Introduction
The incidence of thyroid cancer recently has
increased by 3% in the USA. The primary pathological type is
papillary thyroid carcinoma (PTC) (1), with most cases presenting with an
indolent clinical course. Furthermore, recurrent or metastatic PTC
subsets treated with radioactive iodine ablation, surgery and
thyroid-stimulating hormone suppression usually exhibit a favorable
prognosis; however, a number of patients with PTC progress to a
refractory state, or even succumb to PTC (2). Therefore, identifying molecular
factors associated with PTC pathogenesis is essential to developing
molecular therapeutics that can inhibit disease progression.
Smad-ubiquitination regulator 2 (SMURF2), a
C2-WW-HECT type E3 ubiquitin ligase, is a neural developmental
precursor and protein 4 subfamily member that mediates protein
degradation via ubiquitination (3).
SMURF2 has been demonstrated to exert tumor suppressive effects on
normal cells by controlling genome stability (4), whereas it has been shown to inhibit
the proliferation of colorectal cancer cells by promoting the
ubiquitination of carbohydrate-responsive element binding protein
(5). Additionally, SMURF2 was
revealed to inhibit the metastasis of hepatocellular carcinoma
cells via the ubiquitin-mediated degradation of Smad2 (6). As an E3 ubiquitin ligase, SMURF2 was
shown to interfere with the proliferation, migration, and
tumorigenesis of colon cancer cells by stimulating the
ubiquitination and degradation of special AT-rich sequence-binding
protein-1 (SATB1) (7). Thus, SMURF2
has been revealed to be a potent tumor suppressor that limits tumor
progression and development; nonetheless, its functions in PTC
remain unclear.
Previously, metabolic reprogramming has been
identified as a major cancer hallmark (8). Cancer cells reprogram catabolism
pathways to generate energy and promote cancer progression and
initiation (9). Glutamine and
glucose are vital but unusual energy sources for cancer cells
(10). Critically, cancer cell
metabolism can switch from an oxidative state to a glycolytic state
(Warburg effect). Glycolysis and glutaminolysis aid in rapid cancer
cell proliferation and promote tumor progression, as well as
metabolic reprogramming in PTC (11).
In the present study, the functions of SMURF2 were
investigated in PTC. Initially, SMURF2 expression was examined in
PTC cells by western blotting and polymerase chain reaction (PCR).
Additionally, SMURF2 expression was interfered with to investigate
its effects on major PTC cancer cell processes, including
proliferation, migration and invasion. Finally, the effects of
SMURF2 on PTC cell metabolism were examined.
The findings revealed that altered SMURF2 expression
affected aerobic glycolysis, glutamine breakdown, and key PTC
cancer cell processes in PTC, indicating its key functions in PTC
etiology. Hence, SMURF2 may serve as a novel target for anti-PTC
therapies.
Materials and methods
Tissue specimens and cell culture
PTC specimens
A total of 30 pairs of PTC and distant non-tumorous
tissues, which were 2 cm away from the cancerous lesions and were
confirmed to be tumor-free, were collected from patients treated at
the Department of Thyroid and Breast Surgery, The Second Affiliated
Clinical School of Medicine, Fujian Medical University (Quanzhou,
China) from October 2020 to October 2022. The patients aged 19–68
years (median, 43 years), consisted of 22 women and eight men who
were pathologically confirmed as having PTC and staged according to
the 2022 World Health Organization Classification of Thyroid
Neoplasms criteria (12). No
patient received presurgical treatment, such as chemotherapy or
radiation therapy. The present study was approved by the Ethics
Review Committee of the Second Affiliated Hospital of Fujian
Medical University [approval no. 149 (2021)], and all participants
provided signed informed consent. Fresh tissue specimens were
collected from the surgical room, immediately frozen in liquid
nitrogen, and stored at −80°C. The clinicopathological and
follow-up data were obtained from the medical records of the
patients.
Cell culture
The human TC cell lines, TPC-1 and SW579, were
purchased from Guangzhou Ryder Liankang Biotechnology Co., Ltd. and
were authenticated by performing short tandem repeat profile
analysis. The SW579 cell line was included for comparison and as a
reference, and in the future, relevant alternative cell lines will
be considered for investigation. The cells were cultured in Roswell
Park Memorial Institute (RPMI)-1640 medium supplemented with 5%
fetal bovine serum (FBS; cat. no. SH30087.01) and 1%
penicillin-streptomycin (cat. no. SH30010; both Hyclone; Cytiva) in
a humidified incubator with 5% CO2 and 95% air at
37°C.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total cellular RNA was isolated from the cells
(TPC-1 and sw579) using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) and reverse-transcribed into
complementary DNA (cDNA) using an RNase-free DNase І kit (Promega
Corporation) per the manufacturer's instructions. The obtained cDNA
was subjected to qPCR using 2X SYBR Green qPCR SuperMix
(Invitrogen; Thermo Fisher Scientific, Inc.) and a BioPhotometer
plus Ebend nucleic acid protein analyzer (Eppendorf). The
thermocycling conditions were as follows: 50°C for 2 min, 95°C for
2 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 32
sec. Relative expression was determined using the 2−ΔΔCq
method (13). Standard manufacturer
amplification protocols were followed using SMURF2 forward,
5′-TTGTTGGACGAATAATGGGA-3′ SMURF2 and reverse,
5′-GGATCTACTAACTCCATGTC-3′; GAPDH forward,
5′-GCTCATTTGCAGGGGGGAG-3′ and reverse, 5′-GTTGGTGGTGCAGGAGGCA-3′
primers.
