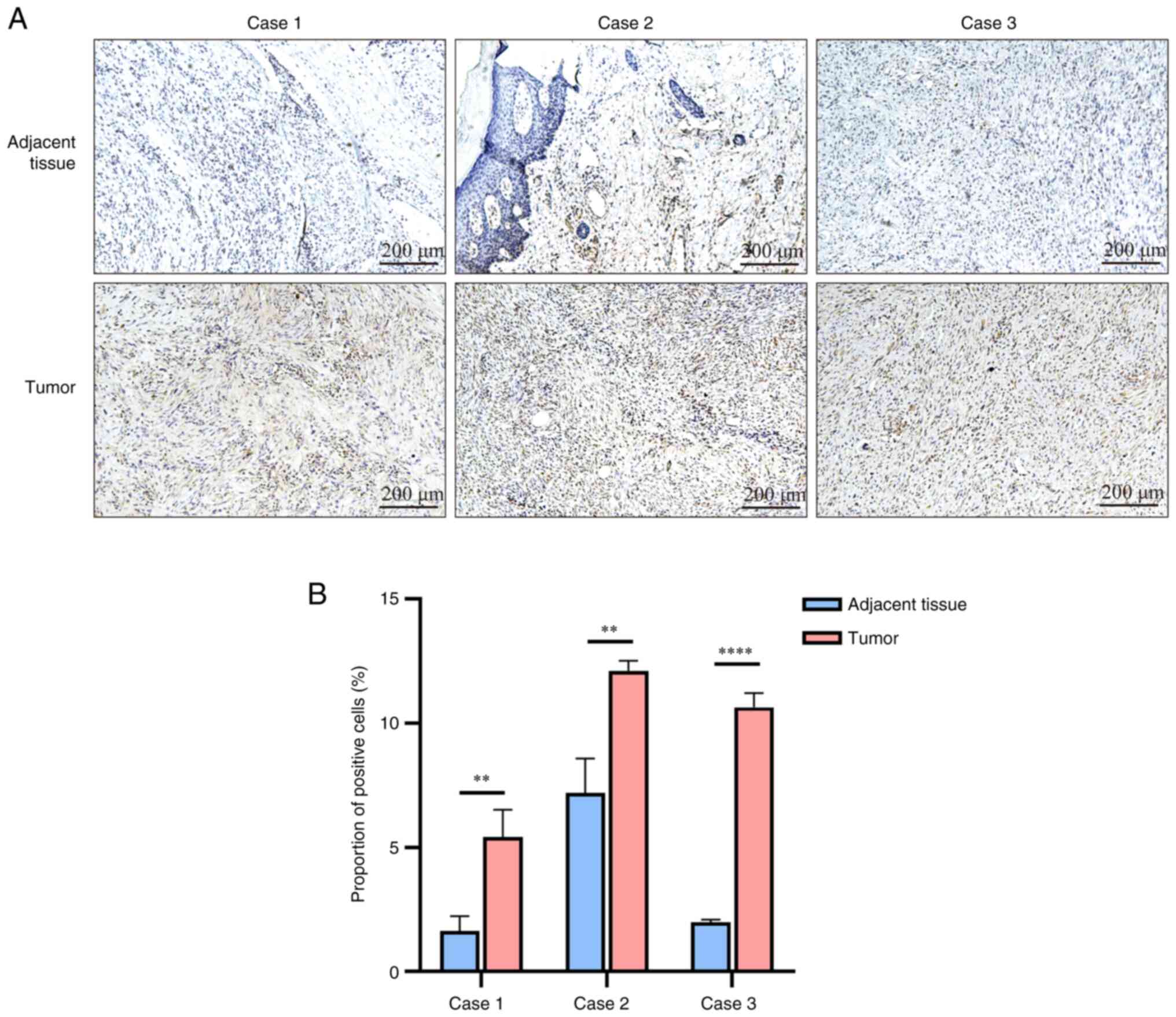
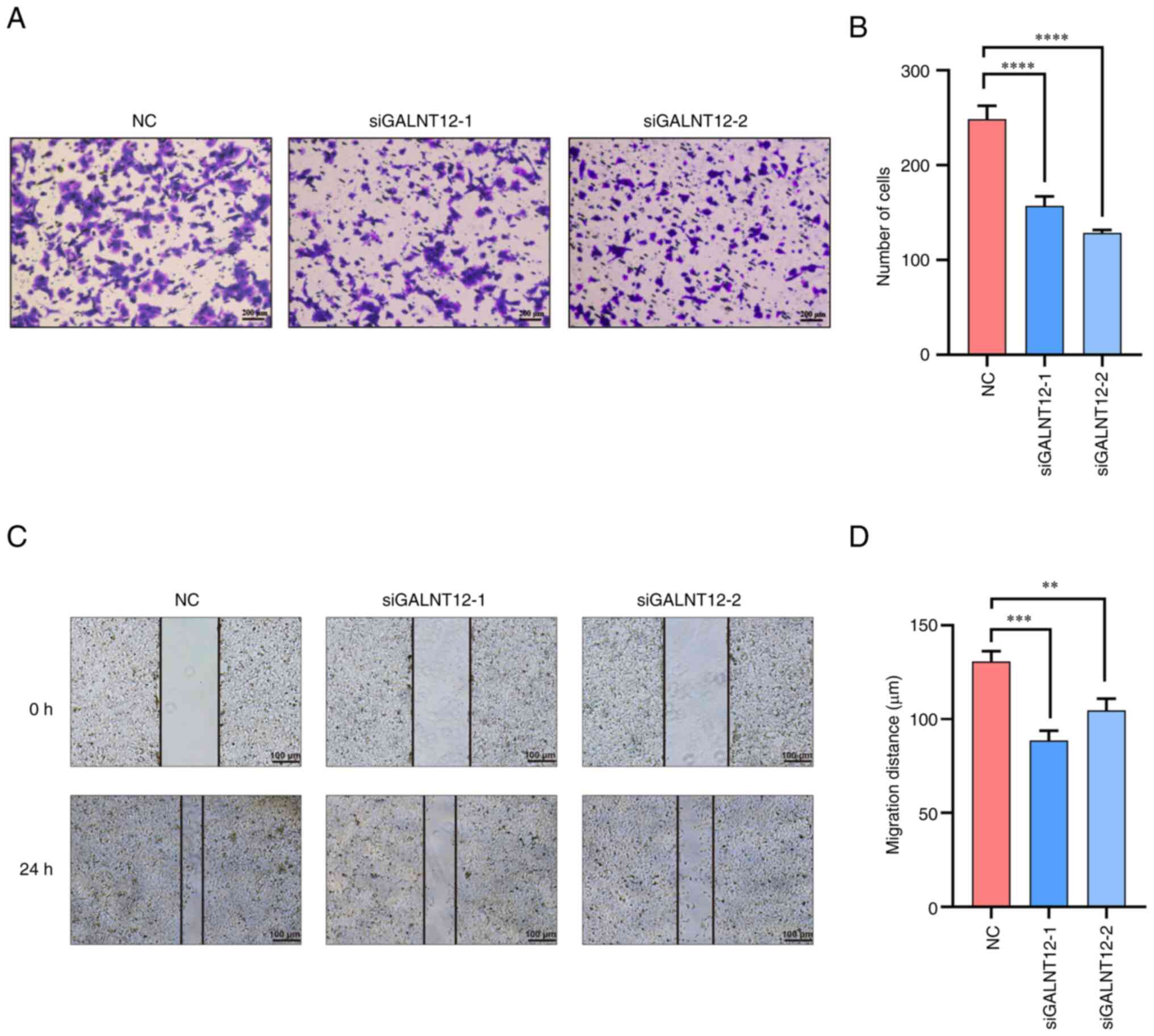
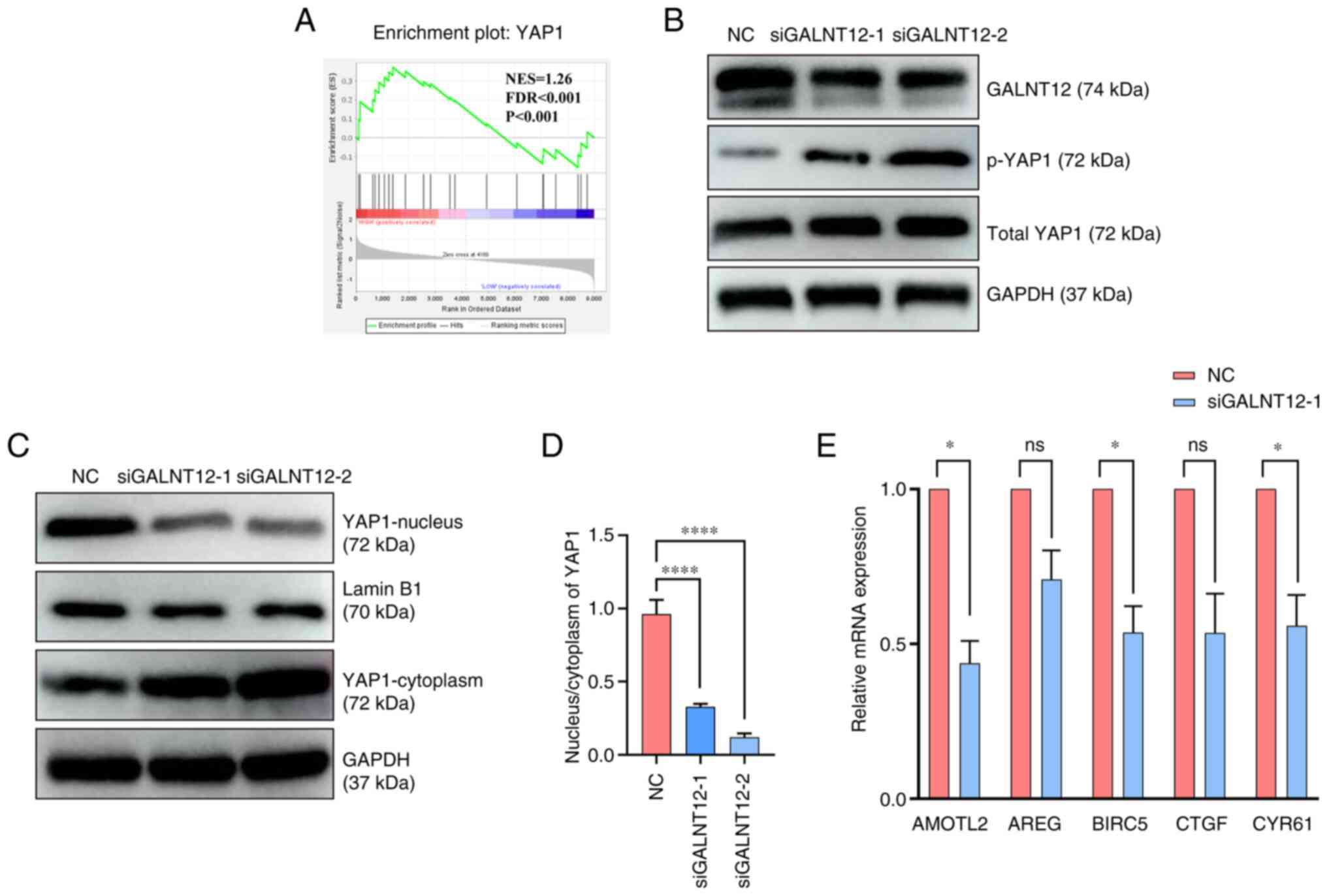
|
1
|
Pankova V, Thway K, Jones RL and Huang PH:
The extracellular matrix in soft tissue sarcomas: Pathobiology and
cellular signalling. Front Cell Dev Biol. 9:7636402021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
American Cancer Society, . What is a soft
tissue sarcoma? American Cancer Society; Atlanta, GA: 2023,
https://cancerstatisticscenter.cancer.org/#!/
|
|
3
|
Howlader N, Noone AM, Krapcho M, Miller D,
Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, et al:
SEER cancer statistics review, 1975–2018. National Cancer
Institute; Bethesda, MD: 2020
|
|
4
|
Fletcher CDM, Bridge JA, Hogendoorn PCW
and Mertens F: WHO classification of tumours of soft tissue and
bone. WHO Classification of Tumours. 4th edition. Vol. 5. IARC
Press; Lyon: 2013
|
|
5
|
Toro JR, Travis LB, Wu HJ, Zhu K, Fletcher
