used to
measure the bands densitometry.
Nuclear protein extraction
Nuclear proteins were extracted using the Nuclear
Protein Extraction kit (cat. no. R0050; Beijing Solarbio Science
& Technology Co., Ltd.) according to the manufacturer's
protocols. Cells were washed with 1× PBS, and then cytoplasmic
protein extraction reagent was added and the mixture was incubated
on ice for 10 min. Cells were centrifuged (12,000 × g for 10 min at
4°C) and the supernatant was collected to obtain cytoplasmic
proteins. Subsequently, nuclear protein extraction reagent was
added to the remaining cell sediment. This mixture was incubated on
ice for 10 min, and then the supernatant was collected after
centrifugation (12,000 × g for 10 min at 4°C) to obtain nuclear
proteins.
Small interfering (si)RNA
knockdown
HT-1080 cells (5×105) were seeded in
6-well plates before transfection. At 70% confluence, cells were
transfected with 100 pmol siRNA against GALNT12 (siGALNT12-1 and
siGALNT12-2) or non-targeting control siRNA (Table SII; TsingKe Biological Technology)
using Lipo6000 (Beyotime Institute of Biotechnology) in Opti-MEM
(Gibco; Thermo Fisher Scientific, Inc.). After 8 h at 37°C,
high-glucose DMEM supplemented with 10% fetal bovine serum
(Shanghai ExCell Biology, Inc.) was added. After 48 h, transfected
cells were used for further experiments.
Cell Counting Kit-8 (CCK-8) assay
Cell viability was assessed using a CCK-8 assay.
HT-1080 cells were seeded in 96-well plates (2×103
cells/well) and transfected with GALNT12 siRNAs and negative
control siRNAs. After transfection, 10 µl CCK-8 reagent (Beyotime
Institute of Biotechnology) was added to each well and incubated
for 2 h in the dark. Absorbance at 450 nm was measured using a
SpectrumMax Plus384 microplate reader (Molecular Devices, LLC).
EdU assay
HT-1080 cells (2×105 cells per well) were
seeded in 6-well plates and allowed to adhere at 37°C for 24 h.
Cells were incubated with EdU reagent (Wuhan Servicebio Technology
Co., Ltd.), fixed with 4% paraformaldehyde, permeabilized with
Triton X-100 and stained following the manufacturer's protocol.
Nuclei were counterstained with 5 µg/ml Hoechst 33342 (Wuhan
Servicebio Technology Co., Ltd.) for 15 min at room temperature.
Cells were imaged using a Nikon inverted fluorescence microscope
(Nikon ECLIPSE Ti2; Nikon Corporation).
Wound healing assay
HT-1080 cells were seeded in 6-well plates
(3×105 cells/well) and grown to 100% confluence. A
sterile pipette tip was used to create vertical scratches across
the cell monolayer and the cells were than cultured with fresh
serum-free medium at 37°C. Images were captured at 0 and 24 h
post-scratch using a Nikon inverted light microscope (Nikon ECLIPSE
Ti2). Cell migration distance was judged by comparing the distance
between the initial scratch (0 h) and the scratch at the 24-h
observation point, which was measured using ImageJ/Fiji software
(2.15.0; Github).
Transwell assay
Transwell chambers (pore size, 8 µm; Corning, Inc.)
were prepared with 500 µl serum-free DMEM in the upper chamber and
3×105 HT-1080 cells/well. The lower chamber contained
500 µl DMEM with 20% FBS. After culturing at 37°C for 24 h, cells
were fixed in 4% paraformaldehyde for 15 min, stained with 0.1%
crystal violet for 10 min at room temperature, imaged under a light
microscope (Nikon ECLIPSE Ti2) and counted.
Gene set enrichment analysis
(GSEA)
GSEA (version 4.2.3, http://www.gsea-msigdb.org/gsea/index.jsp) was
performed on the GSE2553 dataset to identify key enriched pathways.
Normalized Enrichment Score (NES), P-value and false discovery rate
(FDR) are the main observables for enrichment analysis (cut off
values: NES >1, P<0.05, FDR <0.25).
Statistical analysis
GraphPad Prism (version 9.0; Dotmatics) was used for
data analysis. Paired and unpaired Student's t-test, Fisher's exact
test and one-way ANOVA (followed by Turkey's Honest Significant
Difference test) were used to analyze statistical differences
between groups. P<0.05 was considered to indicate a
statistically significance difference.
Results
GALNT12 is upregulated in fibroblastic
and myofibroblastic tumors
To explore gene expression level changes in
fibrosarcoma, the GSE24369 and GSE21124 public datasets from GEO
were analyzed. A total of 308 and 578 differentially expressed
genes (DEGs) in GSE24369 and GSE21124 were identified, respectively
(P<0.05; log2 fold change >1.5). Comparisons
between these datasets identified 15 common DEGs (Fig. 1A). Among these, fatty acid binding
protein 3, GALNT12, protein regulator of cytokinesis 1 and CDC28
protein kinase regulatory subunit 2 were highly expressed in tumors
(Fig. 1B). Based on the statistical
significance observed and the previous literature (19–23),
GALNT12 was selected for further analysis. GALNT12 mRNA levels were
significantly increased in the tumor groups in both datasets when
compared with the control group (P<0.01; Fig. 1C). In the GSE21124 dataset, GALNT12
demonstrated significantly increased expression among GALNT family
members GALNT7, GALNT10 and GALNT12 when compared with the control
group (P<0.01; Fig. 1D).
