Introduction
Lung cancer is the leading cause of cancer-related
morbidity and mortality worldwide, with ~2.2 million new cases and
accounting for ~1.8 million deaths in 2020 (1). Lung cancer is classified into two main
groups according to the histological type: i) Small cell lung
cancer (SCLC) and ii) non-(N)SCLC (2). NSCLC is the most prevalent worldwide
histological type, accounting for ~85% of lung cancer cases
(2). The most common subtypes are
adenocarcinoma (40%), squamous cell carcinoma (25–30%) and large
cell carcinoma (5–10%) (2). In most
cases, patients with lung cancer are in an advanced or metastatic
stage at the time of diagnosis (3).
The treatment strategy has evolved from empirical
chemotherapy to a personalized approach based on improved
histological and molecular characterization of NSCLC primary tumors
(4). Currently, the management of
patients with NSCLC consists of the determination of epidermal
growth factor receptor (EGFR), anaplastic lymphoma kinase, ROS
proto-oncogene 1 and BRAF status, as they receive US Food and Drug
Administration- and European Medicines Agency-approved targeted
therapies (5). In the same way,
several new drugs have been developed with an immunotherapy focus
on immune checkpoint inhibitors. The most significant clinical
improvements have occurred with monoclonal antibodies against
programmed cell death protein 1 and programmed cell death protein
ligand 1 (6). However, despite the
great advances made in targeted therapies and immunotherapy,
platinum-based chemotherapy treatments continue to be used in
different situations and are still used as indicated therapies in
international guidelines (7,8).
Platinum-based chemotherapy has proven efficacy;
however, it is associated with a high prevalence of adverse events,
particularly hematological effects, which affect the quality of
life of patients (9,10). One of the most common hematological
effects caused by platinum-based chemotherapy is anemia, a
condition that develops when the blood produces a lower-than-normal
amount of healthy red blood cells. Platinum-based chemotherapy can
cause anemia, as these drugs can damage the kidneys, leading to
less erythropoietin production. As a result, the bone marrow gets
less of a stimulus to make new red blood cells, so it produces
fewer of them (11,12). Thus, studies on new biomarkers that
can identify adverse reactions to treatment and their mechanisms
are increasingly important, and the study of microRNAs
(miRNAs/miRs) is widely used nowadays.
miRNAs are small single-chain molecules containing
~22 noncoding nucleotides capable of regulating gene expression
(13–15). They are highly stable in
extracellular fluids (16) and are
specifically expressed in different tissues or pathological states;
therefore, they have been widely studied as possible diagnostic,
therapeutic and prognostic biomarkers (17). A review of miRNAs in lung cancer
revealed numerous studies (228 studies) on miRNAs as diagnostic
biomarkers and their associations with treatment response (18). The review discussed the possible
miRNAs that may influence treatment, particularly immunotherapy.
However, for platinum-based chemotherapy, the review included only
four studies on cisplatin and one on carboplatin (18). Furthermore, the studies on miRNAs
associated with cisplatin treatment did not assess the inherent
toxicities of these treatments (18).
Prognostic biomarkers should aid in the decision to
justify highly toxic chemotherapy. Our research group previously
performed a scope review that assessed the possible associations
between adverse reactions to chemotherapy and circulating miRNAs in
patients with NSCLC; however, only one article was identified that
met the inclusion criteria, which indicated a substantial lack of
data in this area (19). To the
best of our knowledge, the present study is the first to evaluate
plasma miRNAs as possible biomarkers of adverse hematological
reactions in patients with NSCLC.
Methods
Patient selection and hematological
adverse reactions
This study was conducted at the Clinical Oncology
Department of the Hospital de Clínicas, Universidade de Campinas
(HC-UNICAMP; Campinas, Brazil), a large tertiary teaching hospital,
from February 2018 to December 2020. The present study was a
case-control study. The inclusion criteria were as follows:
Patients aged 18–80 years with primary NSCLC treated with
carboplatin + paclitaxel (6 cycles every 21 days) were recruited.
Patients were excluded if they had a second primary tumor, declined
to participate at any time during the course of the study or did
not provide blood samples for the study.
Blood was collected in tubes containing EDTA as an
anticoagulant and centrifuged at 700 × g for 10 min at 4°C to
separate the plasma. The plasma was aliquoted and stored in a
freezer at −80°C until RNA extraction was performed. Blood samples
were collected before and 15 days after the administration of
carboplatin + paclitaxel. The present study was performed only in
the first cycle of chemotherapy to assess differences in the miRNA
expression of patients with and without chemotherapy. In addition,
after the first cycle, there may be a considerable reduction in
patients due to reasons such as the decrease in Karnofsky
Performance Status (20), adverse
events, deaths and others, which would have made statistical
analysis unfeasible.
Hematological adverse reactions were classified by
levels of hemoglobin, neutrophils, leukocytes, lymphocytes and
platelets, according to the Common Toxicity Criteria for Adverse
Events, version 4 (21).
Subsequently, the patients were divided into a case group (without
anemia) and a control group (with anemia).
Microarray of miRNAs
miRNAs were extracted from the plasma samples
collected from 6 patients with any grade of anemia and 6 patients
with grade 0 anemia (n=6) (Tables
IA and IIA), using the
miRNeasy Serum/Plasma Kit (cat. no. 217184; Qiagen, Inc.). These
patients were selected consecutively when a sufficient number of
patients with and without anemia had been reached to perform the
microarray analysis (6 with anemia and 6 without anemia). After
miRNA extraction, microarray analysis was performed. The samples
went through processes of marking, insertion in the chips,
hybridization for 18 h, washing and scanning. All procedures were
performed using the Affymetrix multi-user platform® (Applied
Biosystems™; Thermo Fisher Scientific, Inc.), which includes the
FlashTag™ Biotin HSR RNA Labeling Kit, Genechip® miRNA 4.0,
GeneChip Hybridization, Wash and Stain Kit and GeneChip
Hybridization Control Kit (cat. nos. 902445, 900720 and 900454,
respectively; Applied Biosystems™; Thermo Fisher Scientific,
Inc.).
 | Table I.Patient and clinical characteristics
of patients with non-small cell lung cancer treated with
carboplatin + paclitaxel. |
Table I.