Western blotting
Total cellular protein was extracted with
radioimmunoprecipitation assay buffer, and the concentration of the
samples was assayed with a bicinchoninic acid protein assay kit
(Nanjing KGI Biological Development Co., Ltd.) according to the
manufacturer's protocol. Sodium dodecyl sulfate-polyacrylamide gel
electrophoresis was performed [Roche Diagnostics (Shanghai) Co.,
Ltd.] using 10% gel to separate proteins, with 20 µg protein loaded
per lane. The proteins were then transferred onto polyvinylidene
fluoride membranes (MilliporeSigma). After blocking at room
temperature for 1 h in 5% non-fat milk, the membranes were
incubated overnight at 4°C with primary antibodies. Subsequently,
the membranes were washed with phosphate-buffered saline
−0.2%Tween-20 (PBS-T) three times and then incubated with a
horseradish peroxidase (HRP)-labeled secondary antibody for 1 h at
room temperature (37°C). Next, the membranes were lightly washed
with PBS-T three times and positive protein signaling was detected
using a chemiluminescence detection system (Invigentech). The
following primary antibody was used: anti-SMURF2 [EP629Y3]
(ab53316; Abcam; 1:1,000). The secondary antibodies used were: Goat
anti-rabbit IgG (H+L) and mouse/human ads-HRP (dilution ratio,
1:20,000; cat. no. 4050-05; Southern Biotech). To evaluate the
effects of overexpressed SMURF2 on glucose metabolism and glutamine
decomposition, western blotting was performed, and key glucose
metabolism enzymes, such as lactate dehydrogenase (LDH) and
hexokinase 2 (HK2), as well as key glutamine metabolism enzymes,
such as alanine/serine/cysteine-preferring transporter 2 (ASCT2)
and glutaminase (GLS1), were detected. LDH rabbit monoclonal
antibody (mAb; cat. no. 3582; Cell Signaling Technology, Inc.;
1:1,000); recombinant anti-HK II antibody (cat. no. ab209847;
Abcam; 1:1,000); recombinant anti-ASCT2 antibody (cat. no.
ab237704; Abcam; 1:1,000); anti-GLS1 antibody (cat. no. ab150474;
Abcam; 1:500).
Transfection
Overexpression plasmids carrying SMURF2 cDNA contain
the plasmid backbone of the plasmid plvx-zsgreen-puro (LMAI Bio).
The plasmid carried the green fluorescence gene zsgreen, enabling
green fluorescence. The SMURF2 small interfering (si)RNA and
negative control targeting cDNA sequences were synthesized by
Ribobio Co., Ltd. The following siRNA sequences targeting SMURF2
were used: si-NC, 5′-TTCTCCGAACGTGTCACGTTT-3′; si-SMURF2-1 sense,
5′-GAACTACGCAATGGGAGCGC-3′; si-SMURF2-2 sense,
5′-TGTCAGGCTCTATGTGAACT-3′; and si-SMURF2-3 sense,
5′CCACACTTGCTTCAATCGAA-3′. The antisense sequences of all the
siRNAs were as follows: si-NC, 5′-AAGAGGCUUGCACAGUGCAAAdTdT-3′;
si-SMURF2-1, 5′-CUUGAUGCGUUACCCUCGCGdTdT-3′; si-SMURF2-2,
5′-ACAGUCCGAGAUACACUUGAdTdT-3′; and si-SMURF2-3,
5′-GGUGUGAACGAAGUUAGCUUdTdT-3′. These DNA sequences were cloned
into the pLKO.1 lentiviral vector (Addgene, Inc.), and the
lentiviruses were packaged and produced in 293T cells, after vector
transfection using Lipofectamine® 2000 (cat. no.
11668019; Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h at
37°C and supernatant was collected The concentration of nucleic
acid used was 10 nM and the generation system used was second
generation. The lentiviral plasmids used for cell transfection were
used at 10 µg in a 10-cm flat cell culture dish and the ratio of
lentivirus and packaging and envelope plasmids was 10:8:6, while
the transfection was for 48 h at 37°C. The supernatant was then
gathered for cell infection. In brief, TPC-1 or SW579 cells were
plated into 24-well cell culture plates (5×104 cells per
well), grown overnight, and then infected with the viral
supernatant or control viral particles at MOI of 10 for 48 h at
37°C and analyzed for the efficiency of these siRNAs. siRNA-3 was
used for further experiments because it showed the highest
interference efficiency. Stably transduced cells were selected
using puromycin-containing RPMI-1640. The screening concentration
was 12 µg/ml and the maintenance concentration was 4 µg/ml).
