Introduction
Breast cancer, a heterogenous type of tumor with the
highest incidence rate among women worldwide, accounting for 31.0%
of all female cancer cases, is accountable for a high incidence of
cancer-associated mortalities (1).
Based on the genome sequence and protein expression levels of human
epidermal growth factor 2 (HER2), progesterone receptor (PR) and
estrogen receptor (ER), breast cancer can be divided into luminal,
HER2-enriched and triple-negative types (2). Triple-negative breast cancer (TNBC),
the most malignant subtype, is linked to a highly unfavorable
prognosis and poor overall survival (OS) rates compared with other
subtypes (3). In addition, the
current therapeutic strategies for TNBC are limited due to a lack
of clear targets and chemotherapy remains the main treatment
modality for TNBC. Although certain aggressive TNBC types are
immunogenic, the majority of patients exhibit limited responses to
immunotherapy (4,5). Therefore, it is essential to explore
new targets for developing novel therapies for TNBC.
Ferroptosis, a unique iron-dependent cell death
process, is distinct from apoptosis, necrosis and autophagy. A key
hallmark of ferroptosis is enhanced generation of intracellular
reactive oxygen species (ROS) and diminished mitochondrial volume
(6). It has previously been
reported that iron metabolism-mediated ROS accumulation promotes
ferroptosis. Furthermore, ferroptosis-related genes, which regulate
ferroptosis, participate in the onset and progression of a number
of malignancies, including breast cancer (7,8). For
instance, glutathione peroxidase 4 (GPX4), a core regulatory gene
involved in ferroptosis, is negatively associated with the
prognosis of breast cancer, as its expression enhances ferroptosis
in cells (9). Analyzing the
expression levels of acyl-CoA synthetase long chain family member
4, which promotes ferroptosis by upregulating intracellular lipids,
can predict the response to neoadjuvant chemotherapy (10). Furthermore, a previous study
reported that the induction of ferroptosis may overcome drug
resistance and is a potential novel therapeutic approach for cancer
(11). Of note, TNBC cells have
been reported to be susceptible to ferroptosis due to their complex
metabolic characteristics and cellular signaling pathways (12). TNBC cells can express high levels of
the xCT cystine/glutamate antiporter, which leads to a reduction in
xCT-associated glutathione levels, thus reducing cell viability and
sensitizing the cells to ferroptosis (13). In addition, it has been reported
that MDA-MB-231 TNBC cells are highly cystine-dependent and
susceptible to ferroptosis (14,15).
However, limited studies have examined the association of
ferroptosis-related genes with TNBC prognosis. In addition, a
specific ferroptosis-related therapeutic target for TNBC has not
been identified to date.
In the present study, genes linked to ferroptosis
were investigated using mRNA expression data and relevant clinical
profiles from The Cancer Genome Atlas (TCGA) database. The
objective of the current study was to provide valuable insight into
guiding clinical decisions for TNBC treatment. Next, the model was
further validated in a Gene Expression Omnibus (GEO) cohort and
functional enrichment analysis was performed to explore the
possible mechanisms of these identified genes in different risk
groups classified using the developed model. In addition, the
immune-related responses in TNBC were evaluated. Finally, the role
of transferrin receptor 2 (TFR2) in TNBC was verified using in
vitro experiments. The present findings further demonstrated
the functions of ferroptosis-related genes and provided novel
targets for potential therapeutic intervention in TNBC.
Materials and methods
Data collection
The clinical information and transcriptome of
individuals with breast cancer were obtained from the TCGA
(http://cancergenome.nih.gov/) database
(added before May 1, 2022). For transcriptome data, after entering
the file download interface (http://portal.gdc.cancer.gov/), ‘TCGA-BRCA’ project
was selected in the Cases parameter. In the Files parameter
section, ‘transcriptome profiling’ was selected in the data
category, ‘gene expression quantification’ in the data type and
‘HTSeq-FPKM’ in the workflow type. For clinical data, ‘clinical’
was selected in the data category and ‘bcr-xml’ in the workflow
type. The TCGA dataset comprised data from 1,096 breast cancer and
112 healthy breast tissue samples. To distinguish the molecular
subtypes of patients with breast cancer, data from The University
of California, Santa Cruz Xena website (https://xenabrowser.net/datapages/) corresponding to
TCGA-BRCA were retrieved. The inclusion criteria for TNBC were
based on the following immunohistochemical results: i) ER, PR and
HER2 were negative; and ii) fluorescence in situ
hybridization was required to be negative when the HER2 level was
2+. After molecular typing screening and excluding cases with a
follow-up time of <90 days, the training dataset comprised 118
individuals with TNBC. In addition, the validation dataset was
accessed at GEO (https://www.ncbi.nlm.nih.gov/geo/). The RNA expression
data with follow-up information of individuals with TNBC (dataset
accession no. GSE25307) were accessed at GEO and comprised data
from 120 tumor samples. In brief, the series matrix file containing
the original data of probe results and clinical information was
downloaded and the probes were converted into the corresponding
gene symbol according to the annotation information by the platform
(GPL5354 SWEGENE H_v2.1.1 55K). General baseline clinical features
of the individuals included in the two databases were analyzed,
which comprised information such as age, tumor staging and
prognosis (Table I). Furthermore,
the data of 259 genes linked to ferroptosis were sourced from the
FerrDb website (http://www.zhounan/org/ferrdb/; Table SI). These genes have also been
reported by a previous study (16).
 | Table I.Clinicopathological parameters of the
patients with triple negative breast cancer in the present
study. |
Table I.
Clinicopathological parameters of the
patients with triple negative breast cancer in the present
study.
| Patient
characteristic | TCGA-BRCA
(n=230) | GSE25307
(n=120) |
|---|
| Type of sample |
|
|
|
Tumor | 118 | 120 |
|
Normal | 112 | 0 |
| Age, years | 55 (29–85) |
|
| Tumor stage |
|
|
| I | 20 |
|
| II | 76 |
|
|
III | 17 |
|
| IV | 2 |
|
|
Unknown | 3 |
|
| T stage |
|
|
| T1 | 28 |
|
| T2 | 74 |
|
| T3 | 12 |
|
| T4 | 3 |
|
|
Unknown | 1 |
|
| N stage |
|
|
| N0 | 75 |
|
| N1 | 29 |
|
| N2 | 8 |
|
| N3 | 6 |
|
| M stage |
|
|
| M0 | 109 |
|
| M1 | 2 |
|
|
Unknown | 7 |
|
| Survival
status |
|
|
|
Deceased | 18 | 57 |
|
Alive | 100 | 63 |
| Median overall
survival, days | 1,606 | 2,777 |
Detection of ferroptosis-related
differentially expressed genes (DEGs)
Ferroptosis-related DEGs in TNBC and healthy breast
tissue samples were detected in TCGA cohort using the ‘limma’ R
package (version 3.46.0) based on the following criteria: i) False
discovery rate (FDR) of 0.05; and ii) log fold-change (FC) >1.
Heatmaps were plotted to visualize DEGs. To assess these genes
further, the Search Tool for the Retrieval of Interacting Genes and
proteins (STRING; http://string-db.org) database was employed for
establishing the interaction network of these identified DEGs.
Cytoscape (version 3.8.2; http://cytoscape.org) was utilized for the
visualization of the specific molecular regulatory association of
DEGs.
Functional enrichment analysis
Ferroptosis-related DEGs in TNBC vs. healthy breast
tissues were subjected to Gene Ontology (GO) (cellular component,
molecular function and biological process) and Kyoto Encyclopedia
of Genes and Genomes (KEGG) enrichment analyses (log FC ≥1; FDR
<0.05) using the ‘clusterProfiler’ R package (version
3.18.1).
Construction of the prognostic
ferroptosis-related gene signature
Univariate Cox regression analysis of OS was
performed to identify ferroptosis-related prognostic DEGs utilizing
the R ‘survival’ package (version 3.5–5). The expression difference
of ferroptosis-related DEGs with prognostic significance in breast
cancer and healthy breast tissue were analyzed through the Human
Protein Atlas (HPA) database, which is an online tool (www.proteatlas.org) that displays immunohistochemistry
results of protein expression patterns in cancer tissues, healthy
tissues and different cell types. The resulting data were
visualized using forest plots. Furthermore, the survival of
patients with TNBC was examined through Kaplan-Meier (KM) Plotter
(http://kmplot.com/analysis/), which
integrates information on patients with TNBC from E-MTAB-365,
E-TABM-43 and a series of GEO datasets (17). Next, least absolute shrinkage and
selection operator (LASSO) Cox regression analysis was conducted
utilizing the function ‘glmnet’ of R (version 4.1–7) to construct a
prognostic model. The coefficients of the normalized expression
level of the three aforementioned prognostic genes were calculated
(Table SII).
Model assessment
The gene expression level was utilized to compute
the risk score and its formula was as follows: Sum (expression
level of each gene × corresponding coefficient). The categorization
of the individuals under study into the high- and low-risk score
groups was performed according to their median risk score.
