Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
eighth most common malignancy worldwide, with ~878,000 new cases
and ~444,000 deaths recorded in 2020 (1). The most common type of HNSCC is oral
squamous cell carcinoma (OSCC), which is characterized by
malignancies that develop in the oral cavity and on the lips. OSCC
had a global annual incidence of 378,000 and 178,000 annual deaths
in 2020 (1). These numbers are
markedly higher than the 300,000 new cases and 145,000 deaths
reported in 2015 (2). Despite the
development of novel therapeutic strategies, the 5-year survival
rate of patients with OSCC is ~70% (3). OSCC mainly occurs on the tongue, lips
and oral floor; appearing as ulcers, lumps or lesions with aberrant
color. Tobacco and alcohol consumption are primary risk factors for
OSCC (4), and other factors, such
as betel quid chewing, diet, irradiation and infection with
high-risk types of human papilloma virus (HPV), have been
implicated. Comprehensive analyses have identified genomic
alterations in HNSCC, including OSCC, some of which are
etiologically specific (5). For
example, HNSCC that develops in smokers commonly shows specific
mutations in TP53 and inactivation of CDKN2A, whereas mutations in
these genes are rarely observed in HPV-positive HNSCC (5). However, the precise molecular
mechanisms underlying the development and progression of OSCC
remain unclear.
In our previous study, we screened for aberrantly
methylated genomic regions in a mouse model of skin SCC induced by
a two-stage chemical carcinogenesis protocol. The results showed
that the methylation level of CpG islands (CpGi) in intron 3 of
TFAP2E was significantly elevated, and the expression levels of
TFAP2E were markedly reduced in mouse skin SCC compared with those
in normal skin (6). TFAP2E encodes
nuclear transcription factor activator protein-2 (AP-2)ε. The AP-2
family consists of five members, TFAP2A, TFAP2B, TFAP2C, TFAP2D and
TFAP2E, all of which share highly conserved structures, such as an
α-helical DNA-binding domain and a helix-span-helix motif. They
have been shown to serve pivotal roles in the regulation of early
development, as well as in carcinogenesis (7,8).
During embryogenesis in mice, TFAP2E is mainly expressed in neural
tissues (9), and is involved in the
development of the olfactory bulb (10) and retina (11). TFAP2E is also expressed during
chondrocyte differentiation in both mice (12) and humans where it regulates the
expression of integrin α10 (13).
Accumulating evidence has suggested that TFAP2E acts
as a tumor suppressor in several types of cancer. In humans, the
TFAP2E gene is located on chromosome 1p34, a genetic region that is
deleted in numerous types of cancer (14). Similar to the observations in mice
(6), hypermethylation of the CpGi
in intron 3 of human TFAP2E has been observed to be associated with
decreased TFAP2E transcription level, and nonresponse to
5-fluorouracil (5-FU) in colorectal cancer (CRC) and gastric cancer
(GC) (15,16). In addition, hypermethylation of
TFAP2E is more frequently detected in urinary genomic DNA obtained
from patients with prostate cancer than from young healthy male
subjects (17). Our previous study
reported that lower levels of TFAP2E expression are significantly
associated with shorter survival in patients with neuroblastoma
(NB) (18). Collectively, these
findings strongly suggested that TFAP2E is a potential tumor
suppressor.
The present study examined the possible role of
TFAP2E in the development or progression of OSCC. For this purpose,
the effects of TFAP2E knockdown on the viability and cell cycle
progression of OSCC cells were analyzed.
Materials and methods
Cell lines and culture conditions
The human gingival cancer cell line Ca9-22 was
obtained from the Japanese Collection of Research Bioresources Cell
Bank and the human tongue cancer-derived cell line HSC-4 was
obtained from the RIKEN BioResource Center. As it has been reported
that some stocks of Ca9-22 are contaminated with MSK-922 (19), short tandem repeat analysis was
performed for the Ca9-22 cell line by BEX Co., Ltd., and it was
confirmed to be authentic. Ca9-22 cells were cultured in minimal
essential medium (Nacalai Tesque, Inc.) supplemented with 600 mg/l
glutamine and 10% heat-inactivated fetal bovine serum (FBS;
Nichirei Biosciences, Inc.). HSC-4 cells were maintained in
RPMI-1640 medium (Nacalai Tesque, Inc.) supplemented with 10%
heat-inactivated FBS. Both media contained 100 IU/ml penicillin
(Thermo Fisher Scientific, Inc.) and 100 mg/ml streptomycin (Thermo
Fisher Scientific, Inc.). Cells were maintained at 37°C in an
atmosphere containing 95% air and 5% CO2.