Transfection was confirmed using a fluorescence microscope
(magnification, ×200).
Proliferation assays
Cells were added to the 96-well plates at a density
of 1×104 cells/well to verify for cell adhesion. The
cells were collected at different time points (0, 24, 48 and 72 h),
and examined for proliferation using the Titer 96AQ detection
reagent (cat. no. G3582; 1:10; Promega Corp.) after incubating for
4 h at 37°C. The absorbance was measured using a microplate reader
(Multiscan MK3; Thermo Fisher Scientific, Inc.) at 490 nm, followed
by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide
(MTT) assay. The solvent used to dissolve the purple formazan was
dimethyl sulfoxide. A wavelength of 490 nm was used to measure
formazan.
Transwell assays
Transwell chambers were purchased from Corning, Inc.
to assess the migration and invasion abilities of PTC cells. The
cells were resuspended in the RPMI-1640 medium containing 10% FBS
at 1×105 cells/ml. Matrigel (Corning, Inc.) was added to
the upper chambers, and, when the gel was set, 100 µl of cell
suspensions were added to each well and cultured for 48 h at 37°C.
The lower chambers contained the RPMI-1640 medium with 20% FBS
(Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, cells
that remained on the filter surface were removed using a cotton
swab, whereas cells that invaded the gel were first fixed in 4%
paraformaldehyde, then fixed in 10% methanol, stained with 0.1%
crystal violet for 10 min at room temperature, and visualized using
an inverted fluorescence microscope (Olympus CKX41; Olympus Corp.).
The number of invasive cells in five random fields (magnification,
×400) was counted and averaged. For migration studies, the cells
were added to the upper chambers without Matrigel, and the rest of
the steps were the same as for invasion studies. The assay was
performed in duplicate and repeated three times.
ELISA
All of the following experimental samples were
obtained from cultured in vitro thyroid cells. Lactate, ATP,
glutamate, α-ketoglutarate and glutamine levels, as well as glucose
consumption were evaluated using ELISA after overexpression of
SMURF2. ELISA kits included lactate (cat. no. WK-SU62; Shanghai
Valan Biotechnology Co., Ltd.), ATP (cat. no. LE-H3380; Hefei Laier
Biotechnology Co., Ltd.), glucose consumption (cat. no.
Keshun-0017; Shanghai Keshun Biotechnology Co., Ltd.), glutamate
(cat. no. 140739; Nanjing Senbeiga Biotechnology Co., Ltd.),
α-ketoglutarate (cat. no. XGE64578; Shanghai Sig Biotechnology Co.,
Ltd.) and glutamine (cat. no. KL-Gln-Hu; Shanghai Kanglang
Biotechnology Co., Ltd.). The relevant experiments were performed
according to the protocols provided in the kits. ELISA was
performed in duplicate and repeated at least twice.
Statistical analysis
SPSS 27 software (IBM Corp.) was used for
statistical analysis. The experiments were performed in triplicate
independently and the resultant data are summarized as the mean ±
SD The paired sample t-test was used for the comparison of clinical
paired sample differences, the independent sample t-test was used
for the difference in mean between the two groups, and one-way
ANOVA analysis was used to compare the difference in mean between
three or more groups. When performing multiple comparisons, the
Tukey-Kramer post hoc test was used. P<0.05 and P<0.01 were
considered to indicate statistically significant differences.