Principal component analysis (PCA) and t-distributed stochastic
neighbor embedding (t-SNE) were performed utilizing the ‘Rtsne’
(version 0.16) and ‘stats’ (version 4.1.1) R packages to assess the
distribution of various groups. Furthermore, the model was examined
concerning its predictive accuracy by plotting the time-dependent
receiver operating characteristic (ROC) curve using the
‘survivalROC’ R package (version 1.0.3.1).
Survival analyses
Survival analysis of the high- and low-risk groups
was performed using the ‘survminer’ (version 0.4.8) and ‘survival’
(version 3.5–5) R packages. The link between risk scores and
clinicopathological characteristics in individuals with TNBC was
examined using univariate and multivariate Cox analyses. The
clinical benefits of risk scores were determined through decision
curve analysis (DCA). The R package ‘regplot’ (version 1.1) was
utilized to develop a nomogram integrating TNBC clinicopathological
features and risk scores for the prediction of the OS of
individuals at diverse time-points.
Gene set enrichment analysis
(GSEA)
GSEA was performed to identify relevant DEGs between
the high- and low-risk groups employing the R package ‘edgeR’
(version 3.34.0) according to the following criteria: i) FDR
<0.05; and ii) log2 FC ≥1. In addition, KEGG analysis provided
further insight into the potential mechanisms of the risk signature
in TNBC. P<0.05 and FDR q<0.05 were considered to indicate a
statistically significant difference.
Immune analyses
To assess the immune cell types in patients with
TNBC in the high- and low-risk groups, the CIBERSORT,
CIBERSORT-ABS, EPIC, XCELL and MCPCOUNTER algorithms were used. The
role of the ferroptosis-related risk signature in the immunotherapy
response prediction of individuals with TNBC was examined using the
‘ggplot2’ (version 3.4.2) and ‘ggpubr’ (version 0.5.0) R packages.
Single-sample GSEA (ssGSEA) was performed to evaluate the
association of risk score with potential immune checkpoints.
Cell culture and reagents
Healthy breast epithelial cells (MCF-10A), luminal A
breast cancer cells (T-47D), HER2-enriched breast cancer cells
(SK-BR-3) and TNBC cells (BT-549, MDA-MB-468, MDA-MB-436 and
MDA-MB-231) were purchased from Procell Life Science &
Technology Co., Ltd., and luminal A breast cancer cells (MCF-7) and
luminal B breast cancer cells (BT-474) were purchased from The Cell
Bank of the Type Culture Collection of The Chinese Academy of
Sciences. MDA-MB-231 and MDA-MB-468 cells were cultured in Gibco's
Leibovitz's L-15 medium with 10% FBS (Thermo Fisher Scientific,
Inc.). BT-549 cells were cultured in Gibco's RPMI-1640 basic medium
(Thermo Fisher Scientific, Inc.) containing 0.01 mg/ml insulin
(Beijing Solarbio Science & Technology Co., Ltd.) and 10% FBS.
T-47D cells were cultured in Gibco's DMEM (Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS. MCF-7, SK-BR-3,
MDA-MB-436 and BT-474 cells were cultured in Gibco's RPMI-1640
medium containing 10% FBS. MCF-10A cells were cultured in Gibco's
DMEM containing 5% horse serum, 0.5 µg/ml cortisol, 0.01 mg/ml
insulin and 1% non-essential amino acids (Procell Life Science
& Technology Co., Ltd.). MDA-MB-468 and MDA-MB-231 cells were
incubated at 37°C under 100% humidity and 100% O2. The
remaining cell lines were incubated at 37°C with 100% humidity and
5% CO2. The cells used in the present study were
authenticated using short tandem repeat profiling and were
determined to be free of mycoplasma contamination. The ferroptosis
inhibitor ferrostatin-1 (Fer-1; MedChemExpress) was stored at
−80°C.
Transfection and RNA interference
Three different small interfering RNAs (siRNAs)
against TFR2, siRNA negative control (NC; Table SIII) and the GP-transfect-Mate
transfection reagent sourced from Shanghai GenePharma Co., Ltd.
Culture plates (6-well) were utilized for culturing cells
(5×107 cells/l) in their logarithmic growth phase for 24
h to achieve 60% density. siRNA or siNC (5 µl) and transfection
reagent GP-transfect-Mate (4 µl) were added into 300 µl OPTI-MEM
(Gibco; Thermo Fisher Scientific, Inc.) and incubated at room
temperature for 20 min before being added to MDA-MB-436 cells, and
the plate was incubated 48 h or 72 h under 5% CO2 at
37°C. The knockdown efficiency at 48 h or 72 h post-transfection
was examined using reverse transcription-quantitative PCR (RT-qPCR)
and western blot analyses.
RNA extraction and RT-qPCR
analysis
The TFR2 mRNA expression level in MCF-10A, T-47D,
MCF-7, SK-BR-3, BT-474, BT-549, MDA-MB-468, MDA-MB-436 and
MDA-MB-231 cells was assessed using RT-qPCR. Extraction of total
RNA from cells was performed utilizing RNAiso Plus (cat. no. 9108Q;
Takara Biotechnology Co., Ltd.). Total RNA was then
reverse-transcribed into complementary DNA using PrimeScript™ RT
Master Mix (Takara Biotechnology Co., Ltd.) according to the
manufacturer's protocol. qPCR analysis was performed utilizing TB
Green™ Premix EX Taq™ II (Takara Biotechnology Co., Ltd.) in a
GENTIER 96 qPCR instrument (Tianlong). The PCR conditions were as
follows: Initial denaturation at 95°C for 30 sec; and 40 cycles of
95°C for 5 sec and 60°C for 34 sec. The melting curve was generated
under the following conditions: 95°C for 15 sec, followed by 95°C
for 5 sec and 60°C for 34 sec. GAPDH was used as an internal
reference for normalization. The mRNA expression levels were
determined using the 2−ΔΔCq method (18). Table
SIII contains the sequences of the primers utilized.
Cell viability analysis
The density of cells in the logarithmic growth phase
was adjusted to 3×107 cells/l. Cells were seeded into a
96-well plate (100 µl/well, 3×103 cells/well) and the
plate was incubated overnight under 5% CO2 at 37°C.
Cells were then transfected with siRNA or siNC and incubated for
24, 48 and 72 h under 5% CO2 at 37°C. The culture medium
was replaced at each time-point and the cells were then incubated
with 110 µl complete medium containing 10 µl Cell Counting Kit
(CCK)-8 reagent (Beijing Solarbio Science & Technology Co.,
Ltd.) for 90 min at 37°C. The optical density of the reaction
mixture was measured at 450 nm using a microplate reader (Thermo
Fisher Scientific, Inc.).
Lipid peroxidation assay
Cells were cultured in T25 culture flasks to a
density of 80%. Cells were lysed with a western blotting and
immunoprecipitation cell lysis solution (cat. no. P0013; Beyotime
Institute of Biotechnology). Next, the lysate was centrifuged at
12,000 × g for 10 min at 4°C. A BCA protein assay kit (Epizyme) was
utilized for determining the protein concentration in the
supernatant for subsequent calculation of the malondialdehyde (MDA)
content. The MDA content of the samples was determined utilizing a
lipid peroxidation MDA assay kit (Beyotime Institute of
Biotechnology). In brief, 100 µl of sample was incubated with 200
µl MDA assay working solution for 15 min in a water bath at 100°C.
Subsequently, the sample was centrifuged for 10 min at room
temperature at 1,000 × g. The absorbance of 200 µl of the
supernatant at 532 nm was measured using a microplate reader.
Western blot analysis
Extraction of total protein was conducted using a
whole-cell lysis assay kit (cat no. KGP2100; Nanjing KeyGen Biotech
Co., Ltd.), followed by quantification with a BCA protein assay kit
(Epizyme). The proteins (15 µg per lane) were subjected to
separation by 10% SDS-PAGE and then transferred onto a
polyvinylidene fluoride membrane. Membranes were blocked using 8%
skimmed milk in TBS with 0.1% Tween 20 at room temperature for 2 h,
followed by incubation with anti-TFR2 (cat. no. ET7108-21; HUABIO;
1:1,000 dilution), anti-lysyl oxidase (LOX; cat. no. 17958-1-AP;
Proteintech, Inc.; 1:1,000 dilution), anti-solute carrier family 7
member 11 (SLC7A11; cat. no. 12691; Cell Signaling Technology,
Inc.; 1:1,000 dilution), anti-GPX4 (cat. no. ER1803-15; HUABIO;
1:1,000 dilution) and anti-ferritin heavy chain 1 (FTH1; cat. no.
4393; Cell Signaling Technology, Inc.; 1:1,000 dilution) primary
antibodies overnight at 4°C. Next, membranes were incubated with
horseradish peroxidase-conjugated goat anti-rabbit IgG (cat. no.
HA1001; HUABIO; 1:5,000 dilution) or goat anti-mouse IgG (cat. no.