Immunofluorescent analysis demonstrated cytoplasmic localization of
the GALNT12 protein in HT-1080 cells (Fig. 1E).
GALNT12 expression is upregulated in
fibrosarcoma
Analysis of both public and acquired patient data
demonstrated increased GALNT12 mRNA and protein expression levels
in fibrosarcoma. In the GSE2719 GEO dataset, GALNT12 mRNA
expression levels were increased in tumor tissues compared with
normal tissues (P<0.05; Fig.
2A). RT-qPCR results also demonstrated increased GALNT12 mRNA
expression levels in HT-1080 cells compared with normal fibroblasts
(P<0.05; Fig. 2B).
Immunoblotting demonstrated increased GALNT12 protein expression
levels in six fibrosarcoma samples compared with healthy tissue
controls (Fig. 2C).
High GALNT12 expression levels are
associated with poor prognosis in patients with fibrosarcoma
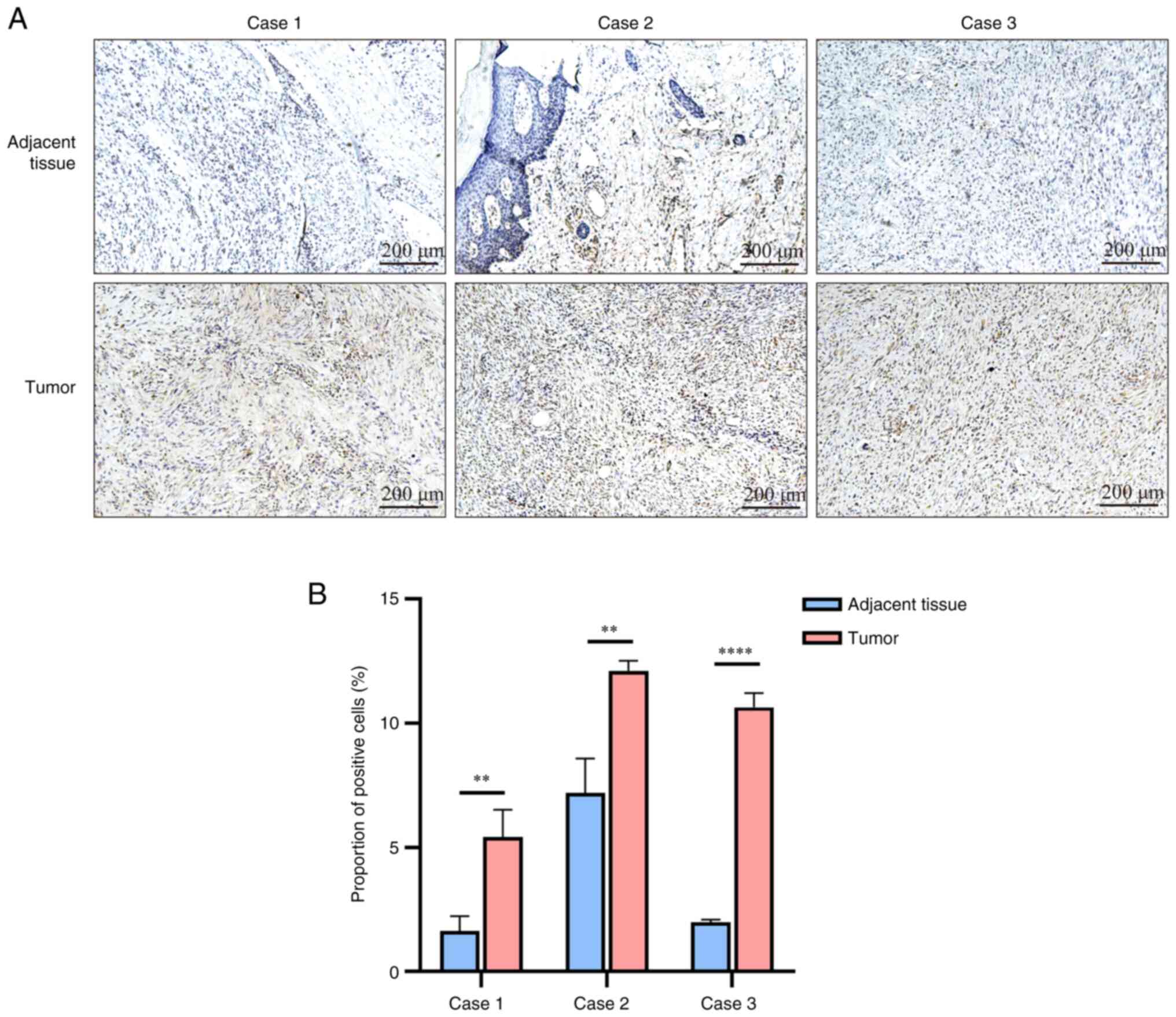
IHC staining demonstrated an increased expression of
GALNT12 in fibrosarcoma tissues compared with adjacent healthy
tissue (Fig. 3A and B). High
GALNT12 expression was significantly associated with larger tumor
size (P=0.002) and advanced T-stage (P=0.027) (Table I).
 | Table I.Relationship between GALNT12
expression and clinical factors. |
Table I.