Patient and clinical characteristics
of patients with non-small cell lung cancer treated with
carboplatin + paclitaxel.
| A, Microarray |
|---|
|
|---|
| Parameter | Non-hematological
adverse reaction (n=6) | Hematological
adverse reaction (n=6) | P-value |
|---|
| Patient and
clinical characteristics |
|
|
|
| Age, years | 62.00±3.57 | 65.33±5.31 | 0.3300a |
| Sex, n (%) |
|
| 0.9999b |
|
Male | 2 (33.33) | 1 (16.67) |
|
|
Female | 4 (66.67) | 5 (83.33) |
|
| Ethnicity |
|
| - |
|
Caucasian | 6 (100.00) | 5 (83.33) |
|
|
Non-caucasian | 0 (0.00) | 1 (16.67) |
|
| Smoker |
|
| 0.9999b |
|
Never | 2 (33.33) | 3 (50.00) |
|
|
Light | 0 (0.00) | 0 (0.00) |
|
|
Moderate | 0 (0.00) | 0 (0.00) |
|
|
Heavy | 4 (66.67) | 3 (50.00) |
|
| Drinker |
|
| 0.9999b |
|
Abstainer | 4 (66.67) | 4 (66.67) |
|
|
Light | 0 (0.00) | 1 (16.67) |
|
|
Moderate | 1 (16.67) | 0 (0.00) |
|
|
Heavy | 0 (0.00) | 1 (16.67) |
|
| Very
heavy | 1 (16.67) | 0 (0.00) |
|
| KPS |
|
| 0.5900b |
|
100 | 5 (83.33) | 4 (66.67) |
|
| 90 | 1 (16.67) | 2 (33.33) |
|
| 80 | 0 (0.00) | 0 (0.00) |
|
| 70 | 0 (0.00) | 0 (0.00) |
|
| 60 | 0 (0.00) | 0 (0.00) |
|
| Histological
type |
|
| 0.9999b |
|
Adenocarcinoma | 3 (50.00) | 4 (66.67) |
|
|
Squamous cell carcinoma | 3 (50.00) | 2 (33.33) |
|
| Stage |
|
| - |
| I | 0 (0.00) | 0 (0.00) |
|
| II | 0 (0.00) | 1 (16.67) |
|
|
III | 1 (16.67) | 0 (0.00) |
|
| IV | 3 (50.00) | 3 (50.00) |
|
|
N/A | 2 (33.33) | 2 (33.33) |
|
|
| B,
Validation |
|
|
Parameter |
Non-hematological adverse reaction
(n=20) | Hematological
adverse reaction (n=26) | P-value |
|
| Patient and
clinical characteristics |
|
|
|
| Age, years | 61.13±5.25 | 65.38±6.91 | 0.0400a |
| Sex |
|
| 0.7900b |
|
Male | 10 (50.00) | 12 (46.15) |
|
|
Female | 10 (50.00) | 14 (53.85) |
|
| Ethnicity |
|
| 0.4700b |
|
Caucasian | 15 (75.00) | 22 (84.60) |
|
|
Non-caucasian | 5 (25.00) | 4 (15.40) |
|
| Smoker |
|
| 0.4000b |
|
Never | 5 (25.00) | 6 (23.10) |
|
|
Light | 7 (35.00) | 0 (0.00) |
|
|
Moderate | 8 (40.00) | 5 (19.20) |
|
|
Heavy | 0 (0.00) | 15 (57.70) |
|
| Drinker |
|
| 0.8700b |
|
Abstainer | 7 (35.00) | 10 (38.45) |
|
|
Light | 7 (35.00) | 8 (30.75) |
|
|
Moderate | 3 (15.00) | 2 (7.70) |
|
|
Heavy | 3 (15.00) | 4 (15.40) |
|
| Very
heavy | 0 (0.00) | 2 (7.70) |
|
| KPS |
|
| 0.6400b |
|
100 | 18 (90.00) | 22 (84.60) |
|
| 90 | 1 (5.00) | 3 (11.55) |
|
| 80 | 0 (0.00) | 1 (3.85) |
|
| 70 | 0 (0.00) | 0 (0.00) |
|
| 60 | 1 (5.00) | 0 (0.00) |
|
| Histological
type |
|
| 0.2000b |
|
Adenocarcinoma | 15 (75.00) | 16 (61.54) |
|
|
Squamous cell carcinoma | 5 (25.00) | 10 (38.46) |
|
| Stage |
|
| - |
| I | 0 (0.00) | 0 (0.00) |
|
| II | 0 (0.00) | 2 (7.69) |
|
|
III | 5 (25.00) | 8 (30.77) |
|
| IV | 10 (50.00) | 11(42.30) |
|
|
N/A | 5 (25.00) | 5 (19.23) |
|
 | Table II.Hematological adverse reactions
parameters of patients with non-small cell lung cancer treated with
carboplatin + paclitaxel. |
Table II.
Hematological adverse reactions
parameters of patients with non-small cell lung cancer treated with
carboplatin + paclitaxel.