Kaplan-Meier survival analysis
To assess the effects of expression levels of TFAP2E
on the survival rate of patients with OSCC, Kaplan-Meier survival
analysis, followed by log-rank test for statistical analysis, was
performed using the online cBioPortal tool (http://www.cbioportal.org). Sample data from 322
patients in The Cancer Genome Atlas (TCGA) HNSCC Firehose Legacy
data set, including gene expression levels in OSCC tissues and
survival period, were obtained from cBioPortal.
Small interfering RNA (siRNA)-mediated
knockdown of TFAP2E
Ca9-22 and HSC-4 cells were seeded and cultured for
24 h before transfecting with 10 nM siRNA using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions at
room temperature. Anti-TFAP2E siRNA (cat. no. s50548; Thermo Fisher
Scientific, Inc.), a control siRNA (si-N/C; cat. no. 4390843;
Thermo Fisher Scientific, Inc.) and anti-TP53 siRNA (cat. no.
sc-29435; Santa Cruz Biotechnology, Inc.) were used in the present
study.
Analysis of cell proliferation
To analyze the cell proliferation rate, cells were
seeded into 96-well culture plates at a density of 5×103
cells/well and transfected with siRNAs after 24 h. After 1, 2, 3, 4
and 5 days, the proliferation of transfected cells was measured
using the standard water-soluble tetrazolium salt (WST)-8 assay
with Cell Count Reagent CF (Nacalai Tesque, Inc.). Briefly, culture
medium was replaced with fresh medium containing 10 µl of WST8
solution, and the cells were cultured for 1 h, followed by
measurement of the absorbance at 450 nm using a plate reader
(Spectra Max ABS plus; Molecular Devices, LLC).
Cell viability was also analyzed by cell staining.
Briefly, cells were seeded into 6-well culture plates at a density
of 5×103 cells/well. A total of 24 h after seeding,
cells were transfected with siRNAs and were cultured for 10 days.
In the middle of culture, i.e. 5 days after siRNAs transfection,
the medium was replaced and siRNA transfection was performed again.
Cells were fixed and stained using a Diff-Quick Stain Kit (Sysmex
Corporation) at room temperature. Briefly, cells were fixed with
99% methanol for 10 min and stained with Diff-Quick solution II for
5 min, followed by washing with distilled water.
For the analysis of drug sensitivity, cells were
seeded into 96-well culture plates at a density of 5×103
cells/well and transfected with siRNAs after 24 h. In addition, 1,
5 and 10 µM cisplatin (CDDP; MilliporeSigma) or 25, 50 and 100 µM
of H2O2 (Nacalai Tesque, Inc.) were added 24
h after the transfection, followed by culture for additional 24 h
and analysis of cell viability using WST-8 assay.
Analysis of cell cycle
distribution
The cells were plated at a density of
2×104 cells/ml in culture dishes 60 mm in diameter (5
ml/dish) and were transfected with siRNAs 24 h later. A total of 3
days after transfection, both floating and attached cells were
collected by centrifugation, washed in PBS and fixed in 70% ethanol
at −20°C overnight for fluorescence-activated cell sorting (FACS)
analysis. The cells were washed in PBS before incubating in PBS
containing 0.1% FBS, 25 µg/ml propidium iodide and 200 µg/ml RNase
A for 15 min at room temperature.
Cell cycle progression was monitored in the presence
of nocodazole, which prevents cells from completing mitosis
(20). The cells were plated and
transfected with siRNAs as aforementioned. After 2 days of culture,
the medium was replaced with fresh medium containing 100 ng/ml
nocodazole (FUJIFILM Wako Pure Chemical Corporation). The cells
were harvested every 3 h and fixed in 70% ethanol at −20°C
overnight. The cells were washed in PBS containing 0.5% FBS,
followed by incubation in PBS containing 0.5% FBS, 2.5 µg/ml
propidium iodide, 200 µg/ml RNase A and anti-phosphorylated histone
H3 serine 28 (p-H3Ser28) conjugated with Alexa 647 (cat. no.
641006; BioLegend, Inc.) for 1 h at room temperature.
All of the aforementioned cells were subjected to
FACS analysis using a Gallios Flow Cytometer (Beckman Coulter,
Inc.) and data analysis was performed using Kaluza Analysis
Software Ver. 2.1 (Beckman Coulter, Inc.). The percentage
distribution of cells in distinct cell cycle phases was calculated
based on Michael H. Fox algorithm (21).
