Introduction
In women, breast cancer (BC) is currently more
widespread than lung cancer, as ~2.26 million new cases (11.7%)
have been reported (1). This makes
it the leading factor contributing to cancer-related fatalities
among women globally (2). Despite
the progressive techniques for early detection and the advancements
in anti-cancer treatments, the reoccurrence and dissemination of BC
continue to present substantial obstacles (3,4).
Predicting BC prognosis is constrained by the limited scope of
existing biomarkers, largely owing to the heterogeneous and complex
nature of tumors (5). Hence, it is
imperative to investigate novel molecular biomarkers in clinical
research to improve prognostic accuracy and facilitate personalized
treatment approaches (6).
ADP-ribosylation factors (ARFs) are small
GTP-binding proteins that have a crucial role in regulating various
cellular processes. Specifically, they function as molecular
switches, activating signaling cascades involved in remodeling the
actin cytoskeleton, altering membrane lipids and facilitating
vesicle formation. In humans, there are six identified ARF proteins
(ARF1 to ARF6), with the exception of ARF2, which is not expressed
in humans. Among these isoforms, ARF1 and ARF6 have been
extensively studied and are considered the most well-characterized.
Traditionally, ARF1 has been associated with the Golgi apparatus,
whereas ARF6 is primarily found in the plasma membrane. However,
recent findings indicate that ARF1 can also be detected at the
plasma membrane in certain cell types (7–9).
ARF isoforms can be classified into three groups:
Class I (ARF1, ARF2 and ARF3), class II (ARF4 and ARF5) and class
III (ARF6). The primary function of ARF1-3 is to regulate the
transport of plasma membrane between the Golgi apparatus and
endoplasmic reticulum (ER) (7,8). By
contrast, the specific functions of ARF4-5 proteins remain unknown.
However, various biochemical assays suggest that both class I and
class II ARFs have crucial roles in the secretion pathway within
the ER-Golgi system (9). ARF6, on
the other hand, primarily participates in the regulation of plasma
membrane transport and the assembly of intracellular actin. In
addition to these roles, ARF6 also has significant physiological
functions in regulating various fundamental biological processes,
such as cytokinesis, cell adhesion, tumor formation, tumor-cell
growth, invasion and metastasis (7,8,10).
To date, the precise roles and prognostic
implications of distinct members within the ARF family in BC have
remained elusive. As a result, in order to shed light on this
matter, the current investigation aimed to evaluate the expression
levels of ARFs in BC tissues in relation to normal breast tissues
by conducting a range of database analyses. Furthermore, the study
also delved into comprehending the particular functions and
prognostic significance of select individual members of the ARF
family in BC.
Materials and methods
RNA-sequencing data and bioinformatics
analysis
The normalized RNA sequencing data and corresponding
clinical characteristics were acquired from The Cancer Genome Atlas
(TCGA) dataset called TCGA BRCA (https://tcga.xenahubs.net). To facilitate this
process, access to the TCGA BRCA dataset was obtained through TCGA.
The samples of BC tissue consisted of 1,097 cases, while 114
samples of healthy breast tissue were procured. For the
high-throughput sequencing data, the fragments per kilobase per
million fragments mapped reads method was utilized to estimate the
levels of transcript expression.
Clinical tissues
Between August 2021 and November 2022, Xingtai
People's Hospital (Xingtai, China) conducted surgical procedures on
patients, from whom a collection of 20 pairs of BC tissues and
adjacent non-tumor tissues was obtained. These patients, who were
randomly selected, 20 females aged 28–65 years, had all been
diagnosed with BC through pathological examination. Prior to
surgery, none of the patients had received any treatment and
written consent was obtained from all participants. Table I provides detailed
clinicopathological information of the patients (age range, 28–65).
The collected tissue samples were frozen in liquid nitrogen and
stored at −80°C. The Ethics Committee of Xingtai People's Hospital
[Xingtai, China; approval no. 2021(035)] authorized the present
study, which adhered to the principles outlined in The Declaration
of Helsinki.
 | Table I.Clinicopathological characteristics
of patients with breast cancer (n=20) in the present study. |
Table I.
Clinicopathological characteristics
of patients with breast cancer (n=20) in the present study.
|
Characteristics | n (%) |
|---|
| Age, years |
|
|
≤50 | 8 (40) |
|
>50 | 12 (60) |
| Tumor size, cm |
|
| ≤3 | 9 (45) |
|
>3 | 11 (55) |
| Grade |
|
| II | 8 (40) |
|
III | 12 (60) |
| TNM stage |
|
| I | 4 (20) |
| II | 8 (40) |
|
III | 8 (40) |
| ER status |
|
|
Positive | 14 (70) |
|
Negative | 6 (30) |
| PR status |
|
|
Positive | 12 (60) |
|
Negative | 8 (40) |
| HER2 status |
|
|
Positive | 6 (30) |
|
Negative | 14 (70) |
| Ki-67, % |
|
|
≤20 | 6 (30) |
|
>20 | 14 (70) |
| Lymph node
status |
|
|
Positive | 9 (45) |
|
Negative | 11 (55) |
Cell lines and cell culture
The MCF7, SK-BR3 and BT549 BC cell lines, which are
derived from humans, were obtained from The Cell Bank of the Type
Culture Collection of The Chinese Academy of Sciences. The MCF10A
mammary epithelial cell line, which is considered normal, was also
acquired from the same source. The SK-BR3, MCF-7 and BT549 cells
were grown in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin solution. The
MCF10A cells, on the other hand, were cultured using MCF-10A cell
medium (Procell Life Science & Technology, Co., Ltd.). All cell
lines were maintained and passaged in a standard cell culture
incubator at a temperature of 37°C and a 5% CO2
environment in an atmosphere with saturated humidity.
University of Alabama at Birmingham
Cancer data analysis Portal (UALCAN) database
Based on the TCGA database, the UALCAN website
(http://ualcan.path.uab.edu/index.html) is a
comprehensive tool for the analysis of cancer data. It offers
detailed information on the mRNA expression levels of ARFs and the
methylation levels of promoters in both BRCA tissues and normal
tissues. Furthermore, UALCAN evaluates the correlations between ARF
expression and clinicopathological parameters. To ascertain
discrepancies between ARF mRNA expression and promoter methylation
levels in BC and healthy controls, the analysis utilized Welch's
t-test. Furthermore, to compare the expression of ARFs among
different tumor substages, the analysis employed one-way ANOVA
followed by Dunnett's multiple-comparison test. P<0.05 was
considered to indicate a statistically significant difference.
ARFs-related gene function enrichment
analyses
The R package DESeq2 (version 1.26.0) (https://cloud.r-project.org/). was used to examine
differentially expressed genes (DEGs) in the present analysis. The
criteria for considering genes as differentially expressed included
a log2 (fold change)>2 threshold and their adjusted P-value of
<0.05. To determine the potential functions of genes associated
with DEGs, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) analyses were conducted. These analyses were
performed in the TCGA database using the R packages org.Hs.eg.db
(version 3.10.0) and clusterProfiler (version 3.14.3).
Reverse transcription-quantitative
(RT-q)PCR
RNA was extracted from the cell lines using the
Total RNA Extraction Kit from Qiagen GmbH. In order to synthesize
cDNA, the HiScript®III First-Strand cDNA Synthesis Kit
(+gDNA wiper) from Vazyme Biotech Co., Ltd. was used, following the
instructions provided by the manufacturer. Subsequently, qPCR was
performed using the ChamQ Universal SYBR qPCR Master Mix, also from
Vazyme Biotech Co., Ltd. All of the kits/mixes mentioned above were
used according to the manufacturer's instructions. The primers
utilized in this process were synthesized by Sangon Biotech Co.,
Ltd. The qPCR program included a pre-denaturation step at 95°C for
30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30
sec. Table II contains the
sequences of the β-actin and ARF primers. The cycle thresholds were
recorded and the 2−∆∆Cq method (11), with β-actin serving as the reference
gene, was used to determine and measure the relative expression
levels of the target genes.
 | Table II.Primer sequences used for
quantitative PCR. |
Table II.