HA1006; HUABIO; 1:5,000 dilution) secondary antibodies at room
temperature for 2 h. Immunoreactive signals were developed
employing an enhanced chemiluminescence solution (Dalian Meilun
Biology Technology Co., Ltd.) and visualized using a gel imaging
system (ChemiDoc MP; Bio-Rad Laboratories, Inc.). The grayscale
values of the protein bands were derived utilizing Gel-Pro analyzer
4.0 (Media Cybernetics, Inc.) to calculate the relative expression
levels of proteins. Anti-GAPDH antibodies (cat. no. EM1101; HUABIO;
1:5,000 dilution) were used to detect GAPDH, which functioned as an
internal control for normalization.
Statistical analysis
A total of 118 patients with TNBC from the TCGA
database were divided into high- and low-risk groups based on their
median risk scores and χ2 test or Fisher's exact test
was used to compare the clinical characteristics of patients with
TNBC in different risk groups. Statistical analyses were conducted
with R packages (version 4.1.3; RStudio, Inc.). Data matrix
construction and data processing were performed using Perl
(www.perl.org). Experimental data were analyzed
using SPSS (version 29.0; IBM Corp.), while figures were produced
using GraphPad Prism (version 8.2.1; Dotmatics). An unpaired t-test
was used for comparative assessment of means between two groups,
whereas multiple groups were compared using one-way ANOVA followed
by Tukey's post-hoc test. All in vitro experiments were
independently repeated three times. Data are presented as the mean
± standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of ferroptosis-related
DEGs in the TCGA TNBC cohort
In the present study, 259 genes were determined to
be associated with ferroptosis by utilizing the FerrDb website. The
ferroptosis-related DEGs across the TNBC and healthy breast samples
obtained from the TCGA database were analyzed (Fig. 1A and B). In contrast to the healthy
breast tissues, 71 of the 256 ferroptosis-related DEGs (27.7%)
varied concerning their expression levels in TNBC samples (34
upregulated and 37 downregulated). Furthermore, the protein-protein
interaction network of DEGs was established through STRING.
Visualization of the detailed regulatory association was achieved
through Cytoscape (Fig. 1C). Next,
the DEGs were analyzed through GO and KEGG pathway enrichment
analyses. GO analysis demonstrated the enrichment of genes involved
in several types of oxidative stress functions (Fig. 1D). In addition, KEGG analyses
indicated the enrichment of genes involved in ROS-related pathways
(Fig. 1E), which were closely
associated with ferroptosis.
Prognostic value of
ferroptosis-related DEGs
Univariate Cox regression analysis demonstrated
that, of the 71 ferroptosis-related DEGs, TFR2, activating
transcription factor 3 (ATF3), dual specificity phosphatase 1
(DUSP1)/MAPK phosphatase 1 (MKP-1), regulator of G protein
signaling 4 (RGS4) and zinc finger protein 36 (ZFP36) demonstrated
prognostic value in TNBC (Fig. 2A).
The present study further analyzed the difference in expression
levels of ferroptosis-related DEGs with prognostic significance in
breast cancer and healthy breast tissue in the HPA database and the
results demonstrated that the expression levels of three genes
(TFR2, ATF3 and ZFP36) were higher in tumor tissue compared with
healthy tissue, which was consistent with the bioinformatics
analysis results (Fig. 2B).
However, DUSP1 and RGS4 could not be found in the HPA database.
Next, the KM Plotter (integrating E-MTAB-365, E-TABM-43 and a
series of GEO datasets) was employed to investigate the prognostic
significance of these genes in patients with TNBC. Increased
expression levels of TFR2, ATF3, RGS4 and ZFP36 were significantly