Reverse transcription-quantitative
RT-qPCR
Ca9-22 and HSC-4 cells were plated at a density of
2×104 cells/ml in culture dishes 60 mm in diameter (5
ml/dish) and were transfected with siRNAs 24 h after seeding. Total
RNA was extracted from cells 2 or 5 days after siRNA transfection,
using RNeasy mini kits (Qiagen GmbH) and cDNA was synthesized by RT
using an iScript cDNA synthesis system (Bio-Rad Laboratories,
Inc.), according to the manufacturers' instructions. qPCR for
TFAP2E and 18S rRNA, which was used as a housekeeping gene
(22), was performed using the
TaqMan Pre-Developed Assay Reagents Hs00698734_m1 and Hs99999901_s1
for TFAP2E and 18S rRNA (Thermo Fisher Scientific, Inc.)
respectively, and Premix Ex Taq Perfect Real Time (Takara Bio,
Inc.). qPCR conditions were as follows: Initial denaturation at
95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C
for 30 sec. qPCR for TP53 was performed using SYBR Premix Ex Taq™
(Takara Bio, Inc.) with the following primers: Sense
5′-CCCCTCCTGGCCCCTGTCATCTTC-3′ and antisense
5′-GCAGCGCCTCACAACCTCCGTCAT-3′. qPCR conditions were as follows:
Initial denaturation at 95°C for 30 sec, followed by 40 cycles at
95°C for 5 sec, 56°C for 10 sec and 72°C for 30 sec. Measurements
were performed in triplicate. Data processing was performed using a
standard curve-based method (23).
A mixture of cDNA generated from the total RNA of Ca9-22 and HSC-4
cells was used to obtain a standard curve for each gene.
Western blotting
Ca9-22 and HSC-4 cells were plated at a density of
2×104 cells/ml in culture dishes 60 mm in diameter (5
ml/dish) and were transfected with siRNAs 24 h after seeding. Cells
were collected 2 days after transfection with siRNAs. For the
analysis of cell cycle-related proteins, cells were treated with
100 ng/ml nocodazole 2 days after the transfection and collected 0,
3, 6, 9 and 12 h after nocodazole addition. Cells were lysed in
RIPA buffer containing a protease and phosphatase inhibitor
cocktail (Nacalai Tesque, Inc.), before passing through a 1-ml
syringe with a 27-G needle. Protein concentrations in the lysates
were measured using a Bio-Rad DC kit (Bio-Rad Laboratories, Inc.).
The lysates containing 10 µg protein were separated by SDS-PAGE on
4–12% gels, followed by electroblotting onto Immobilon-P membranes
(MilliporeSigma). Membranes were blocked with Blocking-one (Nacalai
Tesque, Inc.) overnight at 4°C and incubated with anti-TFAP2E (cat.
no. 29-175; ProSci, Inc.; 1:1,000), anti-Cyclin B1 (D-11; cat. no.
sc-7393; Santa Cruz Biotechnology, Inc.; 1:1,000), anti-histone H3
(cat. no. 9715; Cell Signaling Technology, Inc.; 1:1,000),
anti-p-H3Ser28 (cat. no. 9713; Cell Signaling Technology, Inc.;
1:1,000), anti-GAPDH (cat. no. ab9485; Abcam.; 1:1,000) and
anti-TP53 (DO-1; cat. no. sc-126; Santa Cruz Biotechnology, Inc.;
1:1,000) antibodies at 4°C. After incubation for 24 h, the
membranes were washed in Tris-buffered saline (TBS) containing 0.1%
Tween 20 (TBS-T), followed by incubation with the appropriate
horseradish peroxidase-conjugated secondary antibodies (cat. nos.
NA934V and NA931V; Cytiva; 1:2,000) for 1 h at room temperature.
The membranes were washed extensively with TBS-T and the results
were visualized using Chemi-Lumi-One Super (Nacalai Tesque, Inc.)
and an ImageQuant™ 800 system (Danaher Corp.). Experiments were
performed at least three times and representative blots are shown.
The band density of histone H3 (total H3) and p-H3Ser28 was
semi-quantified using ImageJ (ver. 1.48; National Institutes of
Health) (24) to normalize signal
intensity of p-H3Ser28 to total H3.
Statistical analysis
Statistical analyses to examine the significance of
the differences between two groups were performed using Student's
unpaired t-test. One-way ANOVA followed by post-hoc Tukey's test
was performed to examine the significance of the differences among
multiple groups. All statistical analyses were performed using JMP
software ver. 11.2 (SAS Institute, Inc.). Data are presented as the
mean ± SD of at least three independent experiments. In all
analyses, P<0.05 was considered to indicate a statistically
significant difference.