Primer sequences used for
quantitative PCR.
| Gene | Sequence
(5′-3′) |
|---|
| Β-actin |
|
|
Forward |
CTCCATCCTGGCCTCGCTGT |
|
Reverse |
GCTGTCACCTTCACCGTTCC |
| ARF1 |
|
|
Forward |
ATGGGGAACATCTTCGCCAAC |
|
Reverse |
GTGGTCACGATCTCACCCAG |
| ARF3 |
|
|
Forward |
ATGGGCAATATCTTTGGAAACCT |
|
Reverse |
TGAACCCAATGGTAGGGATGG |
| ARF4 |
|
|
Forward |
CCCTCTTCTCCCGACTATTTGG |
|
Reverse |
GCACAAGTGGCTTGAACATACC |
| ARF5 |
|
|
Forward |
CTCTTTTCGCGGATCTTCGG |
|
Reverse |
TGAAGCCTATGGTTGGGATGG |
| ARF6 |
|
|
Forward |
GGGAAGGTGCTATCCAAAATCTT |
|
Reverse |
CACATCCCATACGTTGAACTTGA |
Tumor immune estimation resource
(TIMER)
The TIMER 2.0 web interface (https://cistrome.shinyapps.io/timer/) is a
user-friendly tool designed for scientific analysis. It offers six
analysis modules that facilitate the comprehensive evaluation of
various immune cell infiltration types and their potential clinical
significance. By utilizing the gene module, ARFs were chosen as the
input and scatterplots were produced to visually depict the
relationship between their expression levels and the extent of
immune infiltration in BC.
cBioPortal
cBioPortal (http://www.cbioportal.org) is a web-based platform
that offers a comprehensive range of cancer genomics data, ensuring
multidimensional visualization and access. The accession date was
Dec 29, 2023. The present study aimed to scrutinize the gene
mutations of ARFs within the context of BRCA, leveraging the
functionalities present in this resource.
GeneMANIA
GeneMANIA (http://www.genemania.org) is an abundantly resourced
website dedicated to the provision of gene data, analysis of gene
lists and the application of a sophisticated prediction algorithm
to prioritize gene function analysis. The utilization of GeneMANIA
was instrumental in the creation of the ARF interaction
networks.
STRING
The STRING online database (https://string-db.org/) is a website for the
investigation of protein-protein interactions (PPIs). Through the
PPI network analysis of STRING, the different expression levels and
potential PPIs of ARFs were identified and assessed.
Statistical analysis
The statistical analysis of the data collected was
performed using GraphPad Prism software (version 8.0; Dotmatics)
and R (version 3.6.3). The transcriptional data and clinical data
for BRCA were obtained from the TCGA database. Comparisons between
or among groups were performed using an unpaired Student's t-test,
paired Student's t-test or one-way ANOVA. Tukey's HSD was used as a
post hoc test following ANOVA. One-way Cox regression analysis was
conducted on these datasets using the R software package
‘Forestplot’ (version 2.0.1). In addition, receiver operating
characteristic (ROC) curves were generated using the ‘pROC’ package
(version 1.18.0) in R. Kaplan-Meier analysis were performed with
log-rank test. Furthermore, multivariate analysis was carried out
using the ‘survival’ package (version 3.2, year 13) in R to assess
various aspects of survival including overall survival (OS),
distant metastasis-free survival (DMFS), post-progression survival
(PPS) and recurrence-free survival (RFS). The correlation between
ARFs expression and immune infiltration was determined using
Spearman's correlation analysis. P<0.05 was considered to
indicate statistical significance.
Results
Expression profile of ARF gene family
in BRCA
Fig. 1A shows the
expression pattern of all ARF gene family members in the TCGA
pan-cancer panel. Various malignancies, such as breast invasive
carcinoma (BRCA), glioblastoma multiforme, glioma, brain lower
grade glioma, lung adenocarcinoma, esophageal carcinoma, stomach
and esophageal carcinoma, kidney renal papillary cell carcinoma,
pan-kidney cohort, colon adenocarcinoma, prostate adenocarcinoma,
stomach adenocarcinoma, head and neck squamous cell carcinoma,
kidney renal clear cell carcinoma, lung squamous cell carcinoma,
liver hepatocellular carcinoma, skin cutaneous melanoma, bladder
urothelial carcinoma, ovarian serous cystadenocarcinoma, pancreatic
adenocarcinoma, testicular germ cell tumors, uterine
carcinosarcoma, acute myeloid leukemia, adrenocortical carcinoma,
kidney chromophobe and cholangiocarcinoma exhibited a significant
upregulation of ARF1, ARF3, ARF4, ARF5 and ARF6. Furthermore, the
mRNA expression levels of the ARF gene family were notably higher
in BRCA samples than in normal mammary samples (P<0.001), as
indicated by both unpaired and paired sample analyses (Fig. 1B and C). This consistency in the
findings was also observed in our tissue sample library and
collection of cell lines (Fig. 1D and
E).
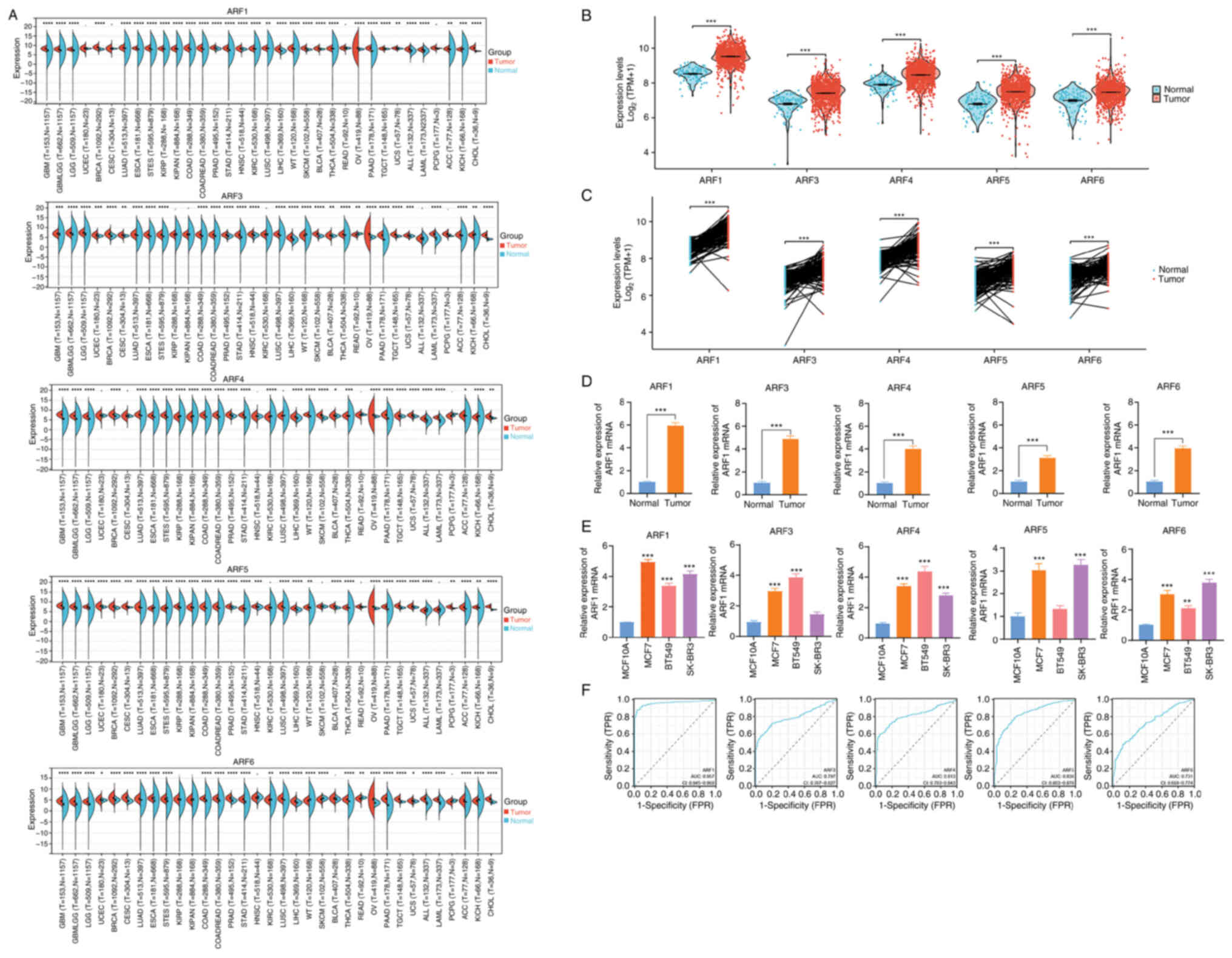 | Figure 1.mRNA expression pattern of the ARF
gene family in pan-cancer and BRCA panels. (A) The expression
pattern of the ARF gene family mRNA was analyzed in the TCGA in
pan-cancer panel. (B) unpaired and (C) paired sample analysis in
the TCGA database. (D) Expression pattern of ARF compared between
BC and normal tissues. (E) The expression pattern of ARF in BC cell
lines and a normal breast cell line was compared. (F) The
diagnostic value of the ARF gene family in BC was assessed by
evaluating ROC curves. *P<0.05, **P<0.01, ***P<0.001,
****P<0.0001 vs. normal/. TCGA, The Cancer Genome Atlas; BC,
breast cancer; ARF, ADP-ribosylation factor; ROC, receiver
operating characteristic; AUC, area under the ROC curve; T, tumor;
N, normal tissue. TPR, true-positive rate; FPR, false-positive
rate. |
Furthermore, in order to assess the diagnostic
efficacy of the ARF gene family in relation to BRCA, an ROC curve
analysis was employed. The variables of ARF1, ARF3, ARF4, ARF5 and
ARF6 revealed a heightened level of accuracy when differentiating
between healthy controls and BC samples (area under the
curve=0.957, 0.797, 0.813, 0.838 and 0.731) (Fig. 1F).