associated with decreased OS in patients with TNBC (Fig. 2C-H). There was no marked association
between DUSP1/MKP-1 expression levels and OS (P=0.058).
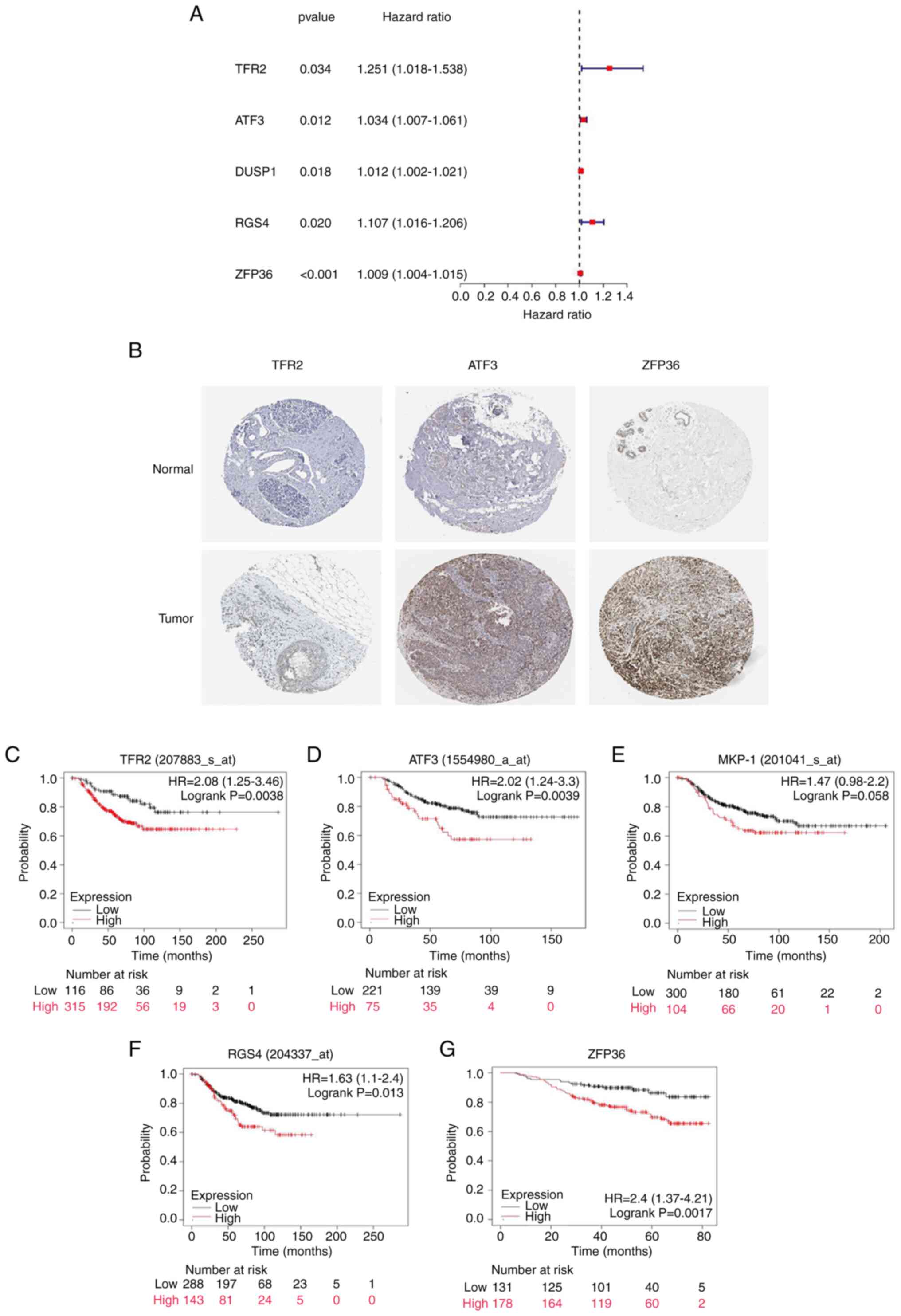 | Figure 2.Evaluation of prognostic
ferroptosis-related DEGs. (A) A total of five ferroptosis-related
genes were identified by univariate Cox regression analysis with
prognostic value (95% CI) in triple-negative breast cancer. (B) The
expression of TFR2, ATF3 and ZFP36 in healthy breast tissue and
breast cancer samples from the Human Protein Atlas database
(magnification, ×100). Association of the expression levels of
ferroptosis-related DEGs with overall survival, including (C) TFR2,
(D) ATF3, (E) MKP-1, (F) RGS4 and (G) ZFP36; information in
brackets represents gene probes. HR, hazard ratio; TFR2,
transferrin receptor 2; ATF3, activating transcription factor 3;
MKP-1, MAPK phosphatase 1; DUSP1, dual specificity phosphatase 1;
RSG4, regulator of G protein signaling 4; ZFP36, zinc finger
protein 36; DEG, differentially expressed gene. |
Construction of a three-gene model
using TCGA cohort and its validation using the GEO cohort
LASSO Cox regression was performed to establish a
prognostic model utilizing the expression of five genes linked to
ferroptosis. Calculation of the risk score was performed as
follows: Risk score=e(0.279× TFR2 expression level + 0.113×
RGS4 expression level + 0.011× ZFP36 expression level)
(Table SII). Regarding the risk
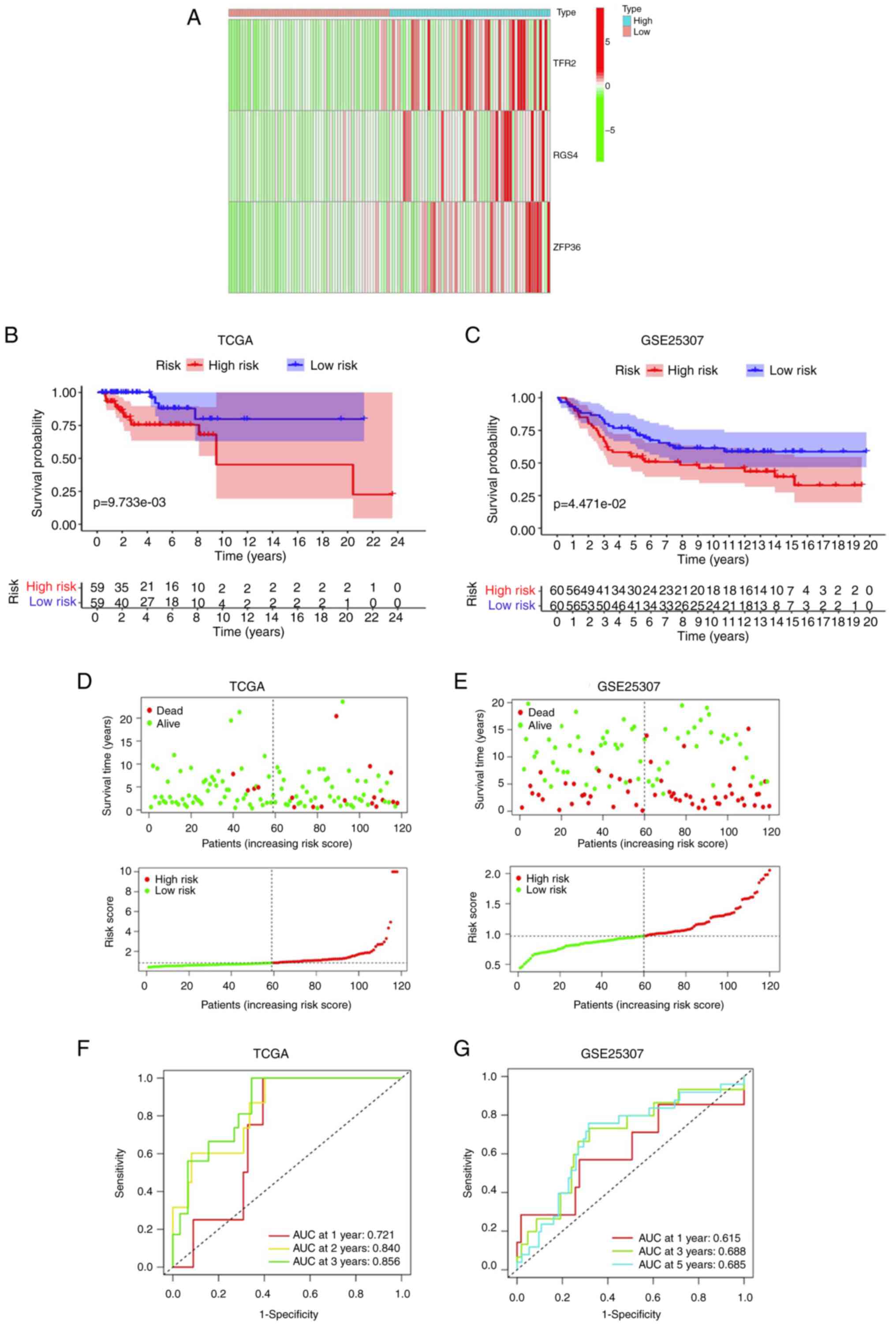
score, a three-gene (TFR2, RGS4 and ZFP36) model was established
for the TCGA cohort. Categorization of patients with TNBC into
high-risk (n=59) and low-risk (n=59) groups was conducted according
to their median risk score and prognostic genes were analyzed
(Fig. 3A). Clinical correlation
analysis demonstrated that patients with TNBC in the high-risk
group exhibited a markedly increased incidence of lymph node
metastases and advanced tumor stages (P=0.085 and P=0.086,
respectively; Table II). KM curve
analysis demonstrated a significantly higher OS in the low-risk
group in comparison with the high-risk group in the TCGA dataset
(Fig. 3B) and also in the GSE25307
dataset (Fig. 3C). The t-SNE and
PCA analyses demonstrated that the individuals in the risk groups
were distinctly separated (Fig.
3D). To validate the prognostic significance of the three-gene
model, the categorization of individuals with TNBC in the GEO
cohort (GSE25307) into risk groups (high and low) was performed
according to their median risk score. Similar PCA and t-SNE results
were demonstrated in the GEO and TCGA cohorts (Fig. 3E). The respective area under the
curve (AUC) values of the three-gene signature model for 1-, 2- and
3-year OS prediction in the TCGA cohort were 0.721, 0.840 and
0.856, respectively, thus indicating a good prognostic value
(Fig. 3F). In addition, the
respective AUC values of the three-gene signature model for 1-, 3-
and 5-year OS prediction in the GSE25307 dataset were 0.615, 0.688
and 0.685, respectively (Fig. 3G).
These findings confirmed the adaptability of the aforementioned
three-gene model in TNBC.
 | Table II.Comparison of clinical
characteristics in low- and high-risk groups based on The Cancer
Genome Atlas cohort. |
Table II.
Comparison of clinical
characteristics in low- and high-risk groups based on The Cancer
Genome Atlas cohort.
|
Characteristics | Low-risk group
(n=59) | High-risk group
(n=59) | P-value |
|---|
| Age, years |
|
| 0.843 |
|
≤60 | 41 (69.5) | 40 (67.8) |
|
|
>60 | 18 (30.5) | 19 (32.2) |
|
| T stage |
|
| 0.387 |
| T1 | 9 (15.3) | 19 (32.2) |
|
| T2 | 40 (15.3) | 34 (57.6) |
|
| T3 | 9 (67.8) | 3 (5.1) |
|