Results
Low TFAP2E expression is related to
poor prognosis of OSCC
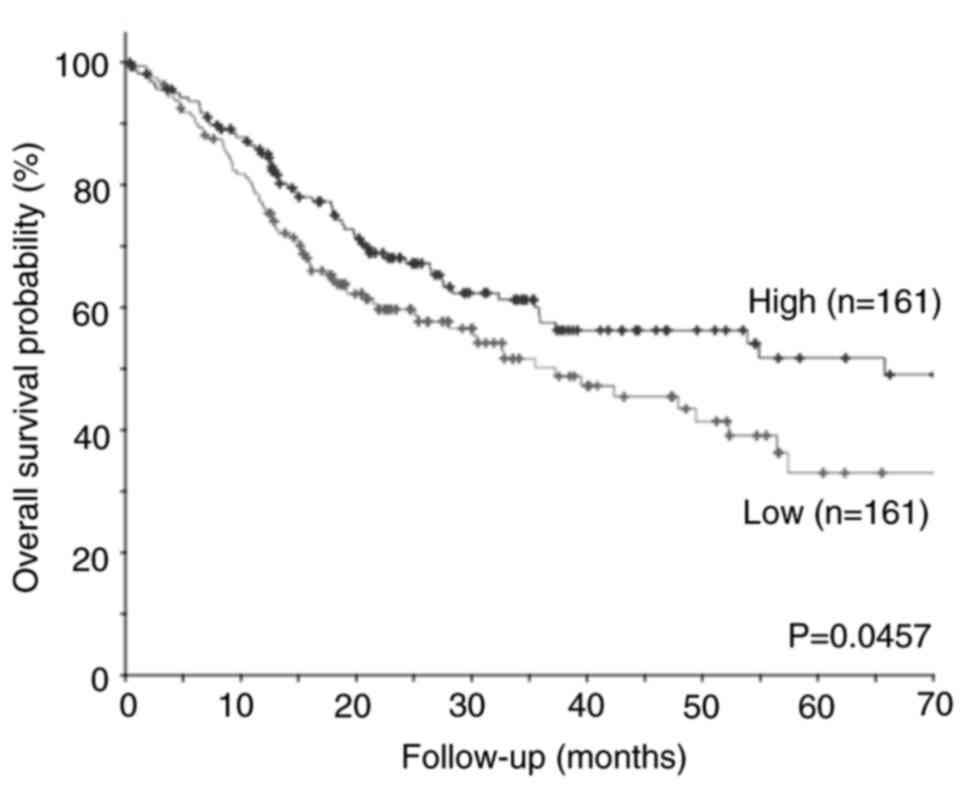
Kaplan-Meier survival analysis of TCGA public
microarray data sets was performed for 322 cases, including 128
cases of OSCC in the oral tongue, 27 in the base of the tongue, 62
in the floor of the mouth, 22 in the buccal mucosa, 7 in the hard
palate, 3 in the lips and 73 in the oral cavity. The patients were
divided to low or high expression groups using the median
expression level of TFAP2E as a cutoff. Kaplan-Meier analysis
showed that low TFAP2E expression was significantly associated with
a shorter overall survival in patients with OSCC (Fig. 1). These results strongly suggested
that TFAP2E plays a suppressive role in the malignant progression
of OSCC.
Knockdown of TFAP2E promotes the
proliferation of OSCC-derived cells
To assess the hypothesis that TFAP2E acts as a tumor
suppressor in OSCC, the present study examined the effects of
TFAP2E knockdown on human OCSS-derived Ca9-22 and HSC-4 cells using
anti-TFAP2E siRNA. RT-qPCR and western blotting confirmed that
Ca9-22 and HSC-4 cells transfected with anti-TFAP2E siRNA exhibited
reduced endogenous TFAP2E expression compared with in the control
group (Fig. 2A and B). The standard
WST-8 cell survival assay showed that TFAP2E knockdown resulted in
a marked increase in cell proliferation (Fig. 2C and D). Consistent with these
results, the number of viable TFAP2E-depleted cells was
substantially greater than that in the control group (Fig. 2E and F).