Results
SMURF2 is downregulated in PTC
tissues
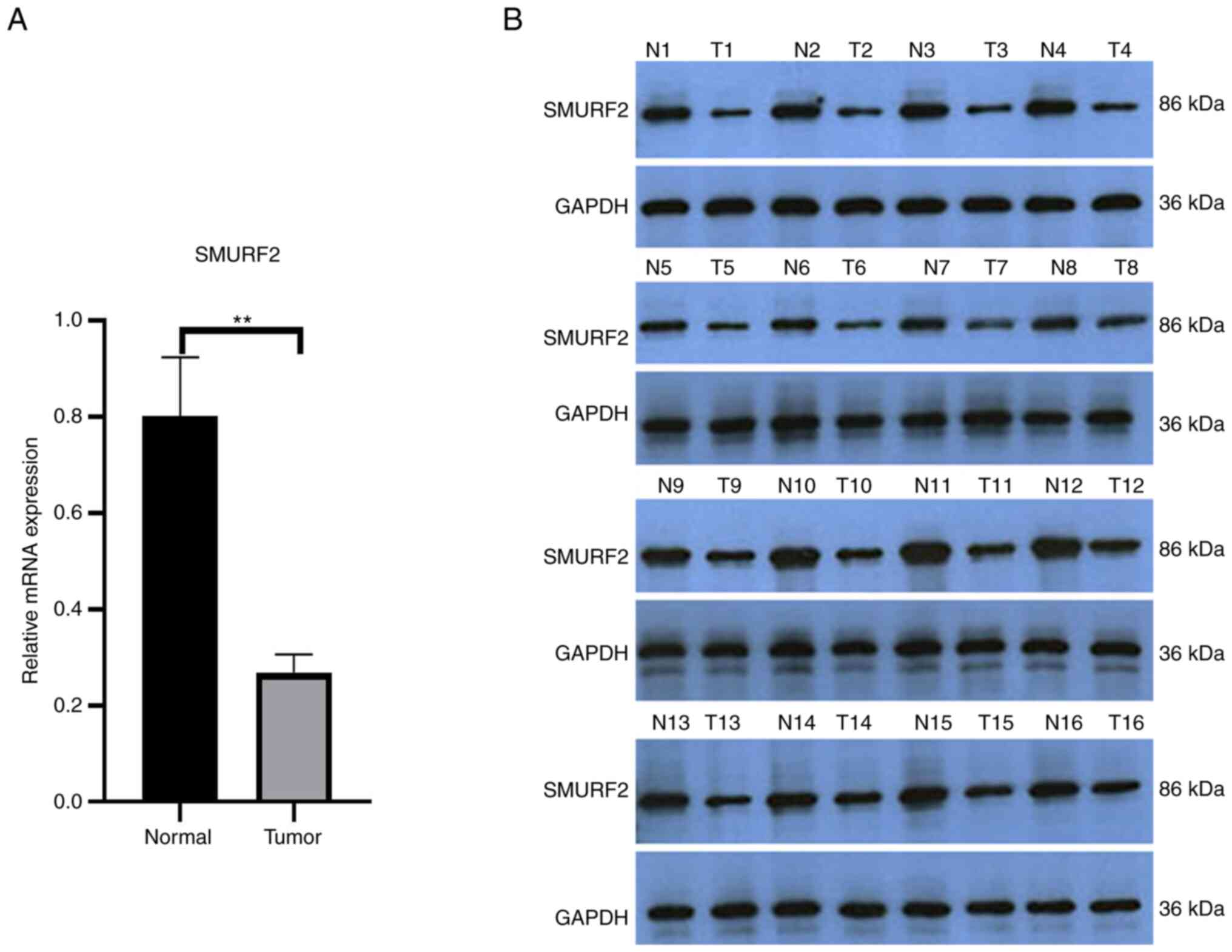
To investigate SMURF2 expression in PTC, RT-qPCR was
performed and SMURF2 mRNA levels were examined in PTC and normal
adjacent tissues (n=30 each). SMURF2 mRNA levels were significantly
lower in the PTC tissues compared with the healthy adjacent tissues
(Fig. 1A). Similarly, western
blotting showed that SMURF2 levels were lower in the PTC tissues
compared with the healthy adjacent tissues (Fig. 1B). Clinical data from the patients
with PTC were examined to determine associations between SMURF2
mRNA expression and clinicopathological characteristics, revealing
that SMURF2 mRNA expression was associated with lymph node
metastasis, but not with age, sex, tumor size, number of lesions or
extrathyroidal extension (Table
I).
 | Table I.Association of SMURF2 mRNA level with
clinical features. |
Table I.
Association of SMURF2 mRNA level with
clinical features.
| Variable | No. of patients | SMURF2 (mRNA) | T-value | P-value |
|---|
| Sex |
|
|
|
|
|
Female | 22 | 0.27±0.00 | −0.583 | 0.565 |
| Male | 8 | 0.27±0.11 |
|
|
| Patient age at
diagnosis |
|
|
|
|
| ≤45 | 18 | 0.28±0.42 | 0.592 | 0.558 |
|
>45 | 12 | 0.27±0.28 |
|
|
| Tumor size (cm) |
|
|
|
|
|
<2 | 12 | 0.27±0.01 | −0.393 | 0.697 |
| ≥2 | 18 | 0.28±0.00 |
|
|
| N stage (AJCC) |
|
|
|
|
| N0 and
Nx | 13 | 0.29±0.00 | 2.614 | 0.014 |
| N1 | 17 | 0.26±0.00 |
|
|
| Gland outside
invasion |
|
|
|
|
| No | 23 | 0.27±0.00 | −0.475 | 0.638 |