| A, Microarray |
|---|
|
|---|
| Parameter | Non-hematological
adverse reaction (n=6) | Hematological
adverse reaction (n=6) | P-value |
|---|
| Hematological
adverse reaction parameters |
|
|
|
| Anemia grade |
|
| 0.0100 |
| 0 | 6 (100.00) | 0 (0.00) |
|
| 1 | 0 (0.00) | 6 (100.00) |
|
| 2 | 0 (0.00) | 0 (0.00) |
|
| 3 | 0 (0.00) | 0 (0.00) |
|
| Leukopenia
grade |
|
| - |
| 0 | 6 (100.00) | 4 (66.67) |
|
| 1 | 0 (0.00) | 1 (16.67) |
|
| 2 | 0 (0.00) | 1 (16.67) |
|
| 3 | 0 (0.00) | 0 (0.00) |
|
| Neutropenia
grade |
|
| - |
| 0 | 6 (100.00) | 4 (66.67) |
|
| 1 | 0 (0.00) | 1 (16.67) |
|
| 2 | 0 (0.00) | 1 (16.67) |
|
| 3 | 0 (0.00) | 0 (0.00) |
|
| Lymphopenia
grade |
|
| - |
| 0 | 6 (100.00) | 5 (83.33) |
|
| 1 | 0 (0.00) | 1 (16.67) |
|
| 2 | 0 (0.00) | 0 (0.00) |
|
| 3 | 0 (0.00) | 0 (0.00) |
|
| Thrombocytopenia
grade |
|
| - |
| 0 | 6 (100.00) | 4 (66.67) |
|
| 1 | 0 (0.00) | 2 (33.33) |
|
| 2 | 0 (0.00) | 0 (0.00) |
|
| 3 | 0 (0.00) | 0 (0.00) |
|
|
| B,
Validation |
|
|
Parameter |
Non-hematological adverse reaction
(n=20) | Hematological
adverse reaction (n=26) | P-value |
|
| Hematological
adverse reactions parameters |
|
|
|
| Anemia grade |
|
| <0.0001 |
| 0 | 20 (100.00) | 0 (0.00) |
|
| 1 | 0 (0.00) | 24 (92.31) |
|
| 2 | 0 (0.00) | 2 (7.69) |
|
| 3 | 0 (0.00) | 0 (0.00) |
|
| Leukopenia
grade |
|
| 0.0800 |
| 0 | 18 (90.00) | 19 (73.07) |
|
| 1 | 2 (10.00) | 2 (7.69) |
|
| 2 | 0 (0.00) | 3 (11.55) |
|
| 3 | 0 (0.00) | 2 (7.69) |
|
| Neutropenia
grade |
|
| 0.1200 |
| 0 | 20 (100.00) | 21 (80.77) |
|
| 1 | 0 (0.00) | 4 (15.38) |
|
| 2 | 0 (0.00) | 1 (3.85) |
|
| 3 | 0 (0.00) | 0 (0.00) |
|
| Lymphopenia
grade |
|
| - |
| 0 | 20 (100.00) | 26 (100.00) |
|
| 1 | 0 (0.00) | 0 (0.00) |
|
| 2 | 0 (0.00) | 0 (0.00) |
|
| 3 | 0 (0.00) | 0 (0.00) |
|
| Thrombocytopenia
grade |
|
| 0.2100 |
| 0 | 19 (95.00) | 20 (76.92) |
|
| 1 | 1 (5.00) | 6 (23.08) |
|
| 2 | 0 (0.00) | 0 (0.00) |
|
| 3 | 0 (0.00) | 0 (0.00) |
|
Validation of selected miRNAs
miR-1273g-3p, miR-3613-5p and miR-455-3p were
selected for validation in a larger cohort of patients, as they are
genes that are most related to the hematopoiesis pathway, according
to mirPath v.3 (22). miRNAs were
extracted from samples collected from 50 patients with and without
anemia using the miRNeasy Serum/Plasma Kit (cat. no. 217184;
Qiagen, Inc.). After extracting the miRNAs from all plasma samples
collected at baseline and on day 15 from all patients included in
the study, cDNA synthesis was performed using the TaqMan™ Advanced
miRNA cDNA Synthesis Kit (cat. no. A28007; Applied Biosystems;
Thermo Fisher Scientific, Inc.) and quantitative (q)PCR using
TaqMan Advanced miRNA Assays (cat. no. A25576; Applied Biosystems;
Thermo Fisher Scientific, Inc.). In addition, qPCR of the
endogenous control, miR-98-3p, was performed for normalization,
which was selected as an endogenous control as its expression was
stable, according to the microarray results of the present study.
Furthermore, miR-39 was selected as an exogenous control to ensure
the quality of the technique, and samples with expression levels
above two standard deviations (SDs) were excluded from the
analysis. The thermocycling conditions were as follows: hold at
95°C for 20 sec followed by 70 cycles at 95°C for 15 sec and then
at 60°C for 60 sec. The sequences of the TaqMan probes used were
are follows: miR-1273g3p (assay ID no. 475626_mat; cat. no.
4440886), 5′-ACCACUGCACUCCAGCCUGAG-3′; miR-3613-5p (assay ID no.
479424_mir; cat. no. A25576), 5′-UGUUGUACUUUUUUUUUUGUUC-3′;
miR-455-3p (assay ID no. 478112_mir; cat. no. A25576),
5′-GCAGUCCAUGGGCAUAUACAC-3′; miR-98-3p (assay ID no. 479223_mir;
cat. no. A25576), 5′-CUAUACAACUUACUACUUUCCC-3′; and miR-39 (assay
ID no. 478293_mir; cat. no. A25576), 5′-UCACCGGGUGUAAAUCAGCUUG-3′.
Thermo Fisher Scientific, Inc. confirms that for the miR-1273g-3p
sequence, the aforementioned ‘assay detects a miRNA target from an
earlier version of miRbase that has been obsoleted in miRbase v.22.
This assay is still available for purchase for continuity of
studies’. At the time that the present study was performed, this
probe matched the Homo sapiens version of its gene target. A
C. elegans probe was used for the miR-39 sequence, as it was
an exogenous process quality control. The miRNA qPCR results were
analyzed using Rotor-Gene™ Q Series software 2.3.5 (23). Each miRNA had its expression
evaluated and relative expressions were quantified using the
2−ΔΔCq method (24). The
aforementioned experiments were performed using samples from the
patients before and 15 days after chemotherapy.
Bioinformatics analysis
miR-455-3p was selected for bioinformatics analysis.
Predicted miRNA target genes were identified using the miRWalk
platform 2.0 (25), which provides
a list of predicted miRNA target genes according to 12 different
algorithms. These genes were selected for unsupervised enrichment
analysis using the Database for Annotation, Visualization and
Integrated Discovery Functional Annotation Bioinformatics
Microarray Analysis software 6.8 (26) to identify the main canonical
signaling pathways involving differentially-expressed miRNAs. To
visualize the networks in which the target genes were involved, all
predicted genes in the matrix were analyzed using GeneMania 3.5.2
(27). The hematopoietic pathway
was selected for further evaluation.