Association between ARFs and cancer
stage and subclass of BC
Data from the UALCAN database demonstrated that the
expression levels of ARF1 (Fig.
2A), ARF3 (Fig. 2B), ARF4
(Fig. 2C) and ARF5 (Fig. 2D) in breast cancer tissues of all
stages was higher than that in normal breast tissues. However, the
expression level of ARF6 in stage 4 BC was not significantly
different from that of normal breast tissue (Fig. 2E). Furthermore, the expression of
ARF1 (Fig. 2F), ARF3 (Fig. 2G), ARF4 (Fig. 2H), ARF5 (Fig. 2I) and ARF6 (Fig. 2J) was higher in almost all molecular
subtypes of BC than in normal breast tissue.
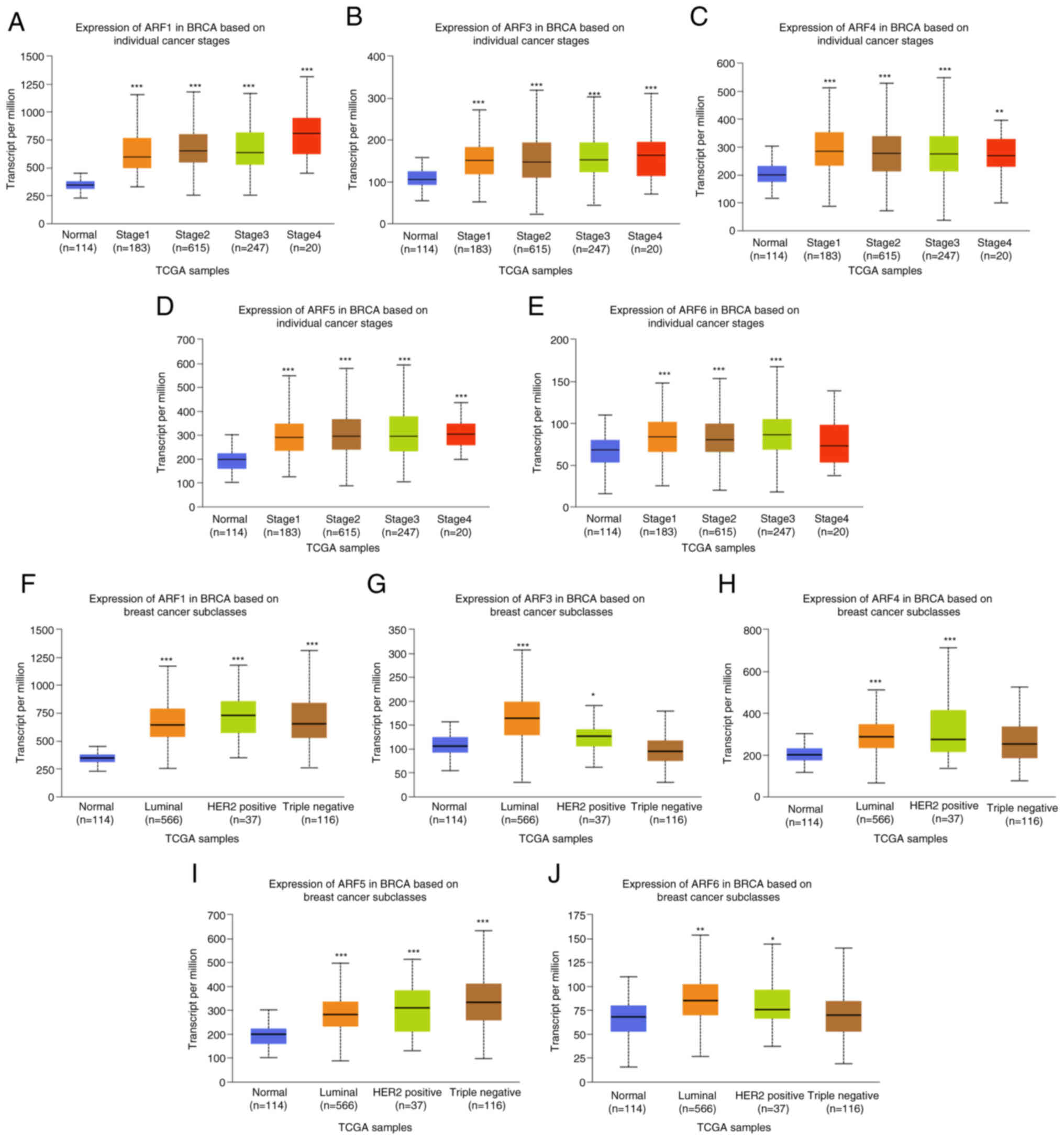 | Figure 2.Expression of ARFs in BRCA was
recorded per million patients of different subgroups. The
expression of ARFs in BC of different stages: (A) ARF1, (B) ARF3
(C) ARF4, (D) ARF5 and (E) ARF6. The expression of ARFs in BRCA of
different subclasses: (F) ARF1, (G) ARF3, (H) ARF4, (I) ARF5 and
(J) ARF6. *P<0.05, **P<0.01, ***P<0.001 vs. normal. TCGA,
The Cancer Genome Atlas; BRCA, breast carcinoma; ARF,
ADP-ribosylation factor; HER2, human EGFR 2. |
Prognostic and diagnostic value of ARF
mRNA expression in patients with BC
In order to determine the value of differently
expressed ARFs in determining the progression of BC, the
association between various ARFs and clinical outcomes was
evaluated using Kaplan-Meier plots (Fig. 3). It was observed that high
expression of ARF1 demonstrated a significant association with
decreased OS (Fig. 3A) and DMFS
(Fig. 3C) among patients with BC.
Furthermore, elevated mRNA expression levels of ARF1 (Fig. 3B), ARF3 (Fig. 3F), ARF4 (Fig. 3J) and ARF6 (Fig. 3R) displayed significant associations
with shorter RFS times in patients with BC. Interestingly, a higher
expression level of ARF5 (Fig. 3O)
and ARF6 (Fig. 3S) predicted a
decrease in DMFS among patients with BC. In addition, a heightened
mRNA expression level of ARF6 (Fig.
3Q) was found to be significantly associated with decreased OS
times in patients with BC, while the other associations of ARFs did
not yield significant results (Fig.
3).