| T4 | 0 (0) | 3 (5.1) |
|
|
Unknown | 1 (1.6) |
|
|
| N stage |
|
| 0.085 |
| N0 | 42 (71.2) | 33 (55.9) |
|
| N1 | 13 (22.0) | 16 (27.1) |
|
| N2 | 2 (3.4) | 6 (10.2) |
|
| N3 | 2 (3.4) | 4 (6.8) |
|
| M stage |
|
| 0.496a |
| M0 | 54 (91.5) | 55 (93.2) |
|
| M1 | 0 (0) | 2 (3.4) |
|
|
Unknown | 5 (8.5) | 2 (3.4) |
|
| Tumor stage |
|
| 0.086 |
| I | 9 (15.3) | 11 (18.6) |
|
| II | 42 (71.2) | 34 (57.6) |
|
|
III | 6 (10.1) | 11 (18.6) |
|
| IV | 0 (0) | 2 (3.5) |
|
|
Unknown | 2 (3.4) | 1 (1.7) |
|
Evaluation of the independent
prognostic value of the three-gene signature
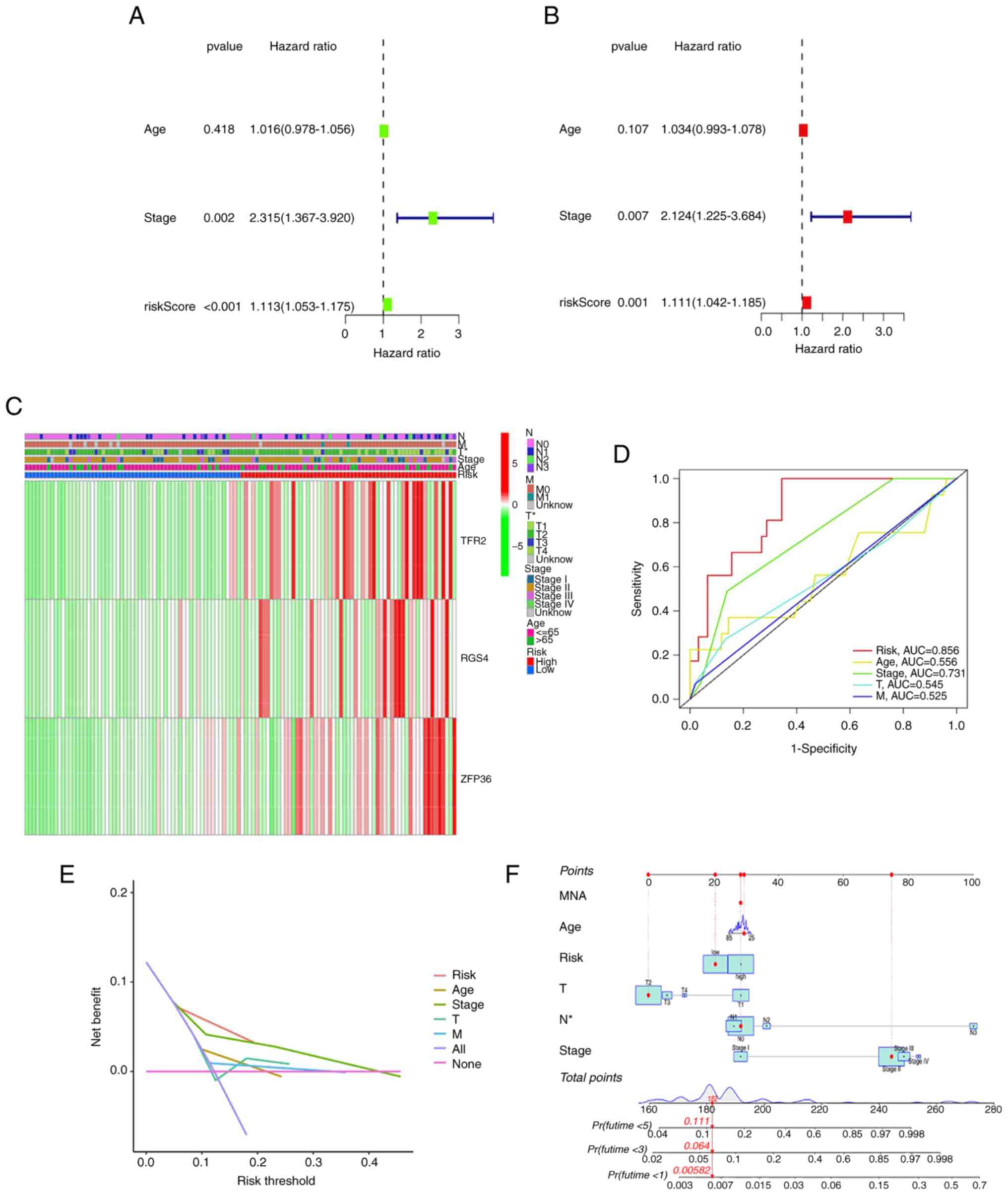
Univariate and multivariate Cox analyses were
conducted to examine the risk scores concerning their independent
prognostic value in predicting the OS of patients with TNBC. The
results indicated the capability of the risk score to function as
an independent prognostic factor in univariate [hazard ratio
(HR)=1.113; 95% confidence interval (CI)=1.053–1.175; P<0.001]
and multivariate (HR=1.111; 95% CI=1.042–1.185; P=0.001) Cox
analyses (Fig. 4A and B). In
addition, multivariate Cox regression analysis demonstrated that
tumor stage could independently function as a prognosis predictor
(P=0.007; Fig. 4B).
A heatmap was utilized to demonstrate the link
between the expression of genes in the prognostic model and
clinical factors (Fig. 4C). Risk
score and clinicopathological factors were examined concerning
their capacity to predict recurrences of TNBC using an ROC curve.
The AUC of the risk score was 0.856, indicative of the improved
capability of the risk score in predicting prognosis in comparison
with the traditional clinicopathological factors (Fig. 4D). DCA demonstrated that the risk
score slightly enhanced the clinical benefit for clinical decisions
(Fig. 4E). Furthermore, the 1-, 3-
and 5-year OS prediction for individuals with TNBC was carried out
by developing a nomogram (Fig. 4F),
which enabled improved individualized prognosis.
GSEA and immune-related function based
on risk scores in TCGA cohort
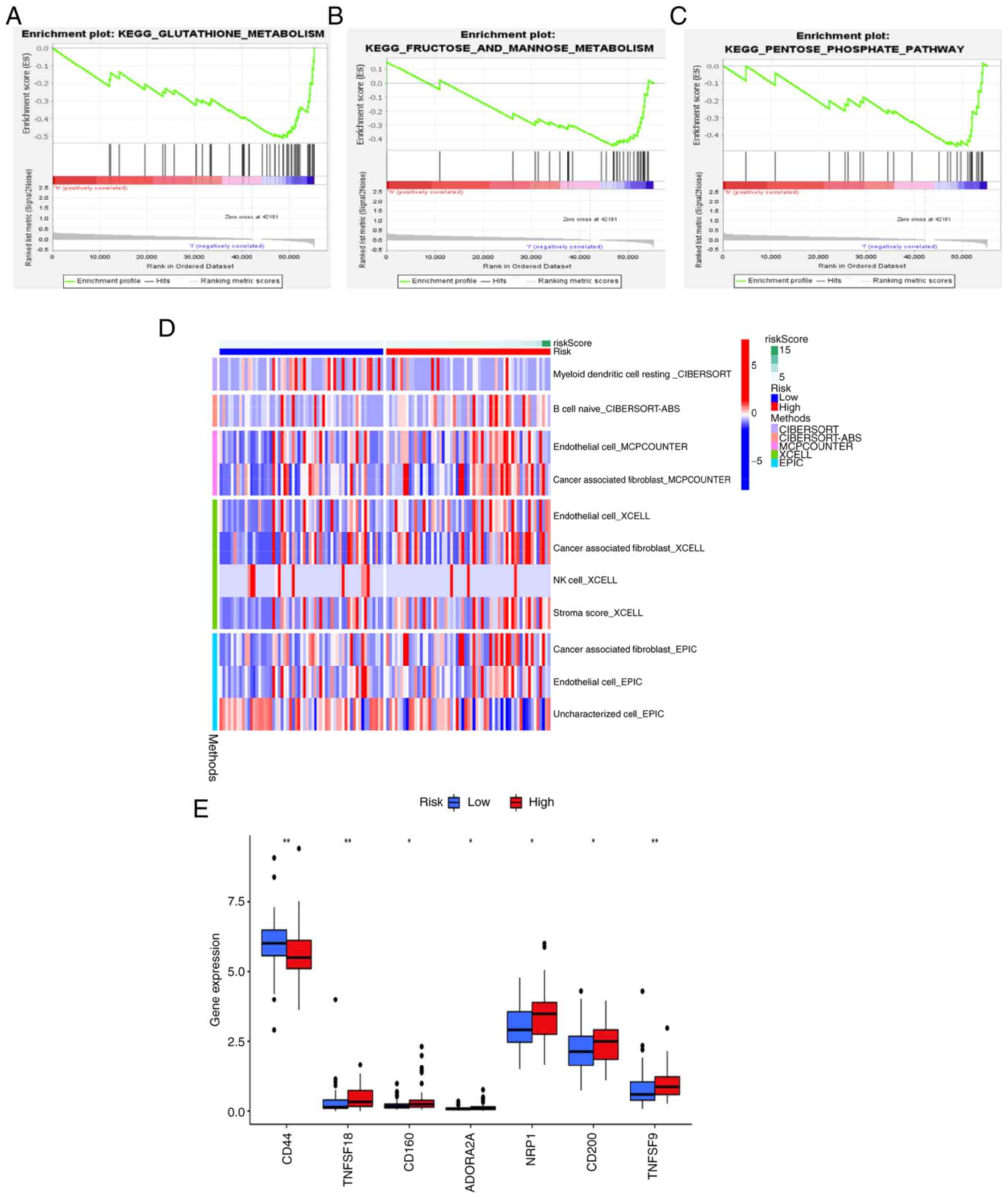
To explore the functions and signaling pathways
linked to the ferroptosis-related risk signature, DEGs were
subjected to GSEA. In low-risk patients, the glutathione
metabolism, fructose and mannose metabolism and pentose phosphate
pathways were enriched (Fig. 5A-C,
respectively). Immunotherapy is an important therapeutic modality
for TNBC. Thus, ssGSEA was conducted to explore the association of
the risk score with the immune status (Fig. 5D). The expression levels of immune
checkpoint molecules were compared across the risk groups and the
results demonstrated that TNF superfamily member (TNFSF)18, CD160,
adenosine A2a receptor, neuropilin-1, CD200 and TNFSF9 were
markedly upregulated, while CD44 was markedly downregulated in the
high-risk group compared with the low-risk group (Fig. 5E). These findings demonstrated
potential ferroptosis-related metabolic pathways and
immunotherapeutic targets of TNBC.
TFR2 knockdown inhibits the
proliferation of TNBC cells
Previous studies have documented the role of RGS4
and ZFP36 in breast cancer (19,20).
Thus, the current study focused on the previously unreported
involvement of TFR2 in TNBC. The expression level of TFR2 in
healthy breast epithelial cells and breast cancer cell lines was
examined. RT-qPCR analysis demonstrated that the TFR2 mRNA levels
were downregulated in healthy and ER-positive breast cancer cells
and upregulated in ER-negative breast cancer cell lines,
particularly in TNBC cell lines (Fig.
6A). The TNBC cell line MDA-MB-436 was subjected to further
analyses, as it exhibited the highest TFR2 mRNA expression levels.