Statistical analysis
The frequencies of clinical/demographic data and
degrees of adverse events related to adverse reactions are
presented as n (%) and descriptive measures are presented as the
mean ± SD. The expression of miRNAs was compared between the
different groups (with vs. without hematological adverse reactions)
at baseline and day 15 using the Mann-Whitney U-test, and between
the same group during the time (D0 vs. D15 with hematological
adverse reactions and D0 vs. D15 without hematological adverse
reactions), using the paired t-test. For comparisons with P<0.05
receiver operating characteristic (ROC) curves were produced. For
the enrichment analysis of the predicted target genes regulated by
miR-455-3p, Fisher's exact test was performed. The significance
level adopted for the present study was 5%. P<0.05 was
considered to indicate a statistically significant difference.
Results
Patient characteristics and
toxicities
For the array, a total of 6 patients with any grade
of anemia and 6 patients with grade 0 anemia (n=6) (Tables IA and IIA) were included; these patients were
representative of the whole cohort, since the age of the groups was
similar and the majority of patients were Caucasians and women when
comparing patients selected for the array and the larger cohort
(Table I). For validation, a total
of 50 patients were included in the present study. These patients
were 50% male with a mean age of 63.5 years; the majority of the
patients were Caucasians and heavy smokers, with adenocarcinoma
stage IV. For the validation step, 4 patients were excluded; thus,
46 subjects were used.
Hematological adverse reactions were prevalent, with
anemia observed in 26 patients (52.0%). The clinical
characteristics and hematological adverse reaction parameters of
patients whose miRNA samples were analyzed using microarray and
patients included in the validation step are presented in Table I.
Microarray results
A total of nine miRNAs were significantly
differentially expressed between patients without and with anemia
(hsa-miR-6779-5p, hsa-miR-3940-5p, hsa-miR-3656, hsa-miR-8072,
hsa-miR-1273g-3p, hsa-miR-6869-5p, hsa-miR-6794-5p, hsa-miR-455-3p
and hsa-miR-3613-5p), in which there was a fold change (FC) >1.5
or FC<-1.5, and P<0.05 (Table
III).
 | Table III.Plasma miRNAs with changes in gene
expression level between patients without (n=6) vs. with (n=6)