| Yes | 7 | 0.28±0.13 |
|
|
| Tumor location |
|
|
|
|
| One
lobe | 24 | 0.27±0.00 | −0.483 | 0.633 |
| More than
one lobe | 6 | 0.28±0.01 |
|
|
SMURF2 affects key PTC cancer
phenotypes
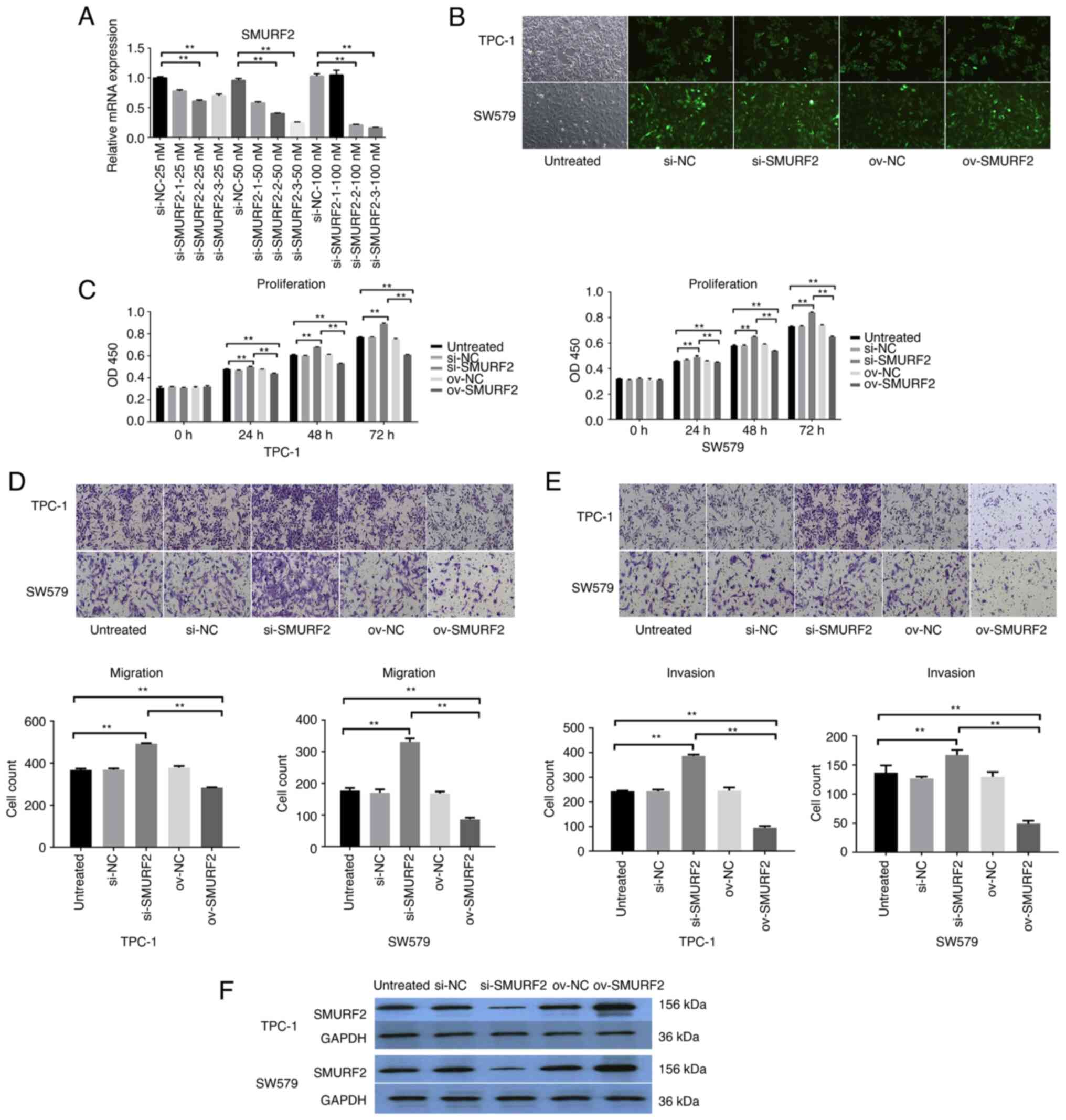
The function of SMURF2 was determined by silencing
the protein using si-SMURF2-1, si-SMURF2-2 or si-SMURF2-3, and
si-NC was used as a negative control. In vitro, SMURF2
silencing efficiency in the cell lines, TPC-1 and SW579, was
assessed using RT-qPCR. The results revealed that SMURF2 expression
was significantly decreased with transfection of si-SMURF2-3
compared with si-NC expression in TPC-1 cells and these cells were
selected for subsequent experiments (Fig. 2A, B and F). Subsequently, the cell
lines were assigned to five groups as follows: Untreated cells,
si-NC, si-SMURF2, SMURF2 overexpression (ov-SMURF2), and
overexpression control (ov-NC) to assess the effect of SMURF2 on
malignant biological function in tumor cells, respectively. Cell
proliferation was examined using MTT assays. Compared with
untreated cells and overexpression control (ov-NC), ov-SMURF2
markedly decreased cell proliferation (Fig. 2C). Moreover, compared with untreated
cells and overexpression control (ov-NC), the ov-SMURF2 cell lines
showed markedly reduced migration and invasion abilities using
Transwell assays (Fig. 2D and E).
Thus, SMURF2 expression was downregulated in PTC cells, whereas its
overexpression inhibited key PTC cancer cell processes.