CDM and Devesa SS: Incidence patterns of soft tissue sarcomas,
regardless of primary site, in the surveillance, epidemiology and
end results program, 1978–2001: An analysis of 26,758 cases. Int J
Cancer. 119:2922–2930. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bahrami A and Folpe AL: Adult-type
fibrosarcoma: A reevaluation of 163 putative cases diagnosed at a
single institution over a 48-year period. Am J Surg Pathol.
34:1504–1513. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gamboa AC, Gronchi A and Cardona K:
Soft-tissue sarcoma in adults: An update on the current state of
histiotype-specific management in an era of personalized medicine.
CA Cancer J Clin. 70:200–229. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Francis M, Charman J, Dennis N, Lawrence G
and Grimer R: Bone and soft tissue sarcomas. UK Incidence and
Survival: 1996 to 2010. Version 2.0. National Cancer Intelligence
Network; Bethesda, MD: 2013
|
|
9
|
Folpe AL: Fibrosarcoma: A review and
update. Histopathology. 64:12–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Phelan CM, Tsai YY, Goode EL, Vierkant RA,
Fridley BL, Beesley J, Chen XQ, Webb PM, Chanock S, Cramer DW, et
al: Polymorphism in the GALNT1 gene and epithelial ovarian cancer
in non-Hispanic white women: The ovarian cancer association
consortium. Cancer Epidemiol Biomarkers Prev. 19:600–604. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khetarpal SA, Schjoldager KT,
Christoffersen C, Raghavan A, Edmondson AC, Reutter HM, Ahmed B,
Ouazzani R, Peloso GM, Vitali C, et al: Loss of function of GALNT2
lowers high-density lipoproteins in humans, nonhuman primates, and
rodents. Cell Metab. 24:234–245. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marucci A, di Mauro L, Menzaghi C,
Prudente S, Mangiacotti D, Fini G, Lotti G, Trischitta V and Di
Paola R: GALNT2 expression is reduced in patients with Type 2
diabetes: Possible role of hyperglycemia. PLoS One. 8:e701592013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Reuter MS, Tawamie H, Buchert R, Hosny
Gebril O, Froukh T, Thiel C, Uebe S, Ekici AB, Krumbiegel M, Zweier
C, et al: Diagnostic yield and novel candidate genes by exome
sequencing in 152 consanguineous families with neurodevelopmental
disorders. JAMA Psychiatry. 74:293–299. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo JM, Zhang Y, Cheng L, Iwasaki H, Wang
H, Kubota T, Tachibana K and Narimatsu H: Molecular cloning and
characterization of a novel member of the UDP-GalNAc:polypeptide
N-acetylgalactosaminyltransferase family, pp-GalNAc-T12. FEBS Lett.
524:211–218. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fernandez AJ, Daniel EJP, Mahajan SP, Gray
JJ, Gerken TA, Tabak LA and Samara NL: The structure of the
colorectal cancer-associated enzyme GalNAc-T12 reveals how
nonconserved residues dictate its function. Proc Natl Acad Sci USA.