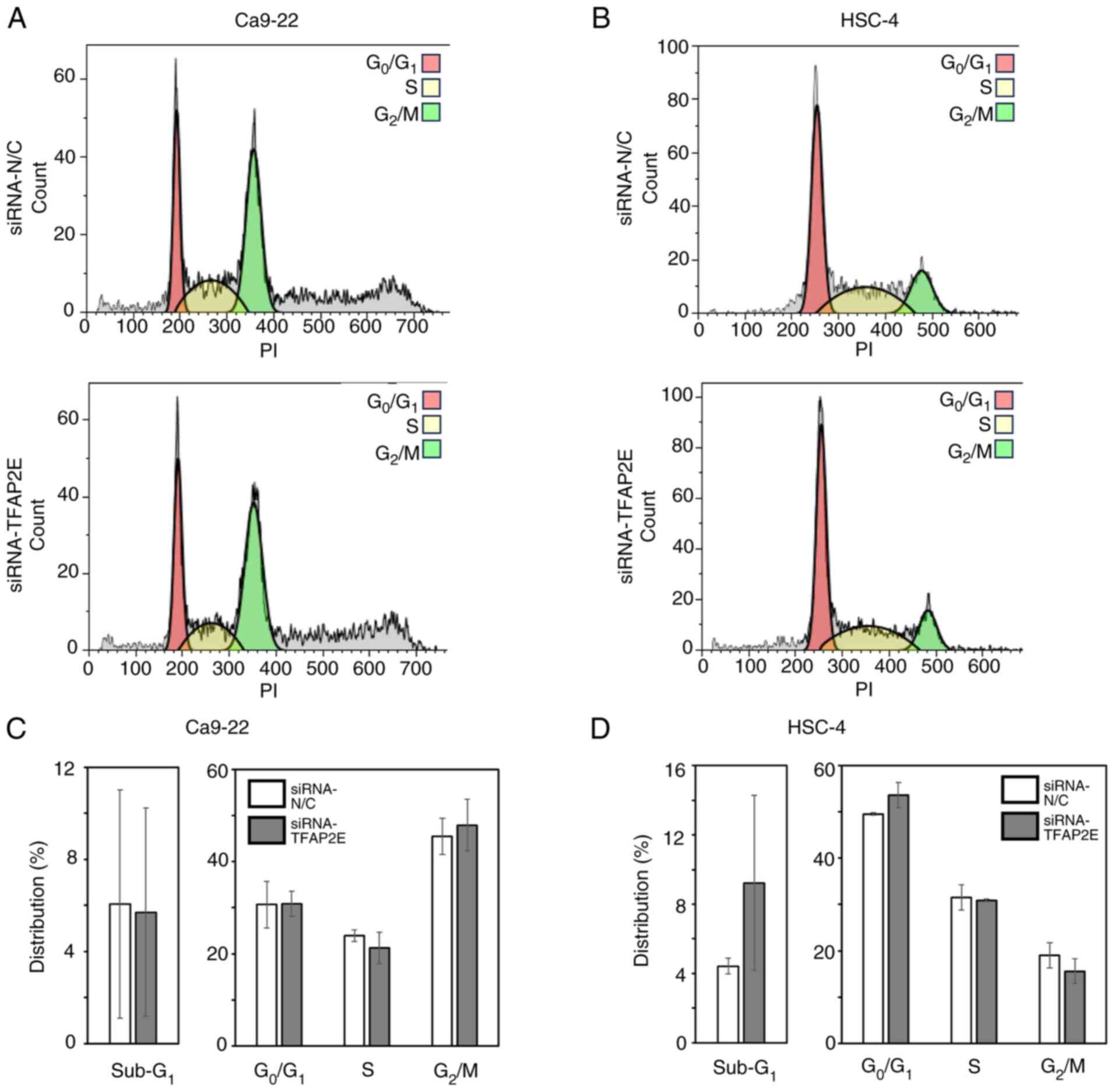
The present study also examined whether TFAP2E
knockdown affected cell proliferation or cell cycle progression by
analyzing the cell cycle distribution of Ca9-22 and HSC-4 cells
transfected with anti-TFAP2E siRNA or control siRNA using FACS.
There were no significant differences in either cell cycle
distribution or the proportion of dead cells, as indicated by
sub-G1 DNA content, between TFAP2E-knockdown and control
cell groups (Fig. 3). In addition,
knockdown of TFAP2E had a negligible effect on the resistance of
the cells to CDDP and H2O2 (Fig. S1), indicating that knockdown of
TFAP2E may accelerate cell growth via some mechanism other than
augmenting stress resistance in the cells.