SMURF2 inhibits the Warburg effect in
PTC cells
The Warburg effect is an anaerobic glycolysis
phenomenon; unlike normal cells, which use oxygen, tumor cells
undergo anaerobic fermentation to meet their energy need,
irrespective of the aerobic environment (14). To further understand SMURF2 function
in PTC, lactate, LDH, HK2 and ATP levels, as well as glucose
consumption in TPC-1 cells were examined. Compared with
empty-vector control cells, the SMURF2 overexpressing cells showed
decreased lactate. Conversely, SMURF2 silencing increased lactate
in the cell lines (Fig. 3A).
Compared with the untreated group and si-NC group, the protein
activities of LDHA and HK2 in the si-SMURF2 group were
significantly enhanced. By contrast, compared with the untreated
group and the ov-NC group, the protein activities of LDHA and HK2
in the ov-SMURF2 group were significantly weakened (Fig. 3B). Furthermore, compared with the
empty vector control cells, cell lines overexpressing SMURF2 showed
higher ATP and lower glucose consumption levels. Conversely, SMURF2
silencing led to the opposite effects (Fig. 3C and D). Therefore, SMURF2 exerted
inhibitory effects on glycolysis in PTC cells.
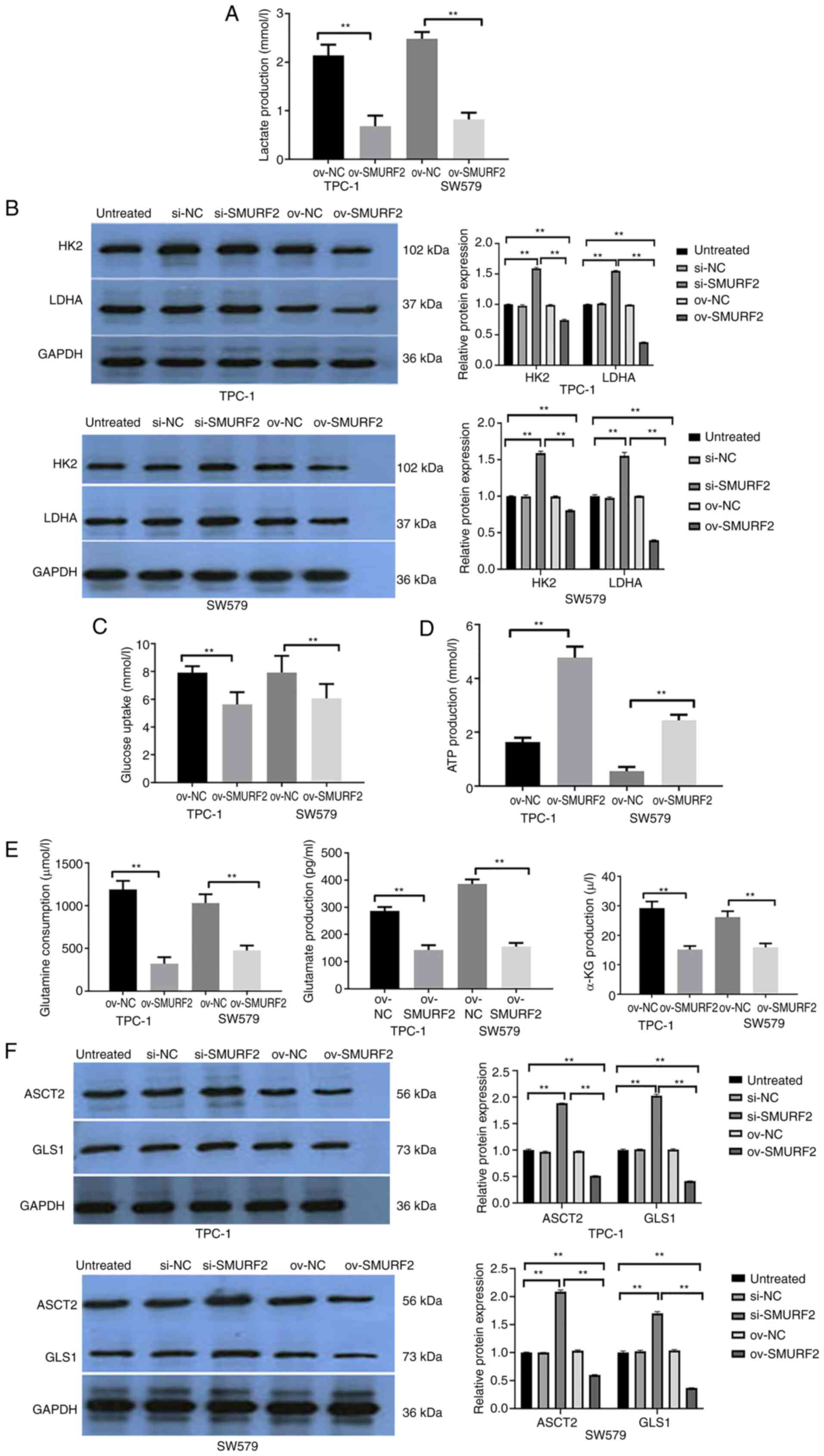 | Figure 3.SMURF2 inhibits the Warburg effect and
glutamine catabolism in papillary thyroid carcinoma. (A) Lactate
production in empty vector control cells, and PTC-1 and SW579 cells
stably overexpressing SMURF2. (B) LDH and HK2 activities in cells
stably overexpressing SMURF2 and empty vector control cells. (C)
Glucose uptake in cells stably overexpressing SMURF2 and empty
vector control cells. (D) ATP content was determined in cells
stably overexpressing SMURF2 and empty vector control cells. (E)
Glutamine consumption, as well as glutamate and α-KG production in
cells overexpressing SMURF2. (F) The key glutaminase enzymes ASCT2
and GLS1 in empty vector cells, and cells stably overexpressing
SMURF2. **P<0.01. SMURF2, Smad-ubiquitination regulator 2; LDH,
lactate dehydrogenase; HK2, hexokinase 2; α-KG, α-ketoglutarate;
ASCT2, alanine/serine/cysteine-preferring transporter 2; GLS1,
glutaminase; ov, overexpressed; NC, negative control; si-, small
interfering RNA. |
SMURF2 affects glutamine metabolism in
PTC cells
In cancer, glutamine catabolism is a characteristic
metabolic pattern observed after metabolic reprogramming (15). To further understand the mechanism
of action of SMURF2, glutamine consumption, glutamate and
α-ketoglutarate production, as well as ASCT2 and GLS1 enzyme
activities in the SMURF2 overexpressing cells were analyzed.
Compared with untreated cells and overexpression control (ov-NC),
the SMURF2 overexpressing cells exhibited significantly decreased
glutamine consumption, and glutamate and α-ketoglutarate production
(Fig. 3E). Furthermore, decreased