TFR2 was knocked down in MDA-MB-436 cells using siRNA and si-TFR2
1# was selected for further experiments, as it exhibited the
highest knockdown efficiency (Fig.
6B). The mRNA and protein expression levels of TFR2 in the
si-TFR2-transfected group were significantly reduced compared with
the NC group (P<0.05; Fig. 6C).
Next, the cell viability impact of knocking down TFR2 was examined
in MDA-MB-436 cells by the CCK-8 assay. TFR2 knockdown
significantly suppressed the proliferation of MDA-MB-436 cells at
24, 48 and 72 h post-transfection (P<0.01; Fig. 6D). These data suggested that TFR2
may function as an oncogenic factor, which was in agreement with
the data acquired through bioinformatics analyses.
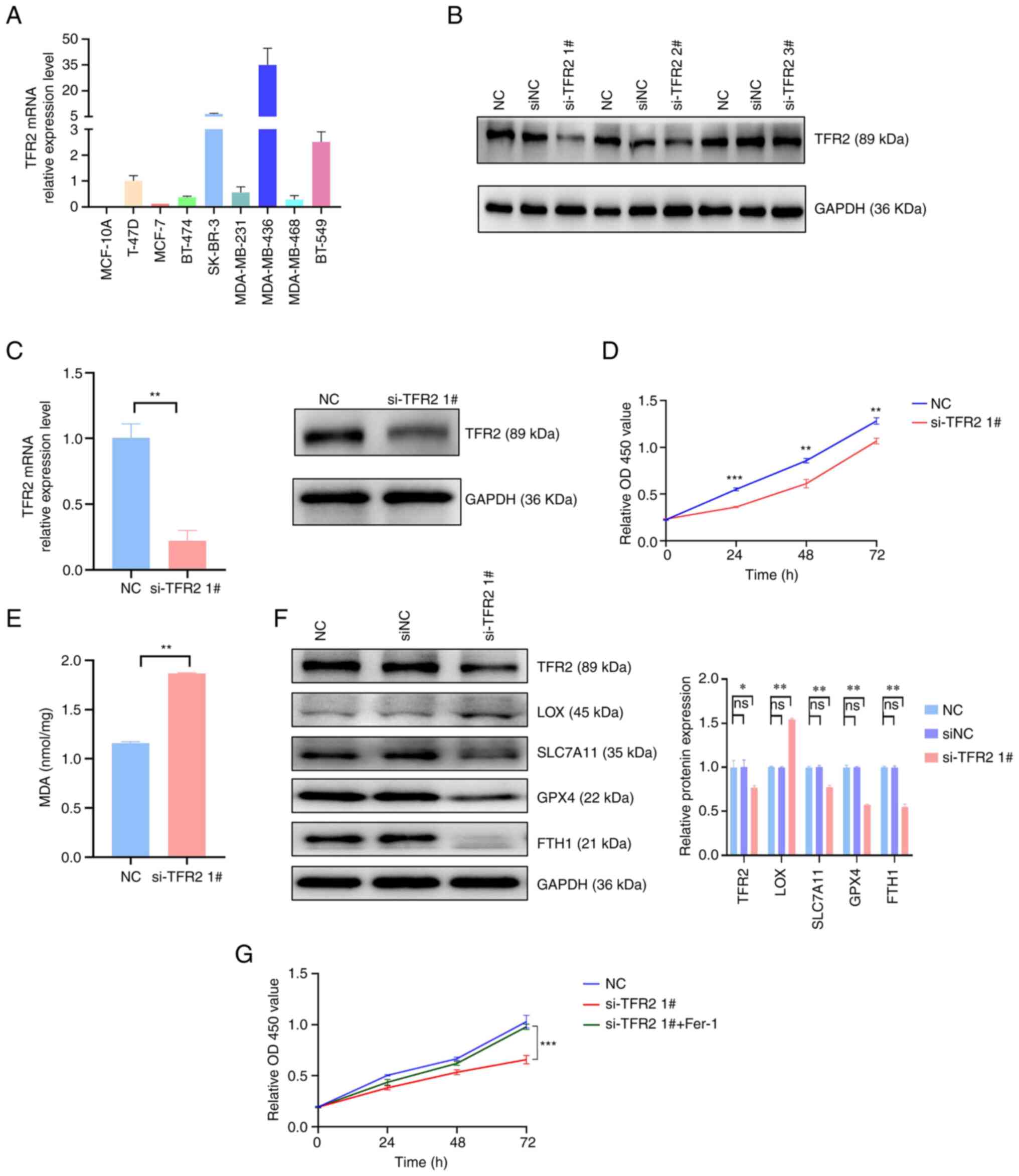 | Figure 6.Knockdown of TFR2 promotes
ferroptosis in the triple-negative breast cancer cell line
MDA-MB-436. (A) The expression levels of TFR2 were detected by
reverse transcription-quantitative PCR in healthy breast and breast
cancer cell lines. (B) Western blot analysis of si-TFR2 knockdown
efficiency. (C) Relative changes in the mRNA and protein expression
levels of TFR2 in MDA-MB-436 cells after transfection with siRNA.
(D) Cell viability of MDA-MB-436 cells was analyzed by a CCK-8
assay at 0, 24, 48 and 72 h after knockdown of TFR2 with siRNA. (E)
Relative changes of MDA levels in MDA-MB-436 cells after knockdown
of TFR2 compared with negative controls. (F) The expression levels
of ferroptosis-related proteins were analyzed by western blot. (G)
The viability of MDA-MB-436 cells was determined using a CCK-8
assay at 0, 24, 48 and 72 h after transfection with siRNA with or
without 0.5 µM ferrostatin-1 treatment. Values are expressed as the
mean ± standard deviation (n=3 independent experiments).
*P<0.05, **P<0.01 and ***P<0.001 vs. NC. CCK-8, cell
counting kit-8; OD, optical density; TFR2, transferrin receptor 2;
NC, negative control; si, short interfering; MDA, malondialdehyde;
LOX, lysyl oxidase; SLC7A11, solute carrier family 7 member 11;
GPX4, glutathione peroxidase 4; FTH1, ferritin heavy chain 1; ns,
not significant. |
TFR2 knockdown promotes ferroptosis in
TNBC cells
The impact of TFR2 knockdown on ferroptosis in
MDA-MB-436 cells was examined. Lipid peroxidation is one of the
characteristics of cellular ferroptosis; therefore, the levels of
lipid peroxidation in MDA-MB-436 cells were examined by analyzing
the MDA levels. TFR2 knockdown significantly increased the MDA
levels in MDA-MB-436 cells (P<0.01; Fig. 6E), which suggested that the
induction of ferroptosis had occurred. Next, the protein expression
levels of ferroptosis-related factors were assessed through western
blotting. Compared with the NC group, the LOX protein expression
levels were significantly elevated, whereas the SLC7A11, GPX4 and
FTH1 levels were significantly reduced in the si-TFR2 transfected
group (P<0.01; Fig. 6F), which
further demonstrated the induction of ferroptosis. Next, the effect
of 0.5 µM ferrostatin-1 on the proliferation of MDA-MB-436 cells
was examined using a CCK-8 assay. Ferrostatin-1 significantly
reversed the decrease in cell viability induced by TFR2 as compared
with the si-TFR2 transfected cells (P<0.05; Fig. 6G). The resulting data were
indicative of the suppressive impact of TFR2 on the viability of
TNBC cells by inducing ferroptosis.
Discussion
It was recently reported that ferroptosis induction
may act as a promising treatment strategy for breast cancer
(21). A previous study highlighted
the role of both experimental compounds and clinical drugs in
inducing ferroptosis in breast cancer cells (22). TNBC, the most malignant subtype of
breast cancer, was previously reported to be intrinsically
sensitive to ferroptosis due to its non-apoptotic characteristics.
The therapeutic effects of a number of types of ferroptosis
inducers on TNBC have been previously evaluated (12,23).
Erastin, the most widely used ferroptosis inducer, increases the
sensitivity of TNBC cells to ferroptosis by upregulating
mitochondrial ROS (24). However,
the application of erastin is limited due to its potentially
serious side effects, such as nephrotoxicity (25). A recent study reported that the
ferroptosis inducer 18-β-glycyrrhetinic acid promoted the death of
TNBC cells through the upregulation of peroxidation (26). However, the association between
genes related to ferroptosis with TNBC and potential biomarkers of
ferroptosis in TNBC has not been completely elucidated to date.
In contrast to a single biomarker-based signature, a
multigene signature model is more accurate for predicting the
prognosis of patients with breast cancer. For instance, Oncotype
Dx® (21-gene signature) and Mammaprint (70-gene
signature) have been widely used in the clinic to evaluate the
favorable impact of chemotherapy in patients with luminal-type
breast cancer (27). A number of
preliminary studies have explored the functions of genes linked to
ferroptosis in breast cancer using a multigene signature model
(16,22,28,29).