anemia in which P<0.05 and FC>1.5 or <-1.5. |
Table III.
Plasma miRNAs with changes in gene
expression level between patients without (n=6) vs. with (n=6)
anemia in which P<0.05 and FC>1.5 or <-1.5.
|
Differentially-expressed miRNA | FC | P-value |
|---|
|
hsa-miR-6779-5p | 1.57 | 0.027 |
|
hsa-miR-3940-5p | −1.64 | 0.039 |
| hsa-miR-3656 | −1.52 | 0.012 |
| hsa-miR-8072 | −1.51 | 0.037 |
|
hsa-miR-1273g-3pa | −2.92 | 0.004 |
|
hsa-miR-6869-5p | −1.99 | 0.040 |
|
hsa-miR-6794-5p | 1.55 | 0.004 |
|
hsa-miR-455-3pa | 1.55 | 0.040 |
|
hsa-miR-3613-5pa | 2.46 | 0.030 |
Validation of the miRNAs
A total of four miRNAs were chosen for expression
measurement: miR-1273g-3p, miR-3613-5p and miR-455-3p as targets,
and miR-98-3p as the endogenous control. The targets were chosen
using bioinformatics analysis, with all nine miRNAs observed in the
microarray. This analysis demonstrated that the three miRNAs were
notably associated with more genes in the hematopoietic pathway
compared with the other six miRNAs differentially expressed on the
microarray analysis. The endogenous control was chosen from the
microarray analysis, where miR-38-3p demonstrated markedly stable
expression between the two groups (FC=−1.04; P=0.97) (Table SI).
In total, 50 patients were enrolled for the
validation; however, four patients were excluded, as miR-39
(quality control) expression was higher than two SDs. Thus, the
expression of the three miRNAs from 46 patients was assessed. The
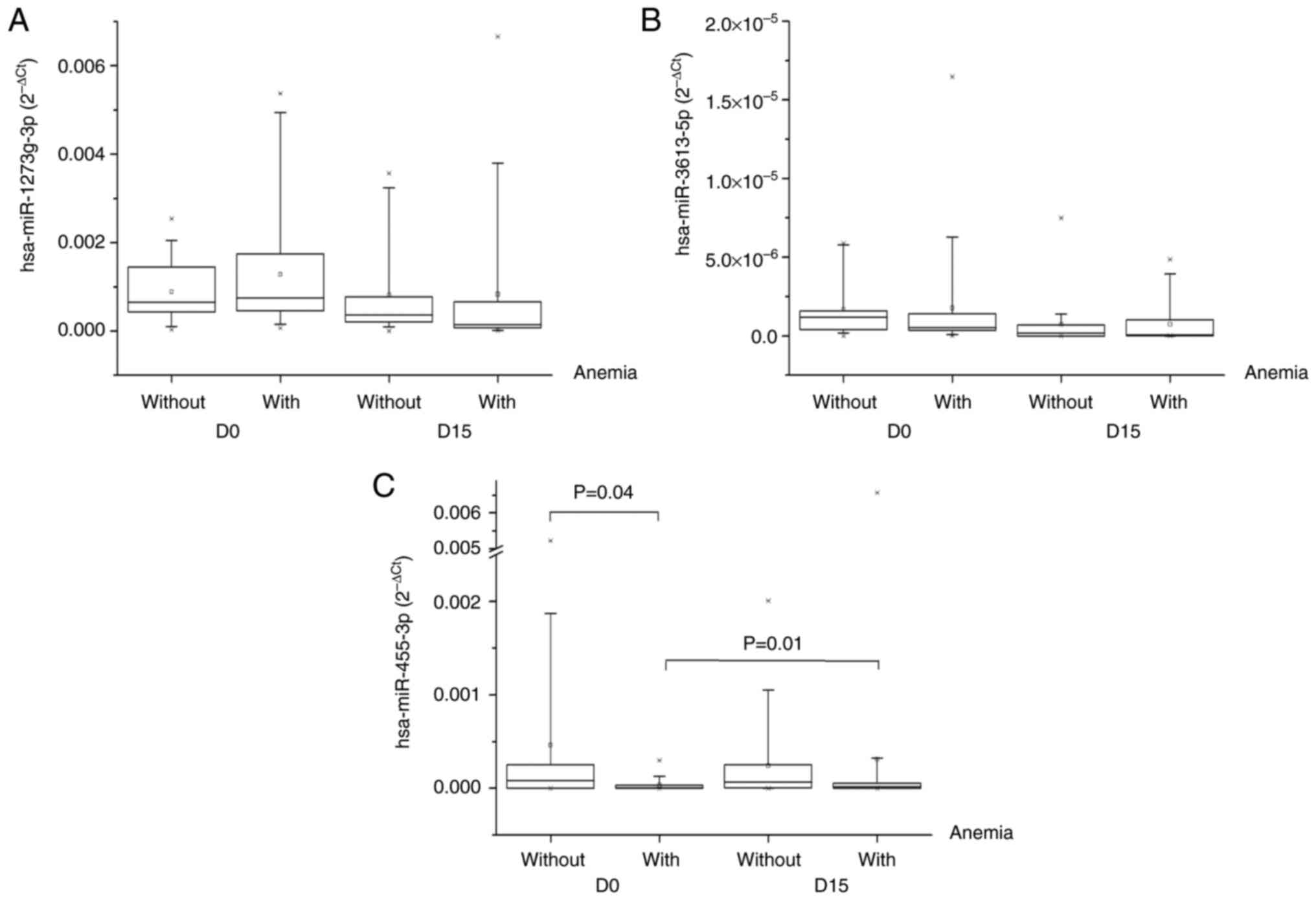
expression of miR1273g-3p and miR-3613-5p did not significantly
differ between the groups (Fig. 1A and
B), whereas miR-455-3p expression was significantly lower
before carboplatin + paclitaxel administration in patients with
anemia than in those without (P=0.04; Fig. 1C). There was no significant
difference in the expression of miR1273g-3p and miR-3613-5p between
the same group during the time (D0 vs. D15 with hematological
adverse reactions and D0 vs. D15 without hematological adverse
reactions), whereas miR-455-3p expression significantly increased
between baseline and day 15 for the group with anemia (P=0.01).
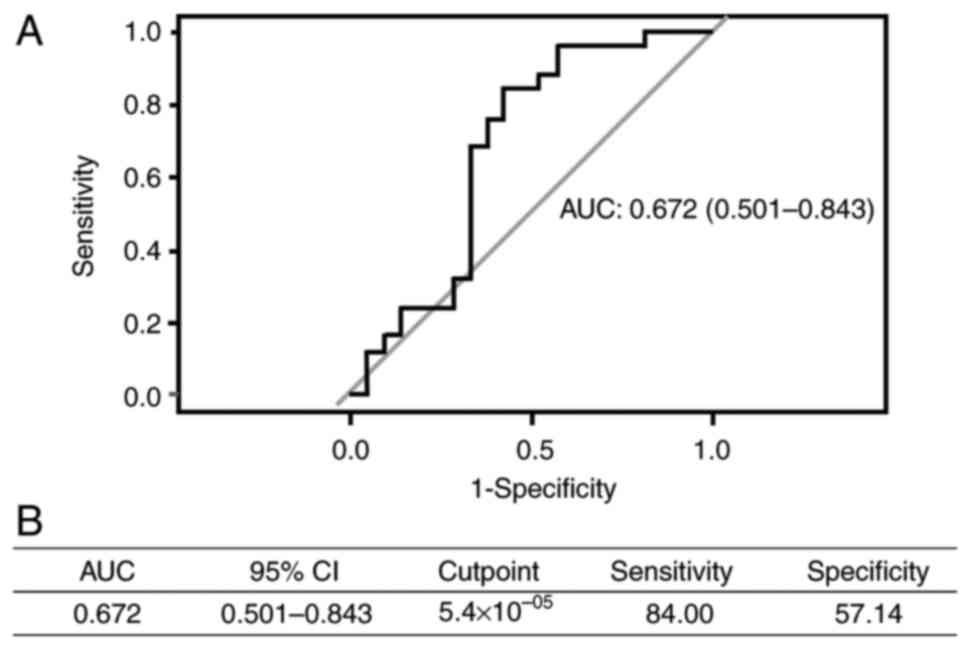
Furthermore, in the predictive performance analysis, miR-455-3p had
an area under the curve (AUC) of 0.672, sensitivity of 84.00% and
specificity of 57.14% (Fig. 2).