Relationship between GALNT12
expression and clinical factors.
| Patient
characteristic | Number of patients
(n) | Low GALNT12
expression (n) | High GALNT12
expression (n) | P-value |
|---|
| Age, years |
|
|
|
|
|
≤35 | 11 | 7 | 4 | 0.080 |
|
>35 | 11 | 2 | 9 |
|
| Sex |
|
|
|
|
|
Male | 15 | 5 | 10 | 0.376 |
|
Female | 7 | 4 | 3 |
|
| Tumor size,
cm3 |
|
|
|
|
|
≤43 | 10 | 8 | 2 | 0.002 |
|
>43 | 12 | 1 | 11 |
|
| Surrounding
invasion of tissues |
|
|
|
|
|
Yes | 10 | 4 | 6 | >0.999. |
| No | 12 | 5 | 7 |
|
| Tumor stage |
|
|
|
|
| T1 | 9 | 7 | 2 | 0.027 |
| T2 | 7 | 1 | 6 |
|
| T3 | 5 | 1 | 4 |
|
| T4 | 1 | 0 | 1 |
|
| Node stage |
|
|
|
|
| N0 | 20 | 8 | 12 | >0.999. |
| N1 | 2 | 1 | 1 |
|
| Metastasis
stage |
|
|
|
|
| M0 | 18 | 8 | 10 | 0.616 |
| M1 | 4 | 1 | 3 |
|
GALNT12 knockdown suppresses
proliferation of HT-1080 cells
The present study investigated the impact of GALNT12
on the biological behavior of fibrosarcoma cells. siRNAs targeting
GALNT12 were used to knock down GALNT12 expression levels in
HT-1080 cells. The efficiency of transfection was confirmed with
RT-qPCR analysis (Fig. 4A).
Subsequently, the CCK-8 assay demonstrated reduced cell viability
in the GALNT12 knockdown groups compared with the negative control
group (Fig. 4B). Moreover, the EdU
assay demonstrated a significant decrease in cell proliferation in
the GALNT12 knockdown groups compared with the negative control
(P<0.05; Fig. 4C and D).
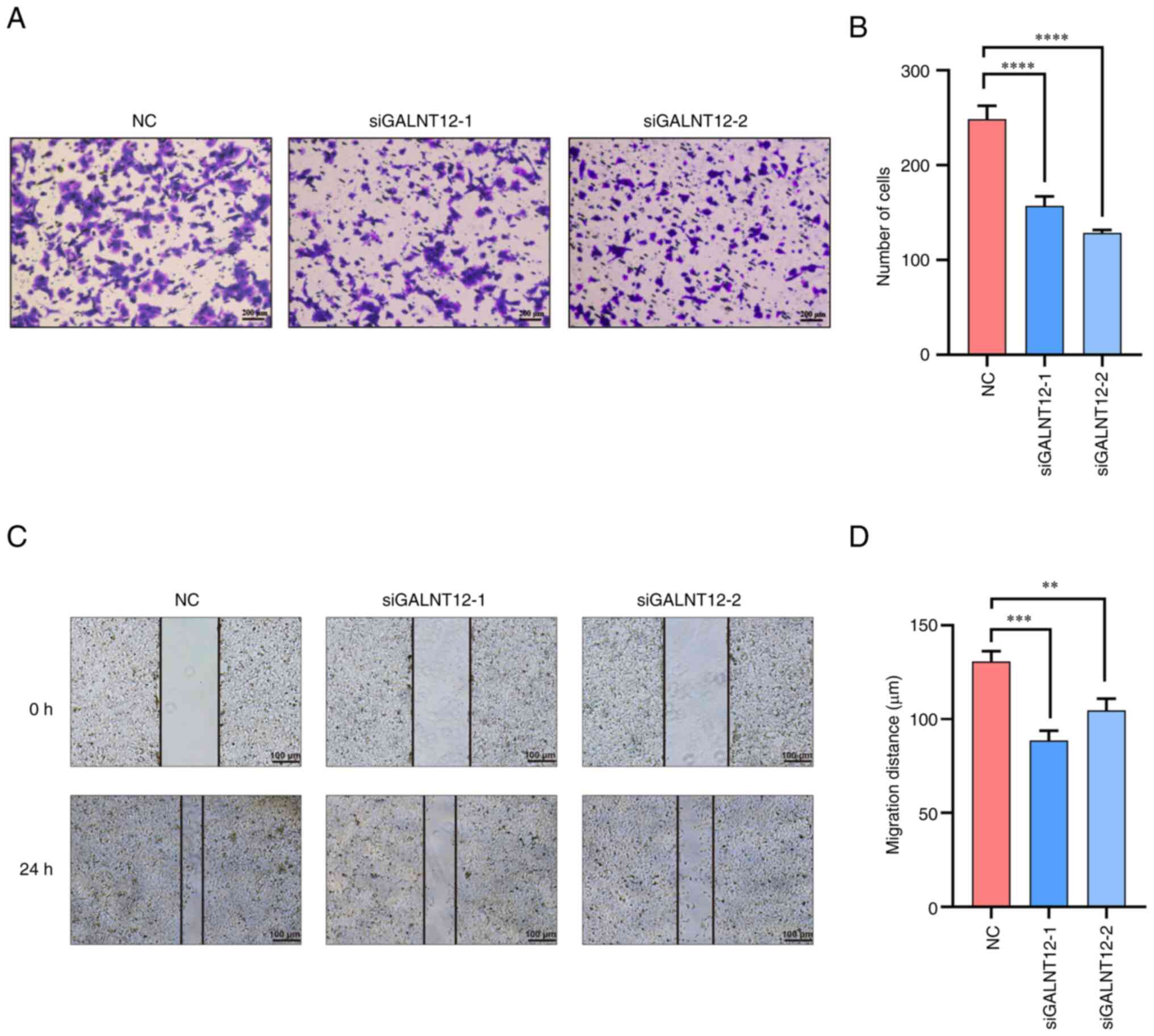
GALNT12 knockdown impairs migration of
HT-1080 cells
To assess the influence of GALNT12 on cell motility,
both wound healing and Transwell assays were used with transfected
HT-1080 cells. The Transwell assay demonstrated a significant
reduction in the number of cells traversing the chamber in the
GALNT12 knockdown groups compared with the control group
(P<0.0001; Fig. 5A and B).
Additionally, the wound healing assay demonstrated a reduced
migration in the GALNT12 knockdown groups compared with the control
group (P<0.01; Fig. 5C and
D).
GALNT12 enhances tumor malignancy by
facilitating YAP1 nuclear localization
Using the GSE2553 dataset from the GEO database,
samples were categorized into high and low GALNT12 mRNA expression
groups. Through GSEA, the YAP1 signaling pathway was identified as