However, the involvement of these regulators in TNBC remains to be
elucidated. In the present study, 259 genes linked to ferroptosis
were retrieved from the FerrDb website and a systematic exploration
of the expression levels of these genes in TNBC was conducted. Of
these 259 genes, 60 (23.4%) varied in expression levels across TNBC
and healthy breast tissues. GO and KEGG analyses demonstrated the
enrichment of these DEGs in several ROS-related pathways, which are
among the main mediators of ferroptosis. Univariate Cox analysis
demonstrated an association of five DEGs with OS of patients with
TNBC. Upregulation of these five genes was associated with poor
prognosis. To further verify these results, KM Plotter was utilized
for examining the association between these genes and prognosis of
TNBC. Upregulation of these five genes was associated with
decreased OS. The resulting data suggested that ferroptosis may
serve a role in the pathogenesis of TNBC.
Next, a prognostic model was established using LASSO
Cox regression. It comprised three ferroptosis-related genes (TFR2,
RGS4 and ZFP36). High- and low-risk categories of patients with
TNBC were established with subsequent categorization performed
according to their median risk score. KM and ROC curves were
employed to validate the prognostic value of the model. Analysis of
risk scores in combination with clinical factors indicated the
capability of the risk score to independently predict survival. The
model demonstrated elevated AUC values concerning the 1-, 3- and
5-year survival prediction relative to the clinical model values.
DCA analysis demonstrated that the model may be able to increase
clinical benefits for treatment decisions. The present study also
developed a nomogram, which may aid the 1-, 3- and 5-year OS
prediction of patients with TNBC. Furthermore, validation of the
prognostic value of the model was performed utilizing the GSE25307
dataset. These findings demonstrated that this model is able to
predict the prognosis of individuals with TNBC.
The prognostic model utilized in the current
research comprised three genes linked to ferroptosis, namely TFR2,
RGS4 and ZFP36. To the best of our knowledge, the present study was
the first to report the combination of these genes as a prognostic
signature for TNBC. RGS4, a maker of ferroptosis (30), regulates the activity of
G-protein-coupled receptors in various types of tumor cell
(31,32), including breast cancer (19). Previous research has reported that
RGS4 overexpression suppresses the proliferation of human breast
cancer cells by proteasome degradation (33). Yau et al (34) reported that RGS4 is one of the
prognostic indicators of early TNBC, thus supporting the findings
of the current study. Furthermore, ZFP36, an RNA-binding protein,
regulates mRNA stability and suppresses ferroptosis. Previous
studies have reported that noncoding RNAs can combine with ZFP36 to
modulate the proliferative and migratory capacities of breast
cancer cells (35,36). Dong et al (37) reported that ZFP36 promoted the
tumorigenesis and progression of breast cancer. TFR2, a driver of
ferroptosis, promotes cellular iron transport. A previous study
reported that TFR2 is present in ~26% of colon cancer types
(38). Furthermore, TFR2 regulated
the cell cycle of colon cancer cells utilizing the ERK pathway
(39). In addition, downregulation
of TFR2 in gastric cancer tissue suggested that the expression of
TFR2 was linked to the survival of patients with gastric cancer
(40). To date, no studies have
been conducted on the potential association between TFR2 and breast
cancer. In the present study, the function of TFR2 in TNBC was
examined through in vitro experiments. TFR2 expression in
TNBC cells was upregulated compared with that in healthy breast
epithelial and ER-positive breast cancer cells. In addition, TFR2
was demonstrated to exert oncogenic effects in TNBC. It was noted
that TFR2 knockdown inhibited TNBC cell proliferation by inducing
ferroptosis. To the best of our knowledge, the current study
appears to be the first to report the role of TFR2 in TNBC. Thus,
TFR2-induced ferroptosis may be a potential future treatment target
for TNBC. However, further research is warranted to fully
understand the specific mechanisms via which TFR2 functions in
TNBC. Of note, in the univariate Cox analysis of the present study,
the HR value of ZFP36 was low and the HR value of DUSP1 was also
similar. The possible reason for the aforementioned results may be
that the TNBC sample size of the present study was limited. In
addition, in the three-gene model, differences in HR values may
also indicate differences in the predictive value of each gene and
the effect of TFR2 with the highest HR value on TNBC proliferation
was verified through in vitro experiments. Further
large-sample analyses and functional experiments may explain the
aforementioned results.
Immunotherapy can be used for the treatment of
aggressive malignancies, including TNBC, which often exhibit
resistance to conventional treatment strategies (4,41).
Immunotherapies, such as immune checkpoint inhibitors (ICIs) (such
as cytotoxic T-lymphocyte-associated protein 4 and programmed cell
death protein 1/programmed death-ligand 1) have yielded positive
results in clinical practice (42).
However, the therapeutic effect of ICIs is limited, as only
one-third of cancer patients show a response to these agents. For
instance, the KEYNOTE-086 study reported that 21.4% of individuals
with TNBC responded to pembrolizumab (43). Prior research has reported that
ferroptosis participates in the remodeling of the tumor immune
microenvironment and has suggested the importance of identifying
novel immunotherapy targets for TNBC (44). Therefore, the present study examined
the association of the risk score with the immune status. A total
of five immune algorithms were used to examine the relative
infiltration levels of immune cells in the samples evaluated. The
expression levels of immune checkpoint molecules (CD44, TNFSF18 and
NRP1) varied across the risk groups (high- and low-). Thus, these
molecules may hold promise as possible immunotherapeutic targets
for TNBC.
Recently, Wu et al (45) reported ferroptosis-related gene
signatures in TNBC. The authors established a
15-ferroptosis-related gene prognostic model using LASSO Cox
regression and TCGA datasets. The levels of certain immune cells
varied across different risk groups in TNBC. Compared with the
study by Wu et al (45), the
current study has revealed novel findings. In addition to
establishing a ferroptosis-related prediction model using a TCGA
dataset, the performance of the model was verified using GEO
datasets, which markedly increased the reliability of the findings.
In addition, the present model determined the differential
expression levels of immune checkpoint molecules, thus suggesting
novel potential immunotherapeutic targets for TNBC. Furthermore,
the data acquired from bioinformatics analyses were confirmed
through in vitro assays. However, the current study has
various limitations. For instance, only data from public databases
were used in the present study. In addition, limited in
vitro experiments were performed. Thus, additional biological
assays and clinical analyses must be performed to confirm these
results. Furthermore, some important genes may be excluded because
the model was developed utilizing genes linked to ferroptosis.
In conclusion, in the present study, a new
predictive signature of three ferroptosis-related genes was
established for accurately predicting TNBC prognosis, which may be
used as a tool for clinical applications. The model developed
revealed the differential expression of immune checkpoint
molecules, providing useful insight into the identification of
treatment targets for TNBC. Furthermore, the present study
demonstrated that TFR2 negatively regulated ferroptosis in TNBC.
Additional studies are required to elucidate the role of TFR2 in
TNBC in the future.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National
Natural Science Foundation of China (grant no. NSFC 82160484),
Guizhou Province Science Plan Program [Qian Ke He Foundation-ZK
(2022) general; grant no. 640] and Zunyi Science and Technology
Project [Zunshi Kehe Hz Zi (2022); grant no. 295].
Availability of data and materials
The data used in the present study are publicly
available from the TCGA (http://cancergenome.nih.gov/), GEO (https://www.ncbi.nlm.nih.gov/geo/), FerrDb
(http://www.zhounan.org/ferrdb/), STRING
(https://string-db.org) and Kaplan-Meier Plotter
(http://kmplot.com/analysis). The
datasets used and/or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
YY and RC designed the study. RC and JD prepared the
original draft of the manuscript. YY and JD performed the
experiments. YY and JD confirm the authenticity of all the raw
data. YFH, WH, LL and DL analyzed the results. YY, RC and JD
designed the tables and figures. RC and YY revised the final
manuscript. All authors have read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sharma M, Turaga RC, Yuan Y, Satyanarayana
G, Mishra F, Bian Z, Liu W, Sun L, Yang J and Liu ZR:
Simultaneously targeting cancer-associated fibroblasts and
angiogenic vessel as a treatment for TNBC. J Exp Med.
218:e202007122021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marra A, Viale G and Curigliano G: Recent
advances in triple negative breast cancer: The immunotherapy era.
BMC Med. 17:902019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu B, Luo F, Sun B, Liu W, Shi Q, Cheng
SY, Chen C, Chen G, Li Y and Feng H: KAT6A Acetylation of SMAD3
regulates Myeloid-derived suppressor cell recruitment, metastasis,
and immunotherapy in Triple-Negative breast cancer. Adv Sci
(Weinh). 8:e21000142021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao W, Wang X, Zhou Y, Wang X and Yu Y:
Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor
immunotherapy. Signal Transduct Target Ther. 7:1962022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yao Y, Shi Y, Gao Z, Sun Y, Yao F and Ma
L: Ferroptosis at the crossroads of tumor-host interactions,
metastasis, and therapy response. Am J Physiol Cell Physiol.