Bioinformatics analysis
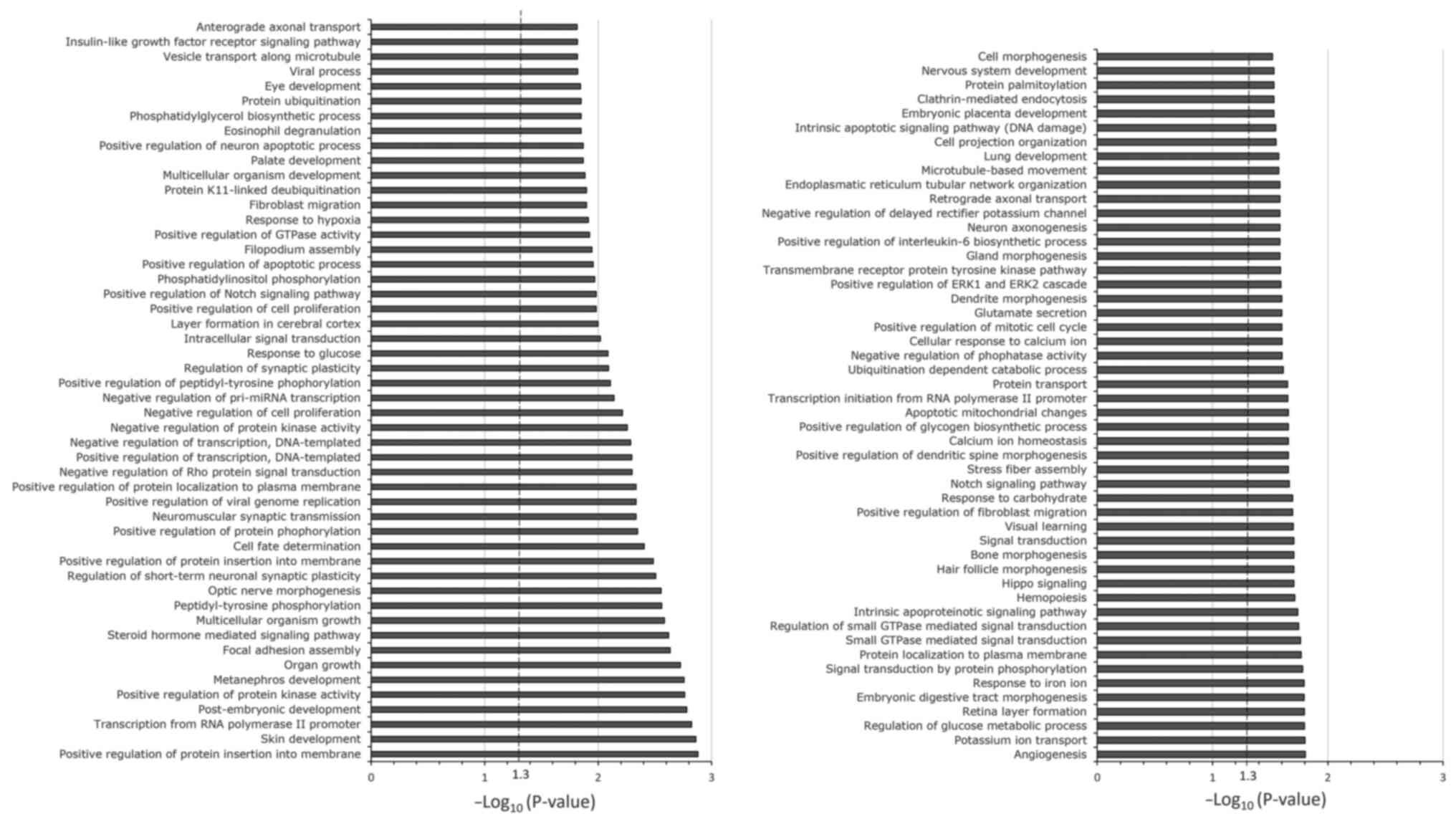
miR-455-3p was selected for bioinformatics analysis.
Gene set enrichment analysis was performed using 9,589 predicted
target genes of miR-455-3p. The top 100 canonical signaling
pathways of the predicted target genes of miR-455-3p are presented
in Fig. 3. Enrichment of the
signaling pathways involved in carboplatin- and paclitaxel-induced
adverse hematological reactions was observed through the
hematopoietic signaling pathway; this is represented by the
hemopoiesis pathway.
Discussion
The present study aimed to identify biomarkers of
adverse hematological reactions induced by chemotherapy with
carboplatin + paclitaxel in patients with NSCLC. Differential
expression of miRNAs was assessed before and 15 days after
treatment.
Microarray analysis of plasma samples from patients
with and without anemia demonstrated the differential expression of
nine plasma miRNAs between the two groups of patients. Among the
nine differentially-expressed miRNAs, miR-1273g-3p, miR3613-5p and
miR-455-3p were selected for further assessment.
In the validation cohort, miR-455-3p demonstrated a
statistically significant differential expression between the
groups, showing lower expression in the group with anemia than in
the group without anemia before chemotherapy with carboplatin +
paclitaxel (P=0.04). An ROC curve was produced to evaluate the
accuracy of predicting anemia. The AUC and specificity were 0.672
and 57%, respectively, with a good sensitivity of ~84%. This
indicated the potential of miR-455-3p as a biomarker for predicting
anemia after treatment with carboplatin + paclitaxel.
However, although the aforementioned findings are
still results, they encourage future studies to include a larger
number of patients and functional analyses of this miRNA. To the
best of our knowledge, there is currently no study that
demonstrates the cost effectiveness of using this protocol before a
chemotherapy regimen to predict anemia, and this could be the next
step of the present work. Thus, if more studies are performed to
confirm the predictive role of miR-455-3p and its cost
effectiveness, this will be clinically valuable and plasma
quantification for patients prior to chemotherapy with carboplatin
+ paclitaxel will be useful, as it allows for the identification of
patients with a prior risk of developing anemia and the opportunity
of making a better choice of treatment for them.
It is important to highlight that miR-455-3p serves
a well-established role as a tumor suppressor in different types of
cancer (28,29). Numerous studies have reported that a
lower expression of miR-455-3p resulted in a worse prognosis for
lung cancer (30), breast cancer
(31), osteosarcoma (32), prostate cancer (28) and esophageal cancer (29). Therefore, these findings, along with
those of the present study, indicate that miR-455-3p may serve as a
biomarker of cancer prognosis in the future, as well as a predictor
of adverse events related to hematological adverse reactions when
the treatment of choice is carboplatin + paclitaxel.
The in-silico analysis of miR-455-3p in the
present study demonstrated enrichment of a canonical pathway
(hemopoiesis) of high importance for hematological adverse
reactions induced by carboplatin + paclitaxel through the
hematopoiesis pathway. Among the genes important for this pathway
are runt-related transcription factor 1 (RUNX1) and T-cell
acute lymphocytic leukemia protein 1 (TAL1). RUNX1 is a
transcription factor that serves an essential role in
hematopoiesis. It is indispensable for the formation of
hematopoietic stem cells, which are precursors of all blood cells,
and for the maturation of T and B lymphocytes (33–35).