ASCT2 and GLS1 levels were observed in TPC-1and SW579 cells
(Fig. 3F). Thus, SMURF2 inhibited
glutamine catabolism in PTC cells.
Discussion
Ubiquitination is a common ATP-dependent cascade
that occurs in eukaryotes (16) and
requires at least the following three enzymes:
Ubiquitin-conjugating enzyme E2, ubiquitin-activating enzyme E1,
and ubiquitin ligase E3 (15).
Ubiquitin ligase E3 is critical, as it targets and degrades
substrates via the ubiquitin-protease system (17) SMURF2, an E3 ubiquitin ligase,
exhibits key functions in malignant tumors, such as preventing
colorectal cancer progression by ubiquitinating the YY1 protein and
decreasing its stability (18).
Additionally, upstream stimulatory factor 2 was revealed to affect
breast malignancy development by inhibiting SMURF2 transcription
(19). Furthermore, SMURF2
transcription inhibition enhanced chemoradiotherapy efficacy in
non-small cell lung cancer (20).
In the present study, it was revealed that SMURF2 expression in PTC
tissues was downregulated. Thus, SMURF2 may act as a tumor
suppressor in PTC cells.
To further confirm the role of SMURF2 in PTC, the
effect of SMURF2 alterations on PTC cell functions and the
metabolism was investigated in TPC-1 cells. SMURF2 silencing
promoted key TPC-1 malignant phenotypes. Furthermore, it inhibited
glucose and glutamine metabolism in TPC-1 cells, which would affect
tumor progression and clinical prognosis in PTC. The underlying
mechanism of SMURF2 glucose and glutamine metabolism inhibition in
TPC-1 cells will be examined in future studies. Current data
suggest that SMURF2 inhibits glycometabolism and glutamine
metabolism in cancer cells. However, the current data warrants
confirmation via in vivo and clinical studies. It is
therefore important to explore how SMUREF2 regulates its downstream
factors in controlling the progression of PTC. Despite its numerous
strengths, there are some limitations to this study. For example,
the present study revealed that SMURF2 inhibits the malignant
biological behavior and metabolism of PTC, however the relationship
between tumor stage and gene expression as well as the results of
enhanced immunohistochemistry will be further explored and improved
in future studies. In addition, the mechanism of the effect of
SMURF2 was not determined. Further studies are therefore required
to determine the underlying regulatory mechanism. In conclusion, in
PTC clinical specimens, SMURF2 expression was low. In vitro,
SMURF2 inhibited the malignant phenotype of PTC cells, which could
affect the clinical prognosis of PTC by mediating metabolic
reprogramming of TPC-1 cells. Therefore, SMURF2 may be a potential
therapeutic target for the treatment of PTC.
Acknowledgements
Not applicable.
Funding
The present study was partially supported by a grant from the
Fujian Provincial Natural Science Foundation (grant no.