116:20404–20410. 2015. View Article : Google Scholar
|
|
16
|
Wang YN, Zhou XJ, Chen P, Yu GZ, Zhang X,
Hou P, Liu LJ, Shi SF, Lv JC and Zhang H: Interaction between
GALNT12 and C1GALT1 associates with galactose-deficient IgA1 and
IgA nephropathy. J Am Soc Nephrol. 32:545–552. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hayashi S, Matsubara T, Fukuda K, Maeda T,
Funahashi K, Hashimoto M, Kamenaga T, Takashima Y and Kuroda R: A
genome-wide association study identifying the SNPs predictive of
rapid joint destruction in patients with rheumatoid arthritis.
Biomed Rep. 14:312021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang J, Wang N and Xu A: miR-10b-3p,
miR-8112 and let-7j as potential biomarkers for autoimmune inner
ear diseases. Mol Med Rep. 20:171–181. 2019.PubMed/NCBI
|
|
19
|
Seguí N, Pineda M, Navarro M, Lázaro C,
Brunet J, Infante M, Durán M, Soto JL, Blanco I, Capellá G and
Valle L: GALNT12 is not a major contributor of familial colorectal
cancer type X. Hum Mutat. 35:50–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Evans DR, Venkitachalam S, Revoredo L,
Dohey AT, Clarke E, Pennell JJ, Powell AE, Quinn E, Ravi L, Gerken
TA, et al: Evidence for GALNT12 as a moderate penetrance gene for
colorectal cancer. Hum Mutat. 39:1092–1101. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zheng Y, Liang M, Wang B, Kang L, Yuan Y,
Mao Y and Wang S: GALNT12 is associated with the malignancy of
glioma and promotes glioblastoma multiforme in vitro by activating
Akt signaling. Biochem Biophys Res Commun. 610:99–106. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Y, Yu M, Yang JX, Cao DY, Zhang Y,
Zhou HM, Yuan Z and Shen K: Genomic comparison of endometrioid
endometrial carcinoma and its precancerous lesions in chinese
patients by high-depth next generation sequencing. Front Oncol.
9:1232019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gibson TM, Wang SS, Cerhan JR, Maurer MJ,
Hartge P, Habermann TM, Davis S, Cozen W, Lynch CF, Severson RK, et
al: Inherited genetic variation and overall survival following
follicular lymphoma. Am J Hematol. 87:724–726. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cates JMM: The AJCC 8th edition staging
system for soft tissue sarcoma of the extremities or trunk: A
cohort study of the SEER database. J Natl Compr Canc Netw.
16:144–152. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu S, Zhang Y, Li Q, Zhang Z, Zhao G and
Xu J: CLDN6 promotes tumor progression through the YAP1-snail1 axis
in gastric cancer. Cell Death Dis. 10:9492019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schindelin J, Arganda-Carreras I, Frise E,
Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S,
Schmid B, et al: Fiji: An open-source platform for biological-image
analysis. Nat Methods. 9:676–682. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu Q, Liu HO, Liu YD, Liu WS, Pan D, Zhang
WJ, Yang L, Fu Q, Xu JJ and Gu JX: Decreased expression of
hepatocyte nuclear factor 4α (Hnf4α)/microRNA-122 (miR-122) axis in
hepatitis B virus-associated hepatocellular carcinoma enhances
potential oncogenic GALNT10 protein activity. J Biol Chem.