However, it is also responsible for the negative regulation of
hematopoietic stem cells and myeloid precursor cells (36). Therefore, the results of the present
study indicate that, due to the decreased expression of miR-455-3p,
there was an increase in the expression of RUNX1. This would lead
to increased negative regulation of hematopoietic stem cells and
myeloid precursor cells, and consequently, a lower production of
blood cells in general, particularly those of the myeloid series.
Furthermore, similar to RUNX1, TAL1 is an important transcription
factor in hematopoiesis. It is responsible for regulating different
processes, with an emphasis on the maturation of lymphoid series
cells (36,37). Reynaud et al (38) reported that increased expression of
TAL1 reduced the differentiation of B lymphocytes by up to 40%, and
the production of B lymphocytes and macrophages by ~20%. This
indicates that the results of the present study agree with the
observation that a reduction in miR-455-3p leads to an increase in
TAL1 expression, consequently leading to a reduction in
lymphocytes.
Therefore, the present study demonstrated that
miR-455-3p has important potential as a predictor of hematological
adverse reactions during treatment with carboplatin + paclitaxel,
which may explain the fact that this miRNA interacts with important
genes involved in the hematopoietic pathway, such as RUNX1
and TAL1. However, although promising, this is still initial
evidence and further investigation is needed so that in the future,
a possible clinical implementation for these findings can be
considered.
There are certain limitations to the analyses
presented. The present study comprised 50 patients, of whom 2
experienced grade ≥2 anemia. This small population with an absence
of severe anemia (grade ≥2) may have generated bias in the results.
However, even with this population, significant differences were
observed.
In conclusion, this present study showed that
patients with lung cancer who were treated with carboplatin and
paclitaxel are mostly male, with a mean age of 63.50 years,
retired, with low education, married, severe smokers, with a
Karnofsky performance status of 100%, who are not eligible for
surgical resection before chemotherapy and who are not eligible for
histopathological diagnosis of adenocarcinoma and stage 4. Among
the adverse reactions evaluated, the high prevalence of anemia
above the degree of 0 was highlighted. The microarray of patients
with and without hematologic adverse reactions found 9 plasma
miRNAs differently expressed among these patients. Of these, 3 were
chosen for validation: miR-1273g-3p, miR-3613-5p and miR-455-3p.
Only miR-455-3p showed a significant reduction in expression
(P=0.04) before carboplatin + paclitaxel administration in patients
with anemia than in those without anemia. Therefore, this miRNA has
potential as a predictor of carboplatin and paclitaxel-induced
haematological adverse reactions and should be evaluated in a
larger number of patients, as well as in functional studies.
Although chemotherapy based on the combination of carboplatin and
paclitaxel has been used in the treatment of lung cancer for
numerous years, hematological adverse reactions are an obstacle to
the efficiency of treatment and, consequently, to patients' quality
of life. The present study highlighted the significance of
hematological adverse reactions induced by carboplatin and
paclitaxel and the importance of identifying new biomarkers for
them. In addition to identifying miR-455-3p as a potential
predictor of hematological adverse reactions, information was
provided for further studies aimed at identifying new biomarkers
and important collaboration for a better understanding of the
etiology of hematological adverse reactions induced by carboplatin
and paclitaxel.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present work was supported by funding from the Coordenação
de Aperfeiçoamento de Pessoal de Nível Superior-Brasil
(CAPES)-finance code 001, Fundação de Amparo à Pesquisa do Estado
de São Paulo – Brasil (FAPESP)-finance code 2022/09438-8 and the
Pharmaceutical Security Nucleus Project, object of Agreement no.
895688/2019, the result of a partnership between the Ministry of
Justice and Public Security of Brazil, through the Fund for the
Defense of Diffuse Rights and the State University of Campinas.
Availability of data and materials
The array data were uploaded to ArrayExpress with
accession no. E-MTAB-13518. The data generated in the present study
may be requested from the corresponding author.
Authors' contributions
PM and ECP conceived the present study. PENSV
designed the study and performed the experiments. PENSV and PM
confirm the authenticity of all the raw data. CSS, LZ, ASB and HMH
selected patients based on the study inclusion criteria,
characterized the type, stage of the tumor and directed treatment,
and reviewed the description and interpretation of clinical data
MWPJr supported on patients' inclusion flow, helping us to reach
the patients that met the inclusion criteria, also on all the
demographics and clinical data analysis and interpretation of
clinical data. MVG helped with the bioinformatical analysis. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
All procedures were authorized by the Research
Ethics Committee of the State University of Campinas (Campinas,
Brazil; approval no. 83196318.8.0000.5404). All patients signed a
consent form approved by the Research Ethics Committee of the State
University of Campinas, where they authorized the use of the
obtained data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Duma N, Santana-Davila R and Molina JR:
Non-Small cell lung cancer: Epidemiology, screening, diagnosis, and
treatment. Mayo Clin Proc. 94:1623–1640. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sateia HF, Choi Y, Stewart RW and Peairs
KS: Screening for lung cancer. Semin.Oncol. 44:74–82. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shroff GS, de Groot PM,
Papadimitrakopoulou VA, Truong MT and Carter BW: Targeted therapy
and immunotherapy in the treatment of non-small cell lung cancer.
Radiol Clin North Am. 56:485–495. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
López-Castro R, García-Peña T,
Mielgo-Rubio X, Riudavets M, Teixidó C, Vilariño N, Couñago F and
Mezquita L: Targeting molecular alterations in non-small-cell lung
cancer: What's next? Per Med. 19:341–359. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Luo W, Wang Z, Tian P and Li W: Safety and
tolerability of PD-1/PD-L1 inhibitors in the treatment of non-small
cell lung cancer: A meta-analysis of randomized controlled trials.
J Cancer Res Clin Oncol. 144:1851–1859. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Planchard D, Popat S, Kerr K, Novello S,
Smit EF, Faivre-Finn C, Mok TS, Reck M, Van Schil PE, Hellmann MD,
et al: Metastatic non-small cell lung cancer: ESMO Clinical
Practice Guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 29 (Suppl 4):iv192–iv237. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ganti AKP, Loo BW, Bassetti M, Blakely C,
Chiang A, D'Amico TA, D'Avella C, Dowlati A, Downey RJ, Edelman M,
et al: Small cell lung cancer, version 2.2022, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw.