pivotal in fibrosarcoma pathogenesis (Fig. 6A). Following GALNT12 knockdown, the
phosphorylation level of YAP1 increased compared with the negative
control group (Fig. 6B). The
nuclear protein extraction assay demonstrated reduced YAP1 nuclear
localization in the siGALNT12-1 and siGALNT12-2 groups compared
with the negative control (Fig. 6C and
D). Furthermore, RT-qPCR analysis demonstrated a significantly
decreased expression of YAP1 downstream target genes, including
CYR61, AMOTL2 and BIRC5, compared with the corresponding negative
control groups (P<0.05) (Fig.
6E). Based on these findings, it could be suggested that
GALNT12 modulates the proliferation and migration capabilities of
the fibrosarcoma cell line HT-1080 by influencing YAP1 nuclear
localization (Fig. 7).
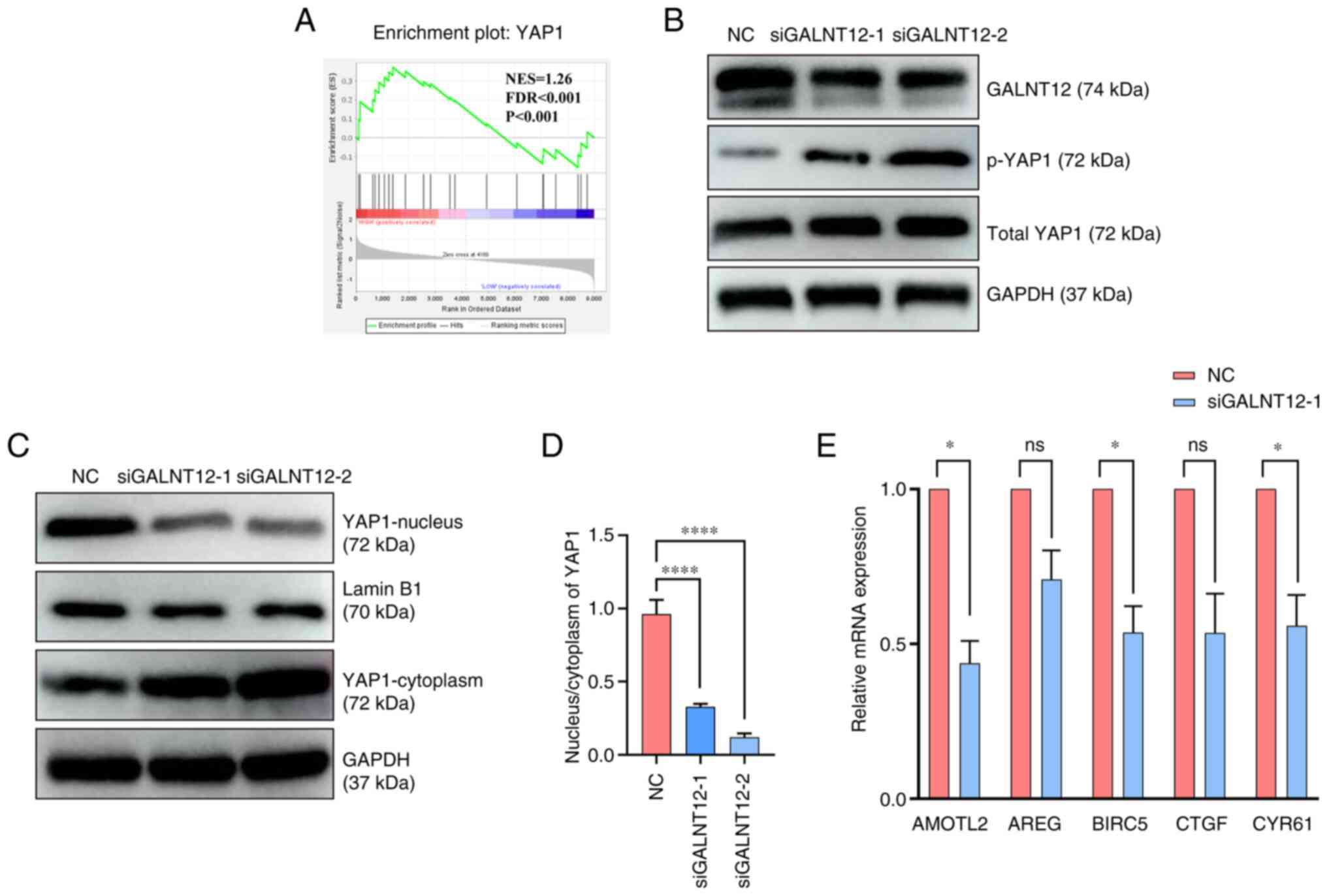 | Figure 6.GALNT12 enhances malignancy by
facilitating YAP1 nuclear localization. (A) Gene Set Enrichment
Analysis was performed on the GSE2553 dataset. (B) Protein
expression levels of GALNT12, YAP1, p-YAP1 and GAPDH were measured
in transfected HT-1080 cells using western blotting. (C) Protein
expression levels of YAP1 in the nucleus and cytoplasm of cells
were measured and (D) the ratio of localization was quantified.
One-way ANOVA (followed by Tukey's Honest Significant Difference
test) was used to analyze the differences between the three groups.
(E) Relative mRNA levels of AMOTL2, AREG, BIRC5, CTGF and CYR61
were detected using reverse transcription-quantitative PCR.
siGALNT12-1 was used for this assay to interfere with the
expression of GALNT12. Unpaired Student's t-tests were used to
analyze the differences between the two groups. Data were presented
as the mean ± standard deviation (n=3). *P<0.05; ****P<0.001.
ns, not significant; si, small interfering; NC, negative control;
GALNT12, polypeptide N-acetylgalactosaminyltransferase 12; YAP1,
yes1 associated transcriptional regulator; p, phosphorylated;
CYR61, cysteine-rich angiogenic inducer 61; CTGF, connective tissue
growth factor; AREG, amphiregulin; AMOTL2, angiomotin-like 2;
BIRC5, baculoviral IAP repeat containing 5; NES, normalized
enrichment score; FDR, false discovery rate. |
Discussion
Overcoming the challenges posed by therapeutic