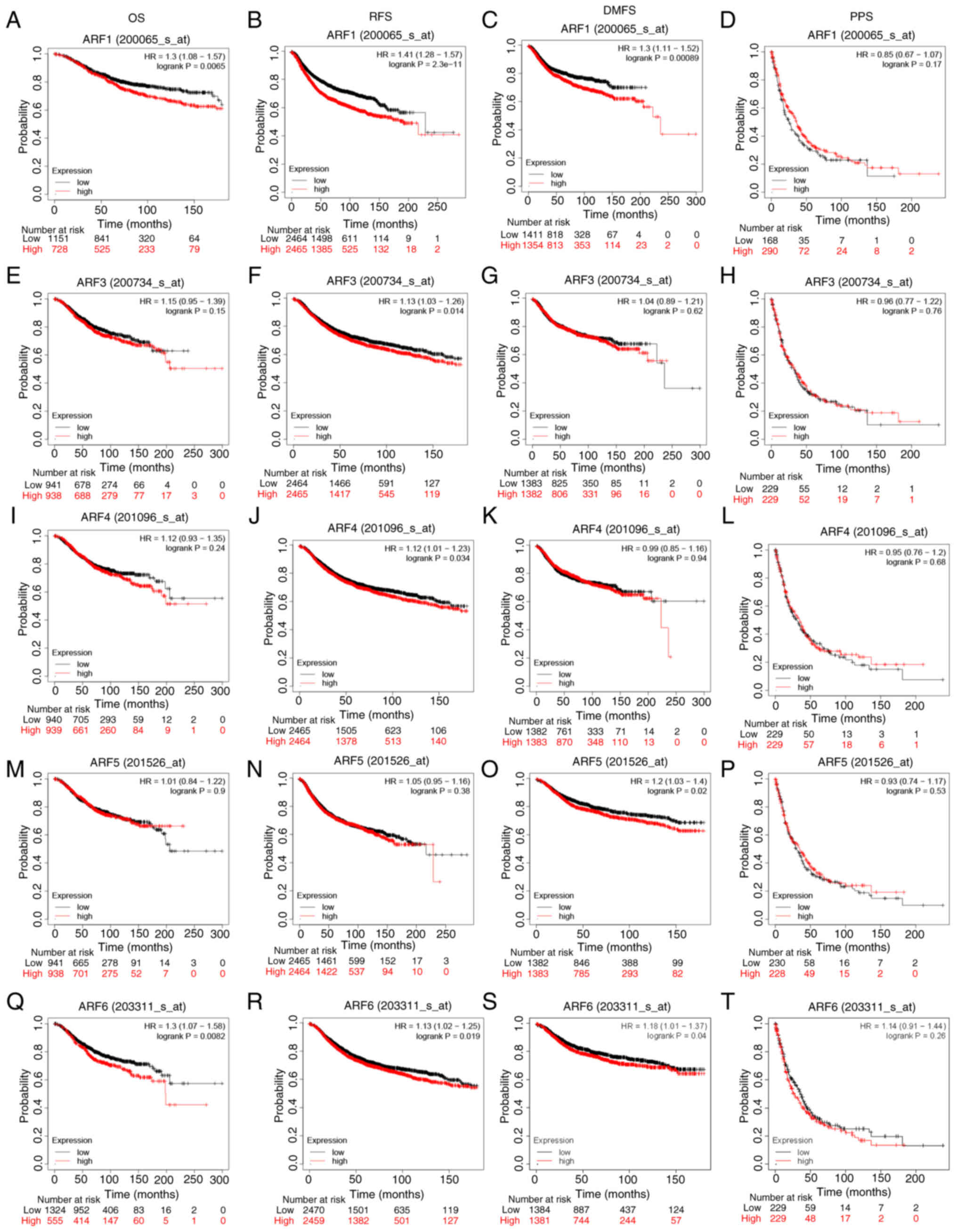 | Figure 3.Kaplan-Meier survival analysis of the
ARF gene family. Association between ARF1 and (A) OS, (B) RFS, (C)
DMFS and (D) PPS. Relationship between ARF3 and (E) OS, (F) PFS,
(G) DMFS and (H) DSS. Association of ARF4 with (I) OS, (J) PFS, (K)
DMFS and (L) DSS. Influence of ARF5 on (M) OS, (N) PFS, (O) DMFS
and (P) DSS. Relationship between ARF6 and (Q) OS, (R) PFS, (S)
DMFS and (T) DSS. HR, hazard ratio; ARF, ADP-ribosylation factor;
OS, overall survival; PFS, progression-free survival; DSS,
disease-specific survival; DMFS, distant metastasis-free
survival. |
Nomograms and calibration curves of
ARFs
In order to assess the predictive performance of
ARFs, nomograms and calibration curves were constructed by
incorporating the expression of ARFs to forecast survival at 1, 3
and 5 years (Fig. 4). The nomogram
(Fig. 4A-E) simplified the
determination of each gene's contribution by leveraging its
expression level. Subsequently, the cumulative score for an
individual patient was obtained by summing up the scores associated
with all the genes. As per the nomogram, ARF3 exerted the most
significant impact on the prognosis of OS. Of note, the calibration
curves for 1-, 3- and 5-year survival displayed a high degree of
concordance between the projected and observed outcomes, suggesting
precise predictions of OS by the nomogram (Fig. 4F-J).
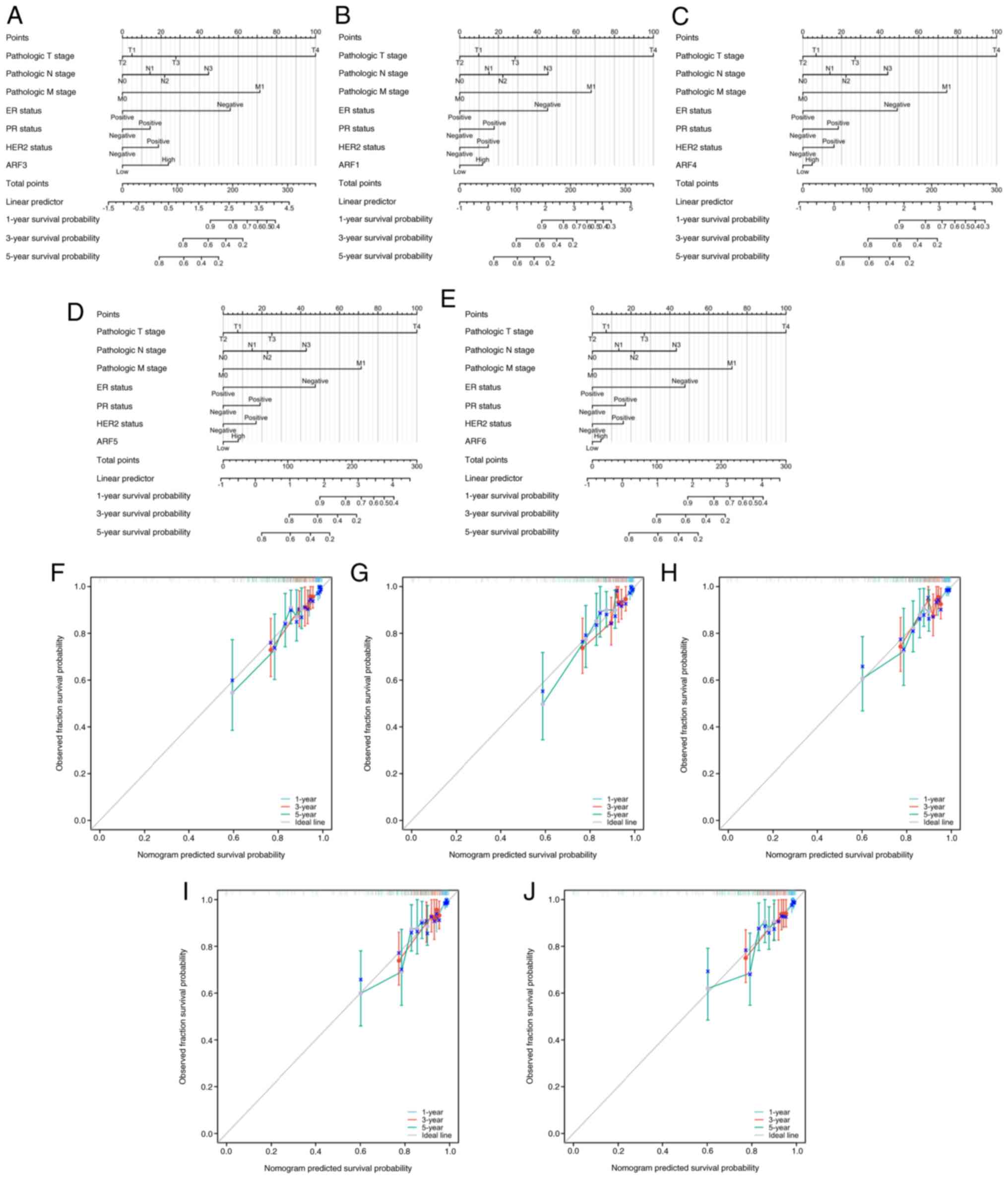 | Figure 4.Nomogram plots of (A) ARF1, (B) ARF3,
(C) ARF4, (D) ARF5 and (E) ARF6 were generated to predict OS of
patients with breast cancer at 1, 3 and 5 years. In addition,
calibration plots of (F) ARF1, (G) ARF3, (H) ARF4, (I) ARF5 and (J)