19:1441–1464. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cepeda V, Fuertes MA, Castilla J, Alonso
C, Quevedo C and Pérez JM: Biochemical mechanisms of cisplatin
cytotoxicity. Anticancer Agents Med Chem. 7:3–18. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Capriotti K, Capriotti JA, Lessin S, Wu S,
Goldfarb S, Belum VR and Lacouture ME: The risk of nail changes
with taxane chemotherapy: A systematic review of the literature and
meta-analysis. Br J Dermatol. 173:842–845. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
NIH National Heart, Lung and Blood
Institute, . What is anemia? https://www.nhlbi.nih.gov/health/anemiaOctober
4–2023
|
|
12
|
Rodgers DM III, Becker PS, Blinder M,
Cella D, Chanan-Khan A, Cleeland C, Coccia PF, Djulbegovic B,
Gilreath JA, Kraut EH, et al: Cancer- and chemotherapy-induced
anemia. J Natl Compr Canc Netw. 10:628–653. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lau NC, Lim LP, Weinstein EG and Bartel
DP: An abundant class of tiny RNAs with probable regulatory roles
in Caenorhabditis elegans. Science. 294:858–862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee RC and Ambros V: An extensive class of
small RNAs in Caenorhabditis elegans. Science. 294:862–864. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pereira TC: Introduction to the world of
microRNAs. Sociedade Brasileira de Genética; Ribeirão Preto,
Brazil: pp. 442015, (In Portuguese).
|
|
18
|
Zhong S, Golpon H, Zardo P and Borlak J:
miRNAs in lung cancer. A systematic review identifies predictive
and prognostic miRNA candidates for precision medicine in lung
cancer. Transl Res. 230:164–196. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vasconcelos PE, Visacri MB, Pincinato EC,
Torso NG, Seguin CS, Zambon L, Barbeiro AS, Junior MW and Moriel P:
miRNAs as biomarkers of adverse drug reactions to platinum-based
agents in patients with non-small-cell lung cancer. Biomark Med.
15:1067–1069. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karnofsky DA and Burchenal JH: The
clinical evaluation of chemotherapeutic agents. MacLeod CM:
Evaluation of chemotherapeutic agents. Columbia University Press;
New York, USA: pp. 1961946
|
|
21
|
U.S Department of Health and Human
Services, . Common Terminology Criteria for Adverse Events (CTCAE).
U.S Department of Health and Human Services, National Institutes of
Health, National Cancer Institute. Version 4.0. 2010, https://evs.nci.nih.gov/ftp1/CTCAE/CTCAE_4.03/Archive/CTCAE_4.0_2009-05-29.xlsMarch
11–2024
|
|
22
|
Vlachos IS, Zagganas K, Paraskevopoulou
MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T and
Hatzigeorgiou AG: DIANA-miRPath v3. 0: deciphering microRNA
function with experimental support. Nucleic Acids Res.
43:W460–W466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rotor-Gene Q 2.3.5-Windows platforms.
https://www.qiagen.com/us/resources/resourcedetail?id=9d8bda8e-1fd7-4519-a1ff-b60bba526b57&lang=enJanuary
30–2024
|
|
24
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dweep H and Gretz N: MiRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
Bioinformatics Resources. Nature Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao Y, Yan M, Yun Y, Zhang J, Zhang R, Li
Y, Wu X, Liu Q, Miao W and Jiang H: MicroRNA-455-3p functions as a
tumor suppressor by targeting eIF4E in prostate cancer. Oncol Rep.
37:2449–2458. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang H, Wei YN, Zhou J, Hao TT and Liu XL:
MiR-455-3p acts as a prognostic marker and inhibits the
proliferation and invasion of esophageal squamous cell carcinoma by
targeting FAM83F. Eur Rev Med Pharmacol Sci. 21:3200–3206.
2017.PubMed/NCBI
|
|
30
|
Gao X, Zhao H, Diao C, Wang X, Xie Y, Liu
Y, Han J and Zhang M: miR-455-3p serves as prognostic factor and
regulates the proliferation and migration of non-small cell lung
cancer through targeting HOXB5. Biochem Biophys Res Commun.
495:1074–1080. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guo J, Liu C, Wang W, Liu Y, He H, Chen C,
Xiang R and Luo Y: Identification of serum miR-1915-3p and
miR-455-3p as biomarkers for breast cancer. PLoS One.
13:e02007162018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yi X, Wang Y and Xu S: MiR-455-3p
downregulation facilitates cell proliferation and invasion and
predicts poor prognosis of osteosarcoma. J Orthop Surg Res.
15:4542020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lam K and Zhang DE: RUNX1 and RUNX1-ETO:
Roles in hematopoiesis and leukemogenesis. Front Biosci (Landmark
Ed). 17:1120–1139. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Okuda T, Nishimura M, Nakao M and Fujita
Y: RUNX1/AML1: A central player in hematopoiesis. Int J Hematol.
74:252–257. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ichikawa M, Yoshimi A, Nakagawa M,
Nishimoto N, Watanabe-Okochi N and Kurokawa M: A role for RUNX1 in
hematopoiesis and myeloid leukemia. Int J Hematol. 97:726–734.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lécuyer E and Hoang T: SCL: From the
origin of hematopoiesis to stem cells and leukemia. Exp Hematol.
32:11–24. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vagapova ER, Spirin PV, Lebedev TD and
Prassolov VS: The Role of TAL1 in hematopoiesis and leukemogenesis.
Acta Naturae. 10:15–23. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Reynaud D, Ravet E, Titeux M, Mazurier F,
Rénia L, Dubart-Kupperschmitt A, Roméo PH and Pflumio F: SCL/TAL1
expression level regulates human hematopoietic stem cell
self-renewal and engraftment. Blood. 106:2318–2328. 2005.
View Article : Google Scholar : PubMed/NCBI
|