inefficacy and the elusive molecular mechanisms in fibrosarcoma
remains a significant hurdle. Identifying effective therapeutic
targets for this condition remains paramount. GALNT has emerged as
an important component in the development of human tumors, exerting
influence over a spectrum of biological processes encompassing
proliferation, invasion, apoptosis, angiogenesis and the
epithelial-mesenchymal transition (EMT) of tumor cells (28–31).
For example, GALNT2 has been implicated in enhancing migration and
proliferation of colorectal cancer cells via AXL signaling
(32). A previous study reported
that GALNT3 suppressed lung cancer by inhibiting myeloid-derived
suppressor cell infiltration and angiogenesis via a TNFR and c-MET
pathway-dependent manner (33).
Additionally, in trophoblast stem cells, blastocyst trophectoderm
and human mammary epithelial cells, GALNT3 O-GalNAc glycosylation
could promote the epithelial phenotype and the upregulation of
GALNT3 expression is associated with the EMT (34). In various types of digestive system
malignancies, including hepatocellular carcinoma, gastric cancer
and esophageal cancer, increased GALNT14 expression is associated
with accelerated tumor growth, reduced chemotherapy responsiveness
and poor patient prognoses (35).
Additionally, upregulation of GALNT7 in prostate cancer induces
O-glycosylation alterations, which ultimately promotes tumor growth
(36). GALNT5 and VVL-binding
glycans have been identified as drivers of cholangiocarcinoma
metastasis through the AKT/ERK signaling pathway (37). Furthermore, GALNT12 variants are
associated with increased susceptibility to colorectal cancer,
demonstrating the significance of glycosylation pathway aberrations
in this disease (20). GALNT12 has
been reported to increase proliferation, invasion and metastasis of
glioma cells via activation of the PI3K/AKT/mTOR signaling pathway
(21). Moreover, Dasgeb et
al (38) suggested that patched
1, ephrin type-B receptor 2, Ret proto-oncogene and GALNT12 may
contribute to the synergistic oncogene-driven malignant
transformation in ulcerating basal cell carcinomas. GALNT3 inhibits
tumor cell growth in lung cancer or promotes the epithelial
phenotype, whereas the majority of other GALNT family members
promote tumor progression (20,21,34–38).