ARF6 were utilized to estimate the accuracy of the nomogram model
for predicting OS at 1, 3 and 5 years. OS, overall survival; ARF,
ADP-ribosylation factor; ER, estrogen receptor; PR, progesterone
receptor; HER2, human EGFR 2. |
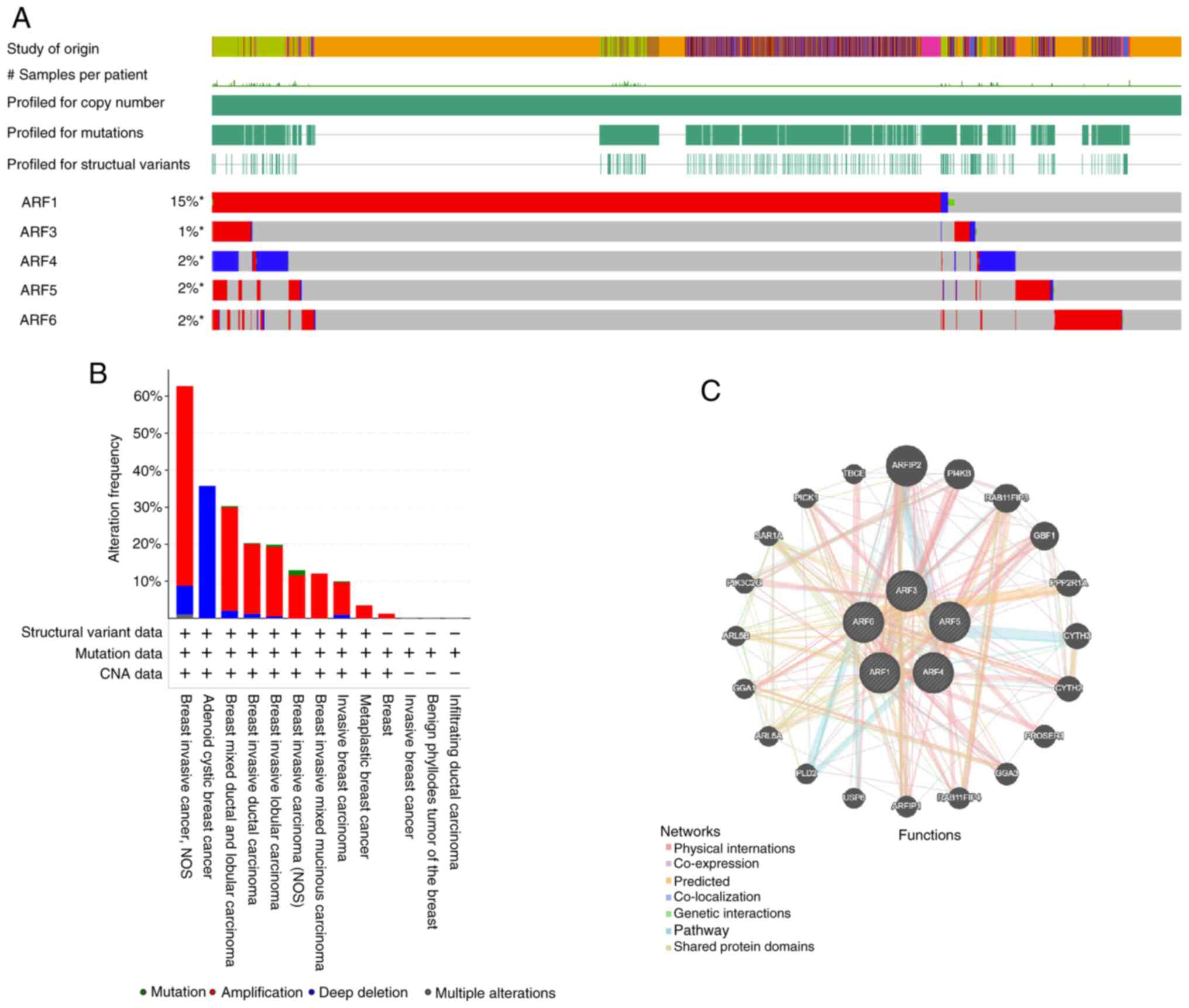
Gene expression changes in ARFs were assessed and an
analysis of ARF gene expression and interaction in individuals
diagnosed with BRCA was then performed. To analyze the influence of
genetic modifications on the expression of ARF family genes in BC,
the cBioPortal web tool was utilized. The examination revealed that
the ARF family members ARF1, ARF3, ARF4, ARF5 and ARF6 exhibited
alterations in 15, 1, 2, 2 and 2% of the BC samples, respectively
(Fig. 5A and B). These analyses
demonstrate that expression of ARFs is significantly amplified in
various BC tissues, such as in breast invasive cancer, non-specific
type, breast mixed ductal carcinoma, breast invasive lobular
carcinoma, and metaplastic BC, further suggesting potential
associations of ARFs with BC.
Following this, a network analysis of
protein-protein interactions (PPI) was conducted using the STRING
website to explore potential interactions among the differentially
expressed ARFs. The PPI network (Fig.
5C) demonstrated the presence of numerous nodes and edges. The
top 10 most related genes to the ARF family were as follows:
ARFIP2, PI4KB, RAB11FIP3, GBF1, PPP2R1A, CYTH3, CYTH2, PROSER1,
GGA5 and RAB11FIP4.
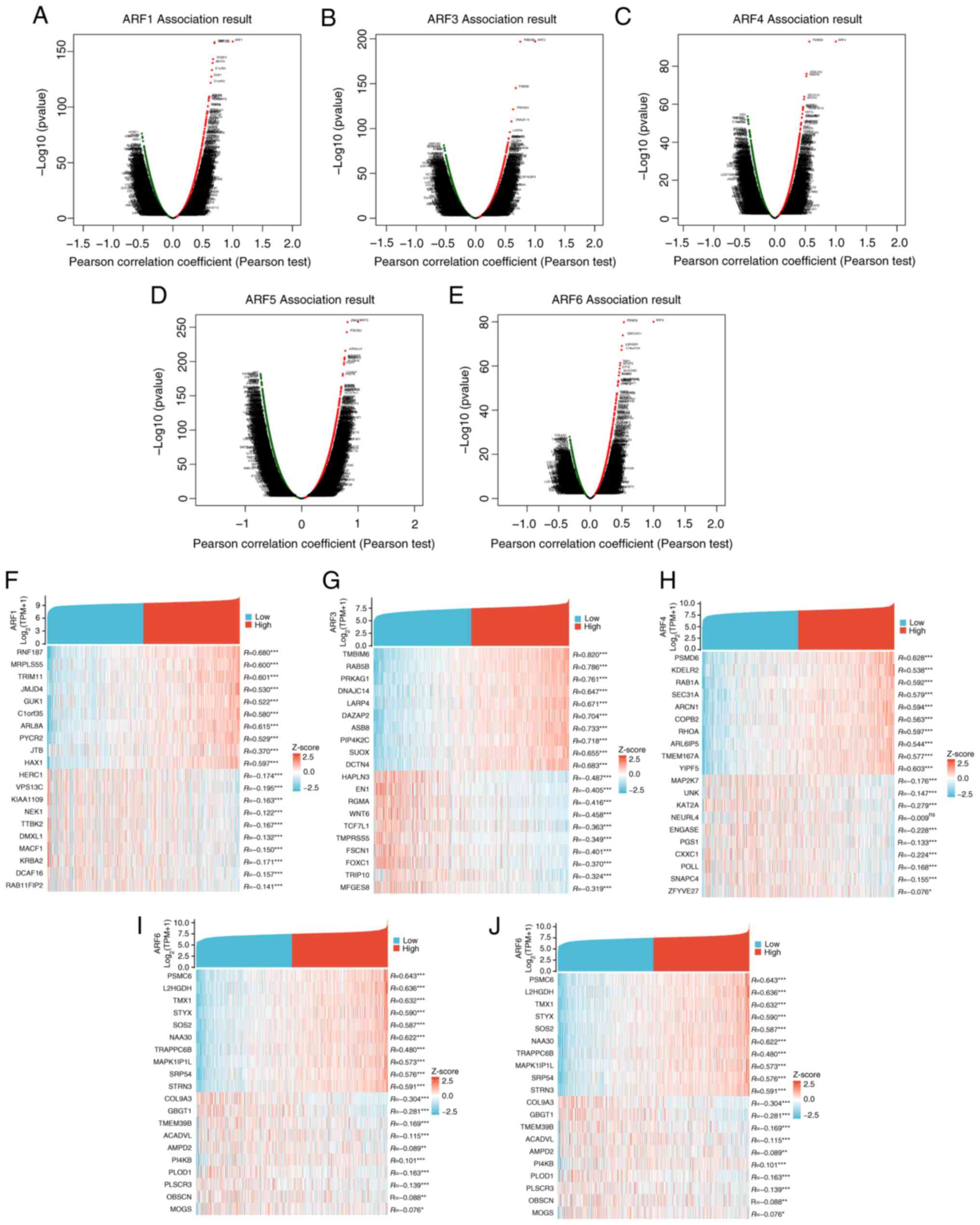
BC TCGA, GO and KEGG analyses of ARFs
and their co-expressed genes
The DEseq2 R software package in the TCGA database
was utilized to examine the expression levels of genes in
individuals with BC displaying either high or low ARF expression.