2021J01251), the Medjaden Academy and Research Foundation for Young
Scientists (grant no. MJA2306089) and the Key Clinical Specialty
Discipline Construction Program of Fujian, P.R.C (Fujian Health
Medicine and Politics [2022]884)
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article
Authors' contributions
GL, WW and LitZ conducted the experiments. JL and HS
analyzed the results. GL and LihZ prepared and revised the
manuscript. LihZ made significant contributions to data analysis
and interpretation. LitZ, YY and WQ designed the present study. YY
and WQ revised and discussed the manuscript. JC, HD and XC made
substantial contributions to interpretation of data. GL and WW
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics Review
Committee of the Second Affiliated Hospital of Fujian Medical
University [approval no. 149 (2021)], and all participants provided
signed informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lim H, Devesa SS, Sosa JA, Check D and
Kitahara CM: Trends in thyroid cancer incidence and mortality in
the united states, 1974–2013. JAMA. 317:1338–1348. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pu WL, Shi X, Yu PC, Zhang MY, Liu ZY, Tan
L, Han P, Wang Y, Ji D, Gan H, et al: Single-cell transcriptomic
analysis of the tumor ecosystems underlying initiation and
progression of papillary thyroid carcinoma. Nat Commun.
12:60582021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ilic N, Tao YL, Boutros-Suleiman S, Kadali
VN, Emanuelli A, Levy-Cohen G and Blank M: SMURF2-mediated
ubiquitin signaling plays an essential role in the regulation of
PARP1 PARylating activity, molecular interactions, and functions in
mammalian cells. FASEB J. 35:e214362021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Blank M, Tang Y, Yamashita M, Burkett SS,
Cheng SY and Zhang YE: A tumor suppressor function of Smurf2
associated with controlling chromatin landscape and genome
stability through RNF20. Nat Med. 18:227–234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li YK, Yang DQ, Tian N, Zhang P, Zhu YM,
Meng J, Feng M, Lu Y, Liu Q, Tong L, et al: The ubiquitination
ligase SMURF2 reduces aerobic glycolysis and colorectal cancer cell
proliferation by promoting ChREBP ubiquitination and degradation. J
Biol Chem. 294:14745–14756. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song DQ, Li SY, Ning LX, Zhang SC and Cai
Y: Smurf2 suppresses the metastasis of hepatocellular carcinoma via
ubiquitin degradation of Smad2. Open Med (Wars). 17:384–396. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu L, Dong L, Wang Y, Liu L, Long H, Li H,
Li J, Yang X, Liu Z, Duan G, et al: Reversible regulation of SATB1
ubiquitination by USP47 and SMURF2 mediates colon cancer cell
proliferation and tumor progression. Cancer Lett. 448:40–51. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Z and Zhang H: Reprogramming of
glucose, fatty acid and amino acid metabolism for cancer
progression. Biochim Biophys Acta Rev Cancer. 73:377–392. 2016.
|
|
9
|
Sun LC, Suo CX, Li ST, Zhang HF and Gao P:
Metabolic reprogramming for cancer cells and their
microenvironment: Beyond the Warburg Effect. Biochim Biophys Acta
Rev Cancer. 1870:51–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Zhou Q, Song SL and Tang S:
Integrating metabolic reprogramming and metabolic imaging to
predict breast cancer therapeutic responses. Trends Endocrinol
Metab. 32:762–775. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Coelho RG, Fortunato RS and Carvalho DP:
Metabolic reprogramming in thyroid carcinoma. Front Oncol.
8:822018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cameselle-Teijeiro JM: Changes and
perspectives in the New 2022 WHO classification of thyroid
neoplasms. Rev Esp Patol. 55:145–148. 2022.(In Spanish). PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun T, Liu Z and Yang Q: The role of
ubiquitination and deubiquitination in cancer metabolism. Mol
Cancer. 19:1462020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bedford L, Lowe J, Dick LR, Mayer RJ and
Brownell JE: Ubiquitin-like protein conjugation and the
ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov.
10:29–46. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun TS, Liu ZN and Yang Q: The role of
ubiquitination and deubiquitination in cancer metabolism. Mol
Cancer. 19:1462020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fan Q, Wang Q, Cai RJ, Yuan HH and Xu M:
The ubiquitin system: Orchestrating cellular signals in
non-small-cell lung cancer. Cell Mol Biol Lett. 25:12020.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao QF, Wang SC and Zhang ZY: E3 ubiquitin
ligase SMURF2 prevents colorectal cancer by reducing the stability
of the YY1 protein and inhibiting the SENP1/c-myc axis. Gene Ther.
30:51–63. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tan YW, Chen YJ, Du MG, Peng ZQ and Xie P:
USF2 inhibits the transcriptional activity of Smurf1 and Smurf2 to
promote breast cancer tumorigenesis. Cell Signal. 53:49–58. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chaudhary KR, Kinslow CJ, Cheng HY, Silva
JM, Yu JY, Wang TJ, Hei TK, Halmos B and Cheng SK: Smurf2
inhibition enhances chemotherapy and radiation sensitivity in
non-small-cell lung cancer. Sci Rep. 12:101402022. View Article : Google Scholar : PubMed/NCBI
|