This may be associated with their different molecular structures
and the tumor type. These previous studies collectively suggest
that the GALNT family of proteins may serve an important role in
tumor genesis, influencing key downstream signaling pathways that
culminate in a malignant phenotype (20,21,34–38).
In the present study, these findings were corroborated and it was
demonstrated that an upregulation of GALNT12 occurred in human
fibrosarcoma tissues and the HT-1080 cell line. Notably, the
increased expression of GALNT12 was associated with tumor size and
T-stage, indicating a poor prognosis for patients. In vitro
assays further demonstrated a reduction in HT-1080 proliferation
and migration in the GALNT12 knockdown groups compared with the
controls. This collective evidence supported the hypothesis that
GALNT12 could be a potential therapeutic target for fibrosarcoma.
However, due to the diverse array of tumor types, distinct
expression patterns within each tumor and molecular disparities
among GALNT family members, GALNT proteins likely use varying
molecular mechanisms to drive tumor progression. Therefore, further
research is required to unravel the molecular intricacies
underpinning fibrosarcoma progression.
The activation of the YAP1 signaling pathway has
previously been implicated to foster malignant behaviors such as
cell proliferation, invasion, EMT and metastasis (39–41).
For example, Liu et al (42)
elucidated the role of the signaling axis
ZIP4-miR-373-LATS2-ZEB1/YAP1-ITGA3 in pancreatic cancer metastasis
and EMT plasticity. Ajani et al (43) reported a significant upregulation of
YAP1 in peritoneal carcinomatosis tumor cells, which conferred
cancer stem cell properties and potentially promoted metastasis.
Additionally, in the present study, GSEA demonstrated that
dysregulation of GALNT12 affected the downstream YAP1 signaling
pathway. Knockdown of GALNT12 significantly reduced the nuclear
localization of YAP1, which consequently suppressed the activation
of downstream target genes and impeded tumor cell growth. Thus, it
could be suggested that GALNT12 promoted the growth of the
fibrosarcoma cell line HT-1080 through facilitation of YAP1 nuclear
localization. Further elucidation of the YAP1 signaling pathway
could hold promise for unravelling the molecular underpinnings of
fibrosarcoma.
In summary, the present study highlighted the
increased expression levels of GALNT12 in clinical tumor tissues,
which was associated with worsened patient prognoses. Through In
vitro assays, the pro-carcinogenic role of GALNT12 in the
fibrosarcoma cell line HT-1080 was investigated, including
analyzing the cell viability, proliferation and migration potential
of these cells. The present study demonstrated GALNT12 as an
important oncogenic protein in fibrosarcoma, and offered fresh
perspectives and potential future avenues for targeted therapeutic
interventions to treat this disease.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study received support from the Academic Construction
Funding of Xiangya Hospital, Central South University (grant no.
XK60000691391).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PZ and SY conceptualized the study. WF, JZ, SZ and
YP contributed to specimen collection. SY and WF conducted the
experiments. SY performed data analysis and drafted the manuscript.
SY and WF confirm the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Informed written consent was obtained from all
patients. The study was conducted in accordance with the
Declaration of Helsinki and approved by the Ethics Committee of
Xiangya Hospital, Central South University (ethics approval no.
201603079).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pankova V, Thway K, Jones RL and Huang PH:
The extracellular matrix in soft tissue sarcomas: Pathobiology and
cellular signalling. Front Cell Dev Biol. 9:7636402021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
American Cancer Society, . What is a soft
tissue sarcoma? American Cancer Society; Atlanta, GA: 2023,
https://cancerstatisticscenter.cancer.org/#!/
|
|
3
|
Howlader N, Noone AM, Krapcho M, Miller D,
Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, et al:
SEER cancer statistics review, 1975–2018. National Cancer
Institute; Bethesda, MD: 2020
|
|
4
|
Fletcher CDM, Bridge JA, Hogendoorn PCW
and Mertens F: WHO classification of tumours of soft tissue and
bone. WHO Classification of Tumours. 4th edition. Vol. 5. IARC
Press; Lyon: 2013
|
|
5
|
Toro JR, Travis LB, Wu HJ, Zhu K, Fletcher
CDM and Devesa SS: Incidence patterns of soft tissue sarcomas,
regardless of primary site, in the surveillance, epidemiology and
end results program, 1978–2001: An analysis of 26,758 cases. Int J
Cancer. 119:2922–2930. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bahrami A and Folpe AL: Adult-type
fibrosarcoma: A reevaluation of 163 putative cases diagnosed at a
single institution over a 48-year period. Am J Surg Pathol.
34:1504–1513. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gamboa AC, Gronchi A and Cardona K:
Soft-tissue sarcoma in adults: An update on the current state of
histiotype-specific management in an era of personalized medicine.