Through this analysis, the top 10 genes that exhibited positive or
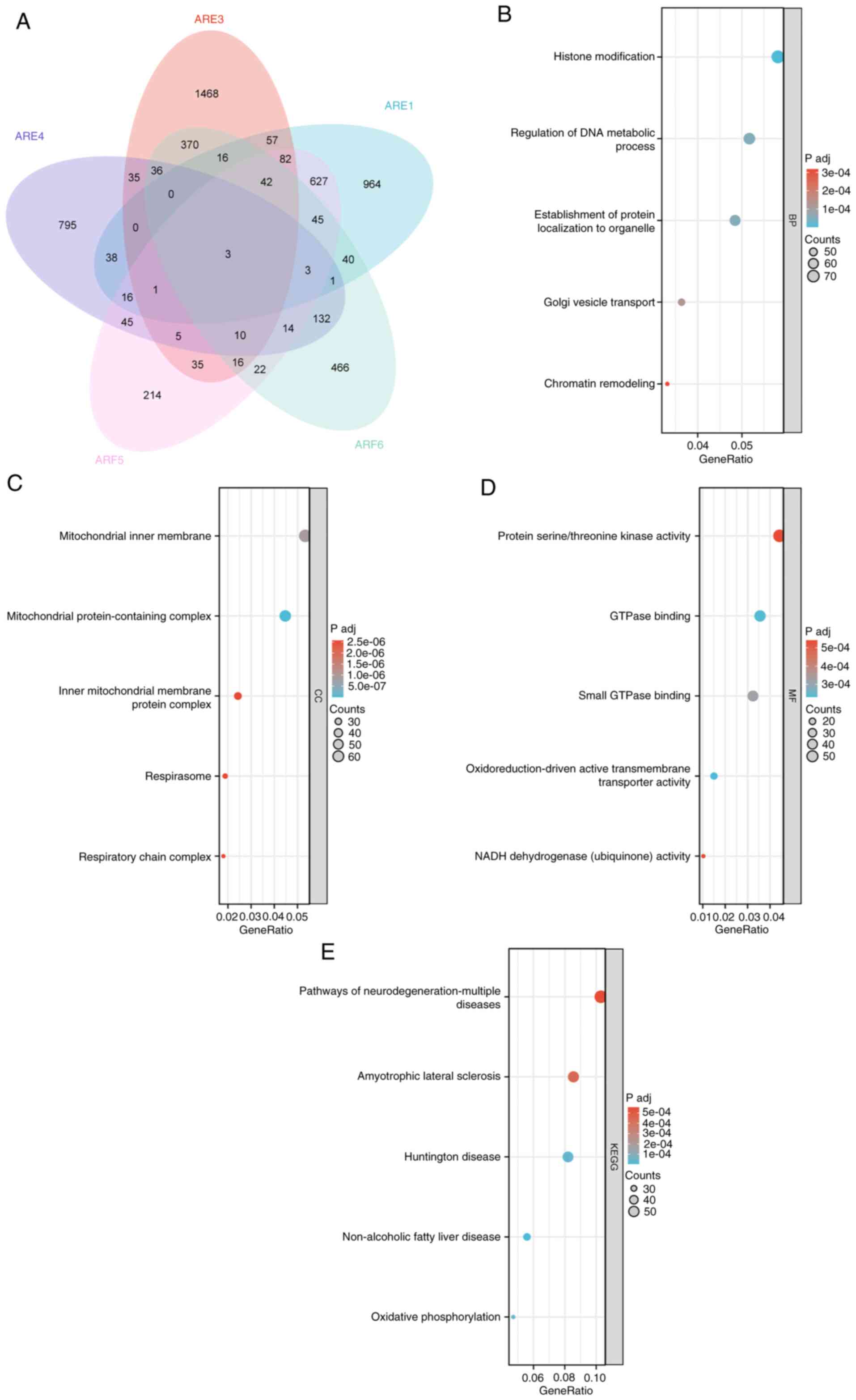
negative correlations with ARFs in BC were determined (Fig. 6). To gain further insight, a Venn
diagram (Fig. 7A) was employed to
analyze the DEGs to show the cross between DEGs of various members
of the ARF family. GO and KEGG enrichment analyses using the top
100 DEGs that were mainly positively correlated with ARFs were
subsequently performed. The condensed information obtained from the
GO analysis encompassed the categories biological process, cellular
component and molecular function. The findings revealed that ARFs
and their interacting genes primarily participated in crucial
processes such as ‘histone modification’, ‘regulation of DNA
metabolic activity’, ‘mitochondrial inner membrane function’,
‘GTPase binding’ and ‘small GTPase binding’ (Fig. 7B-E).