323:C95–C103. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sui S, Xu S and Pang D: Emerging role of
ferroptosis in breast cancer: New dawn for overcoming tumor
progression. Pharmacol Ther. 232:1079922022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh T, Beatty A and Peterson JR: The
AMPK-related kinase NUAK2 suppresses glutathione peroxidase 4
expression and promotes ferroptotic cell death in breast cancer
cells. Cell Death Discov. 8:2532022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sha R, Xu Y, Yuan C, Sheng X, Wu Z, Peng
J, Wang Y, Lin Y, Zhou L, Xu S, et al: Predictive and prognostic
impact of ferroptosis-related genes ACSL4 and GPX4 on breast cancer
treated with neoadjuvant chemotherapy. EBioMedicine. 71:1035602021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu G, Wang H, Li X, Huang R and Luo L:
Recent progress on targeting ferroptosis for cancer therapy.
Biochem Pharmacol. 190:1145842021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Doll S, Proneth B, Tyurina YY, Panzilius
E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A,
et al: ACSL4 dictates ferroptosis sensitivity by shaping cellular
lipid composition. Nat Chemical Biol. 13:91–98. 2017. View Article : Google Scholar
|
|
13
|
Timmerman LA, Holton T, Yuneva M, Louie
RJ, Padró M, Daemen A, Hu M, Chan DA, Ethier SP, van't Veer LJ, et
al: Glutamine sensitivity analysis identifies the xCT antiporter as
a common triple-negative breast tumor therapeutic target. Cancer
Cell. 24:450–465. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen PH, Wu J, Ding CC, Lin CC, Pan S,
Bossa N, Xu Y, Yang WH, Mathey-Prevot B and Chi JT: Kinome screen
of ferroptosis reveals a novel role of ATM in regulating iron
metabolism. Cell Death Differ. 27:1008–1022. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang X, Ding CK, Wu J, Sjol J, Wardell S,
Spasojevic I, George D, McDonnell DP, Hsu DS, Chang JT and Chi JT:
Cystine addiction of triple-negative breast cancer associated with
EMT augmented death signaling. Oncogene. 36:43792017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu L, Tian Q, Jiang S, Gao H, Yu S, Zhou
Y, Yan Y, Ren Y, He J and Wang B: A novel Ferroptosis-Related gene
signature for overall survival prediction in patients with breast
cancer. Front Cell Dev Biol. 9:6701842021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Győrffy B: Survival analysis across the
entire transcriptome identifies biomarkers with the highest
prognostic power in breast cancer. Comput Struct Biotechnol J.
19:4101–4109. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xie Y, Wolff DW, Wei T, Wang B, Deng C,
Kirui JK, Jiang H, Qin J, Abel PW and Tu Y: Breast cancer migration
and invasion depend on proteasome degradation of regulator of
G-protein signaling 4. Cancer Res. 69:5743–5751. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pan QH, Fan YH, Wang YZ, Li DM, Hu CE and
Li RX: Long noncoding RNA NNT-AS1 functions as an oncogene in
breast cancer via repressing ZFP36 expression. J Biol Regul Homeost
Agents. 34:795–805. 2020.PubMed/NCBI
|
|
21
|
Li D, Zhang J and Zhao X: Mechanisms and
molecular targets of artemisinin in cancer treatment. Cancer
Invest. 39:675–684. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peng Y, Yu H, Zhang Y, Qu F, Tang Z, Qu C,
Tian J, Zong B, Wang Y, Ren H and Liu S: A ferroptosis-associated
gene signature for the prediction of prognosis and therapeutic
response in luminal-type breast carcinoma. Sci Rep. 11:176102021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Al-Taie Z, Hannink M, Mitchem J,
Papageorgiou C and Shyu CR: Drug repositioning and subgroup
discovery for precision medicine implementation in triple negative
breast cancer. Cancers (Basel). 13:62782021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An Iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liao M, Qin R, Huang W, Zhu HP, Peng F,
Han B and Liu B: Targeting regulated cell death (RCD) with
small-molecule compounds in triple-negative breast cancer: A
revisited perspective from molecular mechanisms to targeted
therapies. J Hematol Oncol. 15:442022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wen Y, Chen H, Zhang L, Wu M, Zhang F,
Yang D, Shen J and Chen J: Glycyrrhetinic acid induces
oxidative/nitrative stress and drives ferroptosis through
activating NADPH oxidases and iNOS, and depriving glutathione in
Triple-Negative breast cancer cells. Free Radic Biol Med.
173:41–51. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Oliveira LJC, Amorim LC, Megid TBC, de
Resende CAA and Mano MS: Gene expression signatures in early breast
cancer: Better together with clinicopathological features. Crit Rev
Oncol Hematol. 175:1037082022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu ZH, Tang Y, Yu H and Li HD: The role of
ferroptosis in breast cancer patients: A comprehensive analysis.
Cell Death Discov. 7:932021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang W, Xu F, Zhao M and Zhang S:
Ferroptosis regulators, especially SQLE, play an important role in
prognosis, progression and immune environment of breast cancer. BMC
Cancer. 21:11602021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stockwell BR: Ferroptosis turns 10:
Emerging mechanisms, physiological functions, and therapeutic
applications. Cell. 185:2401–2421. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guda MR, Velpula KK, Asuthkar S, Cain CP
and Tsung AJ: Targeting RGS4 Ablates Glioblastoma proliferation.
Int J Mol Sci. 21:33002020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu Y, Zheng M, Wang S, Gao L, Gou R, Liu
O, Dong H, Li X and Lin B: Identification of a five-gene signature
of the RGS gene family with prognostic value in ovarian cancer.
Genomics. 113:2134–2144. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Park HJ, Kim SH and Moon DO: Growth
inhibition of human breast carcinoma cells by overexpression of
regulator of G-protein signaling 4. Oncol Lett. 13:4357–4363. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yau C, Esserman L, Moore DH, Waldman F,
Sninsky J and Benz CC: A multigene predictor of metastatic outcome
in early stage hormone receptor-negative and triple-negative breast
cancer. Breast Cancer Res. 12:R852010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ding Y, Li Y, Duan Y, Wang W, Zheng W,
Cheng W, Qi Y, Feng J, Chen Z, Yu T, et al: LncRNA MBNL1-AS1
represses proliferation and cancer Stem-Like properties of breast
cancer through MBNL1-AS1/ZFP36/CENPA Axis. J Oncol.
2022:99993432022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fang S, Zhao Y and Hu X: LncRNA
ADAMTS9-AS1 restrains the aggressive traits of breast carcinoma
cells via sponging miR-513a-5p. Cancer Manag Res. 12:10693–10703.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dong X, Yang Y, Xu G, Tian Z, Yang Q, Gong
Y and Wu G: The initial expression alterations occurring to
transcription factors during the formation of breast cancer:
Evidence from bioinformatics. Cancer Med. 11:1371–1395. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Calzolari A, Deaglio S, Maldi E, Cassoni
P, Malavasi F and Testa U: TfR2 expression in human colon
carcinomas. Blood Cells Mol Dis. 43:243–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Calzolari A, Larocca LM, Deaglio S,
Finisguerra V, Boe A, Raggi C, Ricci-Vitani L, Pierconti F,
Malavasi F, De MariaR, et al: Transferrin receptor 2 is frequently
and highly expressed in glioblastomas. Transl Oncol. 3:123–134.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao QF, Ji J, Cai Q, Wang C, Shi M, Zhou
CF, Zhu ZG and Zhang J: Low expression of transferrin receptor 2
predict poor prognosis in gastric cancer patients. Kaohsiung J Med
Sci. 36:1014–1020. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cao Y, Chen C, Tao Y, Lin W and Wang P:
Immunotherapy for Triple-Negative Breast Cancer. Pharmaceutics.
13:20032021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xu X, Wang D, Chen W, Li N, Suwinski R,
Rossi A, Rosell R, Zhong J and Fan Y: A nomogram model based on
peripheral blood lymphocyte subsets to assess the prognosis of
non-small cell lung cancer patients treated with immune checkpoint
inhibitors. Transl Lung Cancer Res. 10:4511–4525. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Adams S, Schmid P, Rugo HS, Winer EP,
Loirat D, Awada A, Cescon DW, Iwata H, Campone M, Nanda R, et al:
Pembrolizumab monotherapy for previously treated metastatic
triple-negative breast cancer: Cohort A of the phase II KEYNOTE-086
study. Ann Oncol. 30:397–404. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hsu SK, Li CY, Lin IL, Syue WJ, Chen YF,
Cheng KC, Teng YN, Lin YH, Yen CH and Chiu CC: Inflammation-related
pyroptosis, a novel programmed cell death pathway, and its
crosstalk with immune therapy in cancer treatment. Theranostics.
11:8813–8835. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wu S, Pan R, Lu J, Wu X, Xie J, Tang H and
Li X: Development and verification of a prognostic
Ferroptosis-Related gene model in Triple-Negative breast cancer.
Front Oncol. 12:8969272022. View Article : Google Scholar : PubMed/NCBI
|