CA Cancer J Clin. 70:200–229. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Francis M, Charman J, Dennis N, Lawrence G
and Grimer R: Bone and soft tissue sarcomas. UK Incidence and
Survival: 1996 to 2010. Version 2.0. National Cancer Intelligence
Network; Bethesda, MD: 2013
|
|
9
|
Folpe AL: Fibrosarcoma: A review and
update. Histopathology. 64:12–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Phelan CM, Tsai YY, Goode EL, Vierkant RA,
Fridley BL, Beesley J, Chen XQ, Webb PM, Chanock S, Cramer DW, et
al: Polymorphism in the GALNT1 gene and epithelial ovarian cancer
in non-Hispanic white women: The ovarian cancer association
consortium. Cancer Epidemiol Biomarkers Prev. 19:600–604. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khetarpal SA, Schjoldager KT,
Christoffersen C, Raghavan A, Edmondson AC, Reutter HM, Ahmed B,
Ouazzani R, Peloso GM, Vitali C, et al: Loss of function of GALNT2
lowers high-density lipoproteins in humans, nonhuman primates, and
rodents. Cell Metab. 24:234–245. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marucci A, di Mauro L, Menzaghi C,
Prudente S, Mangiacotti D, Fini G, Lotti G, Trischitta V and Di
Paola R: GALNT2 expression is reduced in patients with Type 2
diabetes: Possible role of hyperglycemia. PLoS One. 8:e701592013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Reuter MS, Tawamie H, Buchert R, Hosny
Gebril O, Froukh T, Thiel C, Uebe S, Ekici AB, Krumbiegel M, Zweier
C, et al: Diagnostic yield and novel candidate genes by exome
sequencing in 152 consanguineous families with neurodevelopmental
disorders. JAMA Psychiatry. 74:293–299. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo JM, Zhang Y, Cheng L, Iwasaki H, Wang
H, Kubota T, Tachibana K and Narimatsu H: Molecular cloning and
characterization of a novel member of the UDP-GalNAc:polypeptide
N-acetylgalactosaminyltransferase family, pp-GalNAc-T12. FEBS Lett.
524:211–218. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fernandez AJ, Daniel EJP, Mahajan SP, Gray
JJ, Gerken TA, Tabak LA and Samara NL: The structure of the
colorectal cancer-associated enzyme GalNAc-T12 reveals how
nonconserved residues dictate its function. Proc Natl Acad Sci USA.
116:20404–20410. 2015. View Article : Google Scholar
|
|
16
|
Wang YN, Zhou XJ, Chen P, Yu GZ, Zhang X,
Hou P, Liu LJ, Shi SF, Lv JC and Zhang H: Interaction between
GALNT12 and C1GALT1 associates with galactose-deficient IgA1 and
IgA nephropathy. J Am Soc Nephrol. 32:545–552. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hayashi S, Matsubara T, Fukuda K, Maeda T,
Funahashi K, Hashimoto M, Kamenaga T, Takashima Y and Kuroda R: A
genome-wide association study identifying the SNPs predictive of
rapid joint destruction in patients with rheumatoid arthritis.
Biomed Rep. 14:312021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang J, Wang N and Xu A: miR-10b-3p,
miR-8112 and let-7j as potential biomarkers for autoimmune inner
ear diseases. Mol Med Rep. 20:171–181. 2019.PubMed/NCBI
|
|
19
|
Seguí N, Pineda M, Navarro M, Lázaro C,
Brunet J, Infante M, Durán M, Soto JL, Blanco I, Capellá G and
Valle L: GALNT12 is not a major contributor of familial colorectal
cancer type X. Hum Mutat. 35:50–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Evans DR, Venkitachalam S, Revoredo L,
Dohey AT, Clarke E, Pennell JJ, Powell AE, Quinn E, Ravi L, Gerken
TA, et al: Evidence for GALNT12 as a moderate penetrance gene for
colorectal cancer. Hum Mutat. 39:1092–1101. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zheng Y, Liang M, Wang B, Kang L, Yuan Y,
Mao Y and Wang S: GALNT12 is associated with the malignancy of
glioma and promotes glioblastoma multiforme in vitro by activating
Akt signaling. Biochem Biophys Res Commun. 610:99–106. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Y, Yu M, Yang JX, Cao DY, Zhang Y,
Zhou HM, Yuan Z and Shen K: Genomic comparison of endometrioid
endometrial carcinoma and its precancerous lesions in chinese
patients by high-depth next generation sequencing. Front Oncol.
9:1232019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gibson TM, Wang SS, Cerhan JR, Maurer MJ,
Hartge P, Habermann TM, Davis S, Cozen W, Lynch CF, Severson RK, et
al: Inherited genetic variation and overall survival following
follicular lymphoma. Am J Hematol. 87:724–726. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cates JMM: The AJCC 8th edition staging
system for soft tissue sarcoma of the extremities or trunk: A
cohort study of the SEER database. J Natl Compr Canc Netw.