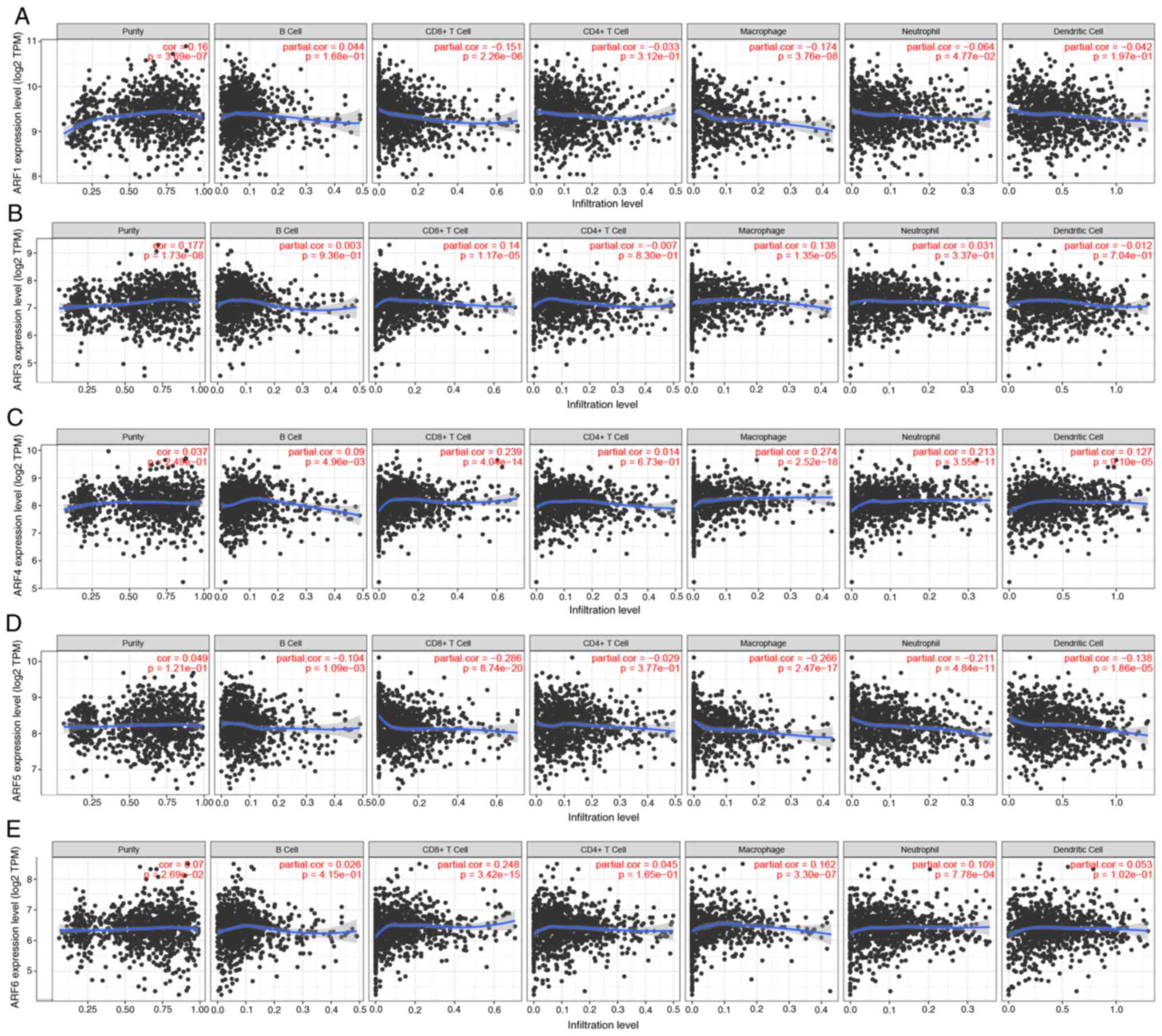
ARF and immune-cell infiltration in
patients with BC
Previous reports have indicated a connection between
the concentration of immune cells and the growth and advancement of
cancer cells (12). The present
study utilized the TIMER database to investigate the link between
ARF members and the invasion of immune cells. The results
demonstrated a significant positive association between the
expression of ARF1 and the invasion of CD8+ T cells, macrophages
and neutrophils in patients with BC (Fig. 8A). Similarly, ARF3 showed a positive
association with the invasion of CD8+ T cells, and a negative
association of macrophages in patients with BC (Fig. 8B). Furthermore, the expression of
ARF4 displayed a positive correlation with the invasion of B cells,
CD8+ T cells, macrophages, dendritic cells and neutrophils in BC
(Fig. 8C). Conversely, the
expression of ARF5 exhibited a significant negative correlation
with B cells, CD8+ T cells, macrophages, neutrophils and dendritic
cells in patients with BC (Fig.
8D). Lastly, the expression of ARF6 in BC exhibited a
significant positive correlation with the invasion of CD8+ T cells,
macrophages and neutrophils (Fig.
8E).
In order to further evaluate the impact of ARFs on
the tumor immune microenvironment (TIME), a Spearman's correlation
analysis was performed to examine the connection between ARFs and
the infiltration of immune cells. The results of this analysis
revealed that there were both positive and negative correlations
between ARFs and various immune cell types. For example, iDC was
negatively correlated with ARF1, ARF3 and ARF5. For instance, a
negative correlation between T cells and all ARFs was consistently
observed (Fig. 9).
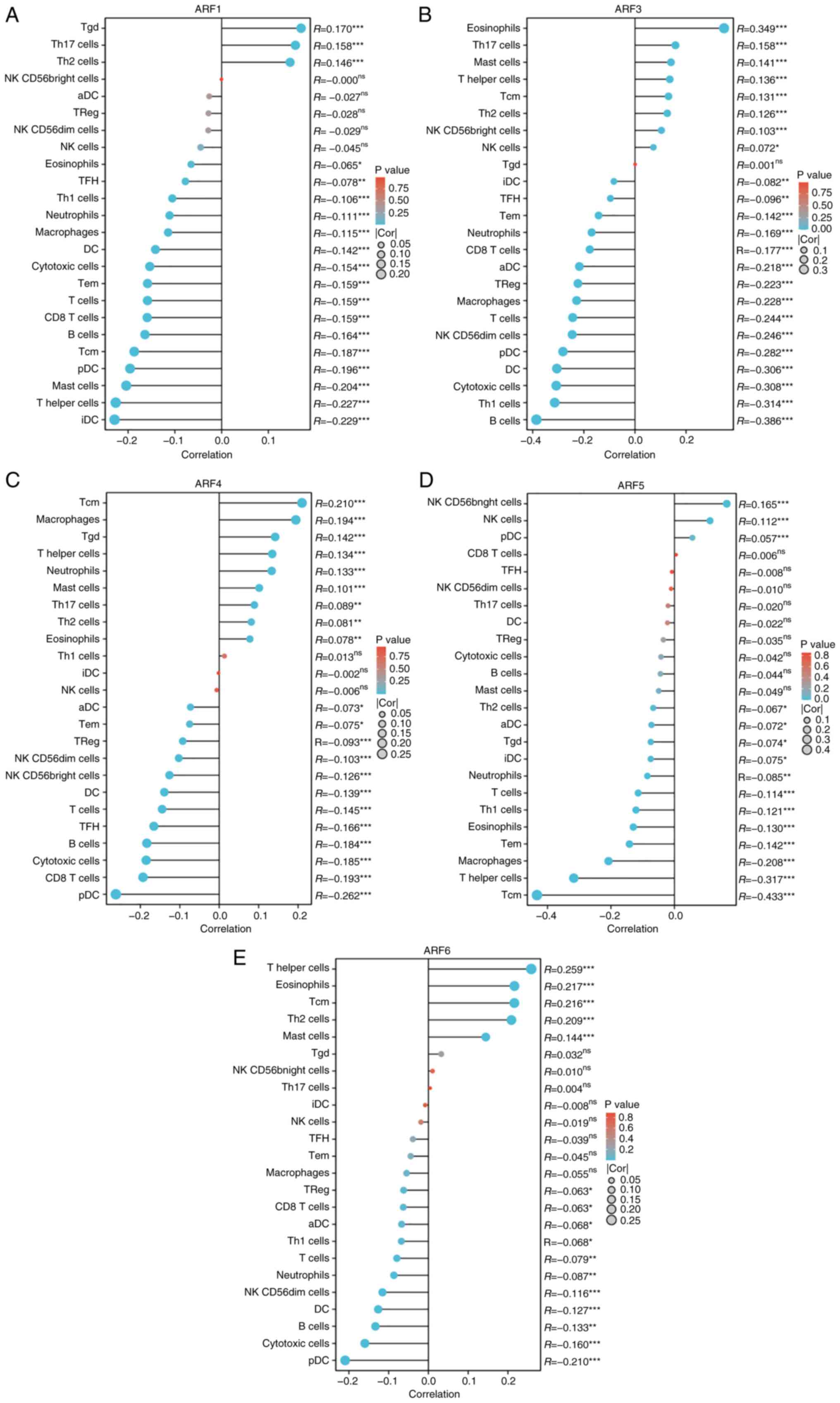 | Figure 9.Expression levels of (A) ARF1, (B)
ARF3, (C) ARF4, (D) ARF5 and (E) ARF6 exhibited a significant
association with the infiltration of immune cells in patients with
BC, as visualized by the use of a Lollipop chart. These findings
were accompanied by statistically significant P-values denoted as
*P<0.05, **P<0.01 and ***P<0.001; ns, no significance. BC
was observed to have a measurable impact on various immune cell
types, including Th, aDC, Treg, TFH, Tgd, NK, iDC and pDC cells.
ARF, ADP-ribosylation factor; BC, breast cancer; Th, T helper
cells; aDC, activated dendritic cells; Treg, regulatory T cells;
TFH, T follicular helper cells; Tgd, γδ T cells; NK, natural killer
cells; iDC, immature dendritic cells; pDC, plasmacytoid dendritic
cells; cor, correlation coefficient; Tcm, central memory T cell;
Tem, effective memory T cell. |
Discussion
Given the limitations of current prognostic
indicators, including estrogen receptor, progesterone receptor,
human EGFR 2, Ki67 and grade, due to the diverse nature of BC, it
becomes imperative to identify new prognostic biomarkers that can
improve individualized therapies. Therefore, the present study
aimed to investigate the potential of ARF family genes as novel
biomarkers for BC prognosis. This research endeavor marks the
pioneering effort in exploring the utility of ARF family genes in
this context.