Knockdown of TFAP2E results in rapid
G2/M transition of OSCC cells
To investigate the mechanisms by which TFAP2E
depletion promotes cell proliferation, the present study analyzed
the cell cycle progression rate. Because anti-TFAP2E siRNA did not
work when cells were treated with a double thymidine block to
achieve synchronization to the late G1 phase (data not
shown), cell cycle progression was analyzed using the previously
reported method for asynchronous cells (25). In this method, cell cycle
progression was monitored in the presence of nocodazole. As
nocodazole prevent cells from completing mitosis, cell cycle
progression rate could be analyzed by measuring the accumulation
rate of G2/M cells. FACS analysis showed that the
proportion of cells in the G0/G1 phase
decreased and that of cells in the G2/M phase increased
in a time-dependent manner (Figs. 4A,
B and S2); however, there were
few significant differences between control and TFAP2E-knockdown
cells. The G2 and M phases could not be distinguished
based on the DNA content of the cells, since cells in both
G2 and M phase have 4N DNA content (tetraploidy);
therefore, the present study also monitored the population of cells
positive for p-H3Ser28, which is a reliable marker of the early M
phase (26). The proportion of
p-H3Ser28-positive cells increased over time, and the rate of
increase was significantly higher in TFAP2E-depleted Ca9-22 cells
compared with that in the control group (Figs. 4C and S3A); similar results were observed in
HSC-4 cells (Figs. 4D and S3B). Consistent with these results,
western blotting demonstrated that the accumulation of p-H3Ser28
and cyclin B1, an alternative molecular marker for the
G2/M phase, occurred earlier in TFAP2E-knockdown cells
than in control cells (Fig. 5).
Collectively, these results suggested that TFAP2E may serve a role
in regulating the G2/M transition in OSCC cells. As a
tumor suppressor gene TP53 is a key molecule in regulation of the
G2/M transition; therefore, the present study examined
whether depletion of TFAP2E increased cell proliferation via
suppressing TP53 expression. The data showed that knockdown of
TFAP2E resulted in downregulation of TP53 at the protein level, but
not at the mRNA level, in both Ca9-22 and HSC-4 cells (Fig. S4A and B). However, knockdown of
TP53 using siRNA suppressed, rather than increased, the viability
of both cells (Fig. S4C-F). These
results indicate that accelerated proliferation of TFAP2E-knockdown
cells could not owe to downregulation of the TP53 protein.
Discussion
It has been reported that TFAP2E acts as a tumor
suppressor in numerous types of cancer. For example,
hypermethylation of the TFAP2E genomic locus and reduced TFAP2E
transcription have been shown to be associated with poor prognosis
and resistance to treatment with 5-FU in patients with CRC and GC
(15,16). Previously, we demonstrated that
TFAP2E depletion in NB-derived cells attenuates the induction of
cell death in response to adriamycin, CDDP or ionizing radiation
(18). These findings indicated
that TFAP2E exerts its tumor-suppressive effect by augmenting the
response to DNA damage response in cancer cells. In the present
study, silencing TFAP2E in OSCC-derived Ca9-22 and HSC-4 cells
increased their proliferation but did not affect their sensitivity
to CDDP or H2O2, indicating that TFAP2E
knockdown did not affect DNA damage response in these cells.
The present study demonstrated that knockdown of
TFAP2E increased the proliferation rate, rather than suppressing
cell death, in OSCC cells. There were no marked differences in cell
cycle distribution pattern at certain time points or in the cell
cycle progression rate from the G0/G1 to
G2/M in the presence of nocodazole between
TFAP2E-knockdown cells and control cells. However, in the presence
of nocodazole, the rate of increase in p-H3Ser28-positive cells was
significantly higher in TFAP2E-depleted cells than that in the
control group. Since p-H3Ser28 is detectable at prophase/early
anaphase during cell cycle progression (26), these observations suggested that
TFAP2E may participate in the regulation of the G2/M
transition, thereby contributing to the attenuation of OSCC cell
proliferation. Cell cycle processes are guarded by cell cycle
checkpoints, which survey DNA damage, DNA replication errors and
incomplete spindle assembly (27,28).
Errors in these processes induce cell cycle arrest or delay,
thereby preventing the accumulation and propagation of genetic
errors during cell division. In the G2 phase, the
checkpoint machinery can be activated by DNA damage, resulting in
the inhibition of cyclin-dependent kinase 1 activity and preventing
entry into mitosis (29). To
clarify the molecular mechanisms by which TFAP2E may act during
cell cycle progression, the expression levels and phosphorylation
status of various functional proteins implicated in this process,
including PLK1, ATM, WEE1 and CDC25, were examined. The results
showed that depletion of TFAP2E did not exhibit a marked effect on
the expression or activation levels of these molecules (data not
shown).
It is well established that the tumor suppressor
molecule TP53 serves a pivotal role in regulating the
G2/M transition. TP53 induces cell cycle arrest at the
G1/S or G2/M phase in response to various
stresses through transactivation of a number of downstream target
genes, including P21/WAF1, GADD45 and 14-3-3s (30). The present results showed that
knockdown of TFAP2E in OSCC cells caused downregulation of TP53 at
the protein level, but not the mRNA level, suggesting that TFAP2E
may contribute to the stability of TP53. Ca9-22 and HSC-4 cells
carry p53 mutations, R248W and R248Q, respectively, which have been
reported to exhibit oncogenic functions (31,32).