16:144–152. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu S, Zhang Y, Li Q, Zhang Z, Zhao G and
Xu J: CLDN6 promotes tumor progression through the YAP1-snail1 axis
in gastric cancer. Cell Death Dis. 10:9492019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schindelin J, Arganda-Carreras I, Frise E,
Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S,
Schmid B, et al: Fiji: An open-source platform for biological-image
analysis. Nat Methods. 9:676–682. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu Q, Liu HO, Liu YD, Liu WS, Pan D, Zhang
WJ, Yang L, Fu Q, Xu JJ and Gu JX: Decreased expression of
hepatocyte nuclear factor 4α (Hnf4α)/microRNA-122 (miR-122) axis in
hepatitis B virus-associated hepatocellular carcinoma enhances
potential oncogenic GALNT10 protein activity. J Biol Chem.
290:1170–1185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang L, Gallup M, Zlock L, Chen YT,
Finkbeiner WE and McNamara NA: Pivotal role of MUC1 glycosylation
by cigarette smoke in modulating disruption of airway adherens
junctions in vitro. J Pathol. 234:60–73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gaziel-Sovran A, Segura MF, Di Micco R,
Collins MK, Hanniford D, Vega-Saenz de Miera E, Rakus JF, Dankert
JF, Shang S, Kerbel RS, et al: miR-30b/30d regulation of GalNAc
transferases enhances invasion and immunosuppression during
metastasis. Cancer Cell. 20:104–118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Beaman EM, Carter DRF and Brooks SA:
GALNTs: Master regulators of metastasis-associated
epithelial-mesenchymal transition (EMT)? Glycobiology. 32:556–579.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liao YY, Chuang YT, Lin HY, Lin NY, Hsu
TW, Hsieh SC, Chen ST, Hung JS, Yang HJ, Liang JT, et al: GALNT2
promotes invasiveness of colorectal cancer cells partly through
AXL. Mol Oncol. 17:119–133. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Park MS, Yang AY, Lee JE, Kim SK, Roe JS,
Park MS, Oh MJ, An HJ and Kim MY: GALNT3 suppresses lung cancer by
inhibiting myeloid-derived suppressor cell infiltration and
angiogenesis in a TNFR and c-MET pathway-dependent manner. Cancer
Lett. 521:294–307. 2021.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Raghu D, Mobley RJ, Shendy NAM, Perry CH
and Abell AN: GALNT3 maintains the epithelial state in trophoblast
stem cells. Cell Rep. 26:3684–3697.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin WR and Yeh CT: GALNT14: An emerging
marker capable of predicting therapeutic outcomes in multiple
cancers. Int J Mol Sci. 21:14912020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Scott E, Hodgson K, Calle B, Turner H,
Cheung K, Bermudez A, Marques FJG, Pye H, Yo EC, Islam K, et al:
Upregulation of GALNT7 in prostate cancer modifies O-glycosylation
and promotes tumour growth. Oncogene. 42:926–937. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Detarya M, Sawanyawisuth K, Aphivatanasiri
C, Chuangchaiya S, Saranaruk P, Sukprasert L, Silsirivanit A, Araki
N, Wongkham S and Wongkham C: The O-GalNAcylating enzyme GALNT5
mediates carcinogenesis and progression of cholangiocarcinoma via
activation of AKT/ERK signaling. Glycobiology. 30:312–324. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dasgeb B, Leila Y, Saeidian AH, Kang J,
Shi W, Shoenberg E, Ertel A, Fortina P, Vahidnezhad H and Uitto J:
Genetic predisposition to numerous large ulcerating basal cell
carcinomas and response to immune therapy. Int J Dermatol Venereol.
4:70–75. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ou C, Sun Z, He X, Li X, Fan S, Zheng X,
Peng Q, Li G, Li X and Ma J: Targeting YAP1/LINC00152/FSCN1
signaling axis prevents the progression of colorectal cancer. Adv
Sci (Weinh). 7:19013802019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yuan Y, Park J, Feng A, Awasthi P, Wang Z,
Chen Q and Iglesias-Bartolome R: YAP1/TAZ-TEAD transcriptional
networks maintain skin homeostasis by regulating cell proliferation
and limiting KLF4 activity. Nat Commun. 11:14722020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim M, Ly SH, Xie Y, Duronio GN,
Ford-Roshon D, Hwang JH, Sulahian R, Rennhack JP, So J, Gjoerup O,
et al: YAP1 and PRDM14 converge to promote cell survival and
tumorigenesis. Dev Cell. 57:212–227.e8. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu M, Zhang Y, Yang J, Zhan H, Zhou Z,
Jiang Y, Shi X, Fan X, Zhang J, Luo W, et al: Zinc-dependent
regulation of ZEB1 and YAP1 coactivation promotes
epithelial-mesenchymal transition plasticity and metastasis in
pancreatic cancer. Gastroenterology. 160:1771–1783.e1. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ajani JA, Xu Y, Huo L, Wang R, Li Y, Wang
Y, Pizzi MP, Scott A, Harada K, Ma L, et al: YAP1 mediates gastric
adenocarcinoma peritoneal metastases that are attenuated by YAP1
inhibition. Gut. 70:55–66. 2021. View Article : Google Scholar : PubMed/NCBI
|