290:1170–1185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang L, Gallup M, Zlock L, Chen YT,
Finkbeiner WE and McNamara NA: Pivotal role of MUC1 glycosylation
by cigarette smoke in modulating disruption of airway adherens
junctions in vitro. J Pathol. 234:60–73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gaziel-Sovran A, Segura MF, Di Micco R,
Collins MK, Hanniford D, Vega-Saenz de Miera E, Rakus JF, Dankert
JF, Shang S, Kerbel RS, et al: miR-30b/30d regulation of GalNAc
transferases enhances invasion and immunosuppression during
metastasis. Cancer Cell. 20:104–118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Beaman EM, Carter DRF and Brooks SA:
GALNTs: Master regulators of metastasis-associated
epithelial-mesenchymal transition (EMT)? Glycobiology. 32:556–579.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liao YY, Chuang YT, Lin HY, Lin NY, Hsu
TW, Hsieh SC, Chen ST, Hung JS, Yang HJ, Liang JT, et al: GALNT2
promotes invasiveness of colorectal cancer cells partly through
AXL. Mol Oncol. 17:119–133. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Park MS, Yang AY, Lee JE, Kim SK, Roe JS,
Park MS, Oh MJ, An HJ and Kim MY: GALNT3 suppresses lung cancer by
inhibiting myeloid-derived suppressor cell infiltration and
angiogenesis in a TNFR and c-MET pathway-dependent manner. Cancer
Lett. 521:294–307. 2021.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Raghu D, Mobley RJ, Shendy NAM, Perry CH
and Abell AN: GALNT3 maintains the epithelial state in trophoblast
stem cells. Cell Rep. 26:3684–3697.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin WR and Yeh CT: GALNT14: An emerging
marker capable of predicting therapeutic outcomes in multiple
cancers. Int J Mol Sci. 21:14912020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Scott E, Hodgson K, Calle B, Turner H,
Cheung K, Bermudez A, Marques FJG, Pye H, Yo EC, Islam K, et al:
Upregulation of GALNT7 in prostate cancer modifies O-glycosylation
and promotes tumour growth. Oncogene. 42:926–937. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Detarya M, Sawanyawisuth K, Aphivatanasiri
C, Chuangchaiya S, Saranaruk P, Sukprasert L, Silsirivanit A, Araki
N, Wongkham S and Wongkham C: The O-GalNAcylating enzyme GALNT5
mediates carcinogenesis and progression of cholangiocarcinoma via
activation of AKT/ERK signaling. Glycobiology. 30:312–324. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dasgeb B, Leila Y, Saeidian AH, Kang J,
Shi W, Shoenberg E, Ertel A, Fortina P, Vahidnezhad H and Uitto J:
Genetic predisposition to numerous large ulcerating basal cell
carcinomas and response to immune therapy. Int J Dermatol Venereol.
4:70–75. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ou C, Sun Z, He X, Li X, Fan S, Zheng X,
Peng Q, Li G, Li X and Ma J: Targeting YAP1/LINC00152/FSCN1
signaling axis prevents the progression of colorectal cancer. Adv
Sci (Weinh). 7:19013802019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yuan Y, Park J, Feng A, Awasthi P, Wang Z,
Chen Q and Iglesias-Bartolome R: YAP1/TAZ-TEAD transcriptional
networks maintain skin homeostasis by regulating cell proliferation
and limiting KLF4 activity. Nat Commun. 11:14722020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim M, Ly SH, Xie Y, Duronio GN,
Ford-Roshon D, Hwang JH, Sulahian R, Rennhack JP, So J, Gjoerup O,
et al: YAP1 and PRDM14 converge to promote cell survival and
tumorigenesis. Dev Cell. 57:212–227.e8. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu M, Zhang Y, Yang J, Zhan H, Zhou Z,
Jiang Y, Shi X, Fan X, Zhang J, Luo W, et al: Zinc-dependent
regulation of ZEB1 and YAP1 coactivation promotes
epithelial-mesenchymal transition plasticity and metastasis in
pancreatic cancer. Gastroenterology. 160:1771–1783.e1. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ajani JA, Xu Y, Huo L, Wang R, Li Y, Wang
Y, Pizzi MP, Scott A, Harada K, Ma L, et al: YAP1 mediates gastric
adenocarcinoma peritoneal metastases that are attenuated by YAP1
inhibition. Gut. 70:55–66. 2021. View Article : Google Scholar : PubMed/NCBI
|