Predictably, depletion of TP53 suppressed, rather than increased,
the proliferation rate of both cell lines, indicating that
downregulation of the TP53 protein could not be responsible for the
increased proliferation rate of TFAP2E-knockdown cells.
Nevertheless, as the stability of mutant TP53 is regulated by
multiple pathways (33), the
observations presented in Fig. S4B
suggest the involvement of TFAP2E in one of these pathways. Further
investigations are required to identify the target molecules of
TFAP2E and to elucidate the mechanisms underlying its regulatory
effect on the cell cycle.
An important limitation of the present study is that
all analyses were done using asynchronous cells. Although the
results suggested that TFAP2E suppressed cell proliferation by
regulating G2/M transition, the possibility that there
are other mechanisms by which TFAP2E affects cell proliferation
cannot be ruled out. To verify this possibility, the analysis may
need to be performed using phase-synchronized cells. Since we
previously observed that anti-TFAP2E siRNA did not work in Ca9-22
and HSC-4 cells when they were treated with a double thymidine
block protocol to synchronize the cell cycle (data not shown), we
are planning to establish TFAP2E stable knockdown cells using short
hairpin RNA. In addition, in our future work, other OSCC cell
lines, such as HSC3 and UM-SCC6, in which anti-TFAP2E siRNA did not
work (data not shown), will be tested. Another limitation of the
present study is that the experiments were performed using only
TFAP2E knockdown cells. Analysis of the effects of TFAP2E
overexpression on OSCC cell function will provide further insight
to understand its role in cell cycle regulation. In the future, we
will establish TFAP2E-overexpressing OSCC cells along with
knockdown cells, and will conduct a comprehensive analysis using
those cells.
In conclusion, the present study showed that TFAP2E
can suppress the proliferation of OSCC cells at least in part
through regulating the G2/M transition. This observation
may explain the reason why patients with OSCC with lower TFAP2E
expression had a shorter survival time.
Supplementary Material
Supporting Data
Acknowledgments
The authors would like to thank Mr. Yushi Arai, Ms.
Mayuko Yano and Mr. Shotaro Yoshida (Nihon University School of
Dentistry) for technical assistance.
Funding
This study was supported in part by KAKENHI (grant no. 22K17028)
to YI, and by grants from the Dental Research Center, Nihon
University School of Dentistry to Kyoko Fujiwara, and the Sato
Fund, Nihon University School of Dentistry to KF.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
RS and KF planned the experiments. RS, KF, ENM, YI,
BY, EMF and YK performed the experiments. RS, KF, TT and SS wrote
the manuscript. KF, ENM, SU, TK, TT and SS confirm the authenticity
of all the raw data. KF, YI, SU, TK, TT and SS contributed to
interpretation of the data. KF, SU, TK, TT and SS critically
revised and approved for the paper for publication. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
National Cancer Institute, . SEER Cancer
Statistics Review (CSR) 1975–2018. NCI; Bethesda, MD: 2021,
https://seer.cancer.gov/csr/1975_2018/
|
|
4
|
Kumar M, Nanavati R, Modi TG and Dobariya
C: Oral cancer: Etiology and risk factors: A review. J Cancer Res
Ther. 12:458–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cancer Genome Atlas Network, .
Comprehensive genomic characterization of head and neck squamous
cell carcinomas. Nature. 517:576–582. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fujiwara K, Ghosh S, Liang P, Morien E,
Soma M and Nagase H: Genome-wide screening of aberrant DNA
methylation which associated with gene expression in mouse skin
cancers. Mol Carcinog. 54:178–188. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kolat D, Kaluzí Nska Z, Bednarek AK and
Zbieta Pluciennik E: The biological characteristics of
transcription factors AP-2α and AP-2γ and their importance in
various types of cancers. Biosci Rep. 39:BSR201819282019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Eckert D, Buhl S, Weber S, Jäger R and
Schorle H: The AP-2 family of transcription factors. Genome Biol.
6:2462005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang HV, Vaupel K, Buettner R, Bosserhoff
AK and Moser M: Identification and embryonic expression of a new
AP-2 transcription factor, AP-2 epsilon. Dev Dyn. 231:128–135.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Feng W, Simoes-de-Souza F, Finger TE,
Restrepo D and Williams T: Disorganized olfactory bulb lamination
in mice deficient for transcription factor AP-2epsilon. Mol Cell
Neurosci. 42:161–171. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jain S, Glubrecht DD, Germain DR, Moser M
and Godbout R: AP-2ε Expression in developing retina: Contributing
to the nolecular diversity of amacrine cells. Sci Rep. 8:33862018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wenke AK, Grä Ssel S, Moser M and
Bosserhoff AK: The cartilage-specific transcription factor Sox9
regulates AP-2e expression in chondrocytes. FEBS J. 276:2494–2504.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wenke AK, Rothhammer T, Moser M and
Bosserhoff AK: Regulation of integrin alpha10 expression in
chondrocytes by the transcription factors AP-2epsilon and Ets-1.
Biochem Biophys Res Commun. 345:495–501. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Giaretti W, Molinu S, Ceccarelli J and
Prevosto C: Chromosomal instability, aneuploidy, and gene mutations
in human sporadic colorectal adenomas. Cell Oncol. 26:301–305.
2004.PubMed/NCBI
|
|
15
|
Ebert MP, Tänzer M, Balluff B,
Burgermeister E, Kretzschmar AK, Hughes DJ, Tetzner R, Lofton-Day
C, Rosenberg R, Reinacher-Schick AC, et al: TFAP2E-DKK4 and
chemoresistance in colorectal cancer. N Engl J Med. 366:44–53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun J, Du N, Li J, Zhou J, Tao G, Sun S
and He J: Transcription Factor AP2ε: A potential predictor of
chemoresistance in patients with gastric cancer. Technol Cancer Res
Treat. 15:285–295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Payne SR, Serth J, Schostak M, Kamradt J,
Strauss A, Thelen P, Model F, Day JK, Liebenberg V, Morotti A, et
al: DNA methylation biomarkers of prostate cancer: Confirmation of
candidates and evidence urine is the most sensitive body fluid for
non-invasive detection. Prostate. 69:1257–1269. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hoshi R, Watanabe Y, Ishizuka Y, Hirano T,
Nagasaki-Maeoka E, Yoshizawa S, Uekusa S, Kawashima H, Ohashi K,
Sugito K, et al: Depletion of TFAP2E attenuates adriamycin-mediated
apoptosis in human neuroblastoma cells. Oncol Rep. 37:2459–2464.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao M, Sano D, Pickering CR, Jasser SA,
Henderson YC, Clayman GL, Sturgis EM, Ow TJ, Lotan R, Carey TE, et
al: Assembly and initial characterization of a panel of 85
genomically validated cell lines from diverse head and neck tumor
sites. Clin Cancer Res. 17:7248–7264. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jordan MA, Thrower D and Wilson L: Effects
of vinblastine, podophyllotoxin and nocodazole on mitotic spindles.
Implications for the role of microtubule dynamics in mitosis. J
Cell Sci. 102((Pt 3)): 401–416. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fox MH: A model for the computer analysis
of synchronous DNA distributions obtained by flow cytometry.
Cytometry. 1:71–77. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Larionov A, Krause A and Miller W: A
standard curve based method for relative real time PCR data
processing. BMC Bioinformatics. 6:622005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thellin O, Zorzi W, Lakaye B, De Borman B,
Coumans B, Hennen G, Grisar T, Igout A and Heinen E: Housekeeping
genes as internal standards: Use and limits. J Biotechnol.
75:291–295. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schneider CA, Rasband WS and Eliceiri KW:
NIH Image to ImageJ: 25 years of image analysis. Nat Methods.
9:971–675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sherman J and Wang R: Rapid profiling of
G2 phase to mitosis progression by flow cytometry in asynchronous
cells. Cell Cycle. 19:2897–2905. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pérez-Cadahía B, Drobic B and Davie JR: H3
phosphorylation: Dual role in mitosis and interphase. Biochem Cell
Biol. 87:695–709. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
De Wever V, Lloyd VC, Nasa I, Nimick I,
Trinkle-Mulcahy M, Gourlay R, Morrice N and Moorhead GB: Isolation
of human mitotic protein phosphatase complexes: Identification of a
complex between protein phosphatase 1 and the RNA helicase Ddx21.
PLoS One. 7:e395102012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Matthews HK, Bertoli C and de Bruin RAM:
Cell cycle control in cancer. Nat Rev Mol Cell Biol. 23:74–88.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Medema RH and Macurek L: Checkpoint
control and cancer. Oncogene. 31:2601–2613. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stark GR and Taylor WR: Control of the
G2/M transition. Mol Biotechnol. 32:227–248. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan W and Chen X: Characterization of
functional domains necessary for mutant p53 gain of function. J
Biol Chem. 285:14229–14238. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ng JW, Lama D, Lukman S, Lane DP, Verma CS
and Sim AY: R248Q mutation-Beyond p53-DNA binding. Proteins.
83:2240–2250. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang J, Liu W, Zhang L and Zhang J:
Targeting mutant p53 stabilization for cancer therapy. Front
Pharmacol. 14:12159952023. View Article : Google Scholar : PubMed/NCBI
|