Bioinformatics analysis is the scientific field that
explores biology through the utilization of informatics, applied
mathematics, computer science and statistics (11). Within this research, a
bioinformatics analysis was conducted to examine the presence of
ARF members in BC, investigate signaling pathways in BC, explore
immune-cell infiltration in the BC microenvironment and assess its
impact on the prognosis of patients with BC. Through analysis of
data acquired from the TCGA database, alongside the examination of
ARF expression levels in tissues collected from patients with BC as
well as BC cell lines, it was identified that the five members of
the ARF family exhibit higher expression in BC as compared to
normal breast tissue. BC has the capability to induce local immune
dysregulation by suppressing innate and adaptive immune responses
(12). The immune microenvironment
of a tumor, known as the TIME, holds significant importance within
the overall tumor microenvironment. It exhibits high levels of
heterogeneity and serves a critical function in both tumor
progression and disease prognosis across different types of cancers
(13). Therefore, achieving an
accurate disease classification based on the TIME is of utmost
importance, not only for assessing prognosis but also for guiding
treatment decisions. Substantial evidence indicates that subsets of
CD8+ T cells have crucial roles in controlling tumors, as evidenced
by the correlation between the quantity of CD8+ T cells present in
the tumor prior to therapy and the response to programmed cell
death 1 treatment (14).
Furthermore, the present results substantiated that there is a
significant association between the heightened expression of the
immune infiltration level of CD8+ T cells and macrophages and ARF
family members, except ARF5. This finding offers valuable insight
into the potential effectiveness of future immunotherapeutic
approaches for BC.
In a study conducted by Lewis-Saravalli et al
(15), it was demonstrated that the
regulation of cell migration in highly invasive cancer cells is
mediated by the interaction between Rac family small GTPase 1 and
insulin receptor substrate of 53 kDa, involving the protein ARF1.
The present investigation confirmed a positive association between
the increased expression of ARF1 and reduced OS, RFS and DMFS in
patients with BC.
Unlike other ARFs, ARF3 has five exons and four
introns. It selectively associates with recruiting the Golgi shell
complex and activating phospholipase D and PI kinase (16,17).
There is accumulating evidence suggesting that ARF3 may have a
crucial role in the development of cancer (18,19). A
previous study indicated that the expression levels of ARF3 are
positively associated with the clinical staging of BC (20). In a study focusing on GC, Chang
et al (18) utilized
microarray assays and identified APF3 as one of the central genes
involved in regulating liver metastasis of gastric cancer. In the
present study, Kaplan-Meier plotter analysis was used to confirm
the significance of high ARF3 expression in the prognosis of human
BC. In addition, the higher the expression of ARF3, the shorter the
RFS in patients with BC observed.
In human glioblastoma cells, previous studies have
shown that ARF4 has a vital role in activating phospholipase D and
inhibiting the generation of reactive oxygen species, making it a
crucial anti-apoptotic protein (21–23).
To further investigate its significance in BC, Kaplan-Meier plotter
analysis was utilized to assess the prognostic relevance of ARF4.
The results indicated a negative association between high ARF4
expression and RFS in patients with BC, highlighting its potential
as a promising biomarker for this condition.
However, the plasma membrane and various endosomes
are the specific locations where ARF6 is found. In addition to its
crucial involvement in membrane trafficking, ARF6 also governs
membrane-associated pathological undertakings, such as the creation
of membrane ruffles, elongation of neurites, as well as cellular
migration and infiltration (24–26).
In a clinical context, heightened expression of ARF6 and the
stimulation of its subsequent signaling pathways have been detected
in diverse tumor categories, correlating with inferior OS rates
(27,28). Examples of such malignancies
encompass BC, lung adenocarcinoma and head and neck cancers
(29–31).
Analysis of ARF family-related proteins identified
ARFIP2, PI4KB, RAB11FIP3, GBF1 and PPP2R1A as proteins with strong
correlations. ARFIP2 regulates epithelial to mesenchymal transition
and autophagy through the PI3K/AKT pathway in hepatocellular
carcinoma (31), whereas PPP2R1A
promotes cancer development through the SRC-JNK-c-Jun pathway
(32), and the ARF family may also
regulate BC development through related mechanisms, which need to
be verified by further studies.
Furthermore, GO analysis was employed to investigate
the biological processes associated with the gene family of ARFs
and their interacting counterparts. The findings revealed the
predominant involvement of ARFs and their interacting genes in
diverse physiological activities, including histone modification,
regulation of DNA metabolic processes, mitochondrial inner membrane
functionality, GTPase binding and small GTPase binding.
Furthermore, KEGG analysis supported the concentration of the gene
family of ARFs and their interacting genes in conditions such as
amyotrophic lateral sclerosis and Huntington's disease. Overall,
these results indicate a potential link between the aberrant
expression of ARFs and its interacting genes and the regulation of
DNA metabolic processes, as well as GTPase binding. Results
analyzed by the ibioportal database showed that there may be
differences in gene expression and deletion of ARFs in different
studies, which may be due to the heterogeneity of BC. As for the
reason for this difference, this may be the focus of further
research efforts by our group.
The present study aimed to investigate the presence
of ARFs in BC, which holds significant importance in predicting the
prognosis of patients with BC. Overexpression of these genes often
contributes to a more aggressive form of BC. All five members of
the ARF gene family are anticipated to serve as crucial biomarkers
for the diagnosis and prognosis prediction of BC. In addition, the
expression levels of ARF gene family members are strongly
associated with the infiltration of immune cells in the TIME of BC.
However, it is important to acknowledge the limitations of the
present study. First, although all available data were utilized,
further experimental verification is still required. Furthermore,
the present study solely focused on exploring the presence of ARF
family genes in BC tissues and failed to examine their expression
in blood samples of patients with BC. By conducting additional
research on the expression of ARFs in the peripheral blood of
patients with BC, an easier and more convenient diagnostic
screening method could potentially be developed. In addition, in
the present study, the expression levels and biological functions
of the ARF gene family in BC were analyzed, but no in-depth study
was conducted on how individual ARF gene family members, such as
ARF1 and ARF3, affect the RFS or OS of patients with BC through
their enriched pathways or their roles in the TIME. The mechanisms
of this part will be explored in depth in the next study.
In conclusion, the present bioinformatics analysis
confirmed the significant overexpression of the ARF gene family in
BC. The expression levels of all five members of the ARF gene
family were associated with BC prognosis and immune infiltration.
Consequently, their overexpression presents an innovative
therapeutic target for BC and offers fresh perspectives on the
effectiveness of immunotherapy in BC treatment.
Acknowledgements
Not applicable.
Funding
This study was funded by the Xingtai City Key Research and
Development Plan (grant no. 2021ZC148), Xingtai Science and
Technology Plan Project (grant no. 2023ZZ104) and the Scientific
Research Fund of Health Commission of Hebei Province (grant no.
20220224).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to restrictions
applied by Xingtai People's Hospital but may be requested from the
corresponding author.
Authors' contributions
SZ, FK and LY conducted the data acquisition and
data analyses, and prepared the figures/tables. SZ, LZ, PP and XL
provided material input, data analysis and assisted with revising
the manuscript. SZ, LZ and LJ designed the implementation of the
research. LZ and LY wrote the manuscript. All authors read and
approved the final version of the manuscript. SZ and LY confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
All procedures performed in the present study
involving human participants were in accordance with The
Declaration of Helsinki (as revised in 2013). The study was
approved by the Institutional Ethics committee of Xingtai People's
Hospital [Xingtai, China; approval no. 2021(035)]. Written informed
consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu H, Qiu C, Wang B, Bing P, Tian G,
Zhang X, Ma J, He B and Yang J: Evaluating DNA methylation, gene
expression, somatic mutation, and their combinations in inferring
tumor tissue-of-origin. Front Cell Dev Biol. 9:6193302021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hunter NB, Kilgore MR and Davidson NE: The
long and winding road for breast cancer biomarkers to reach
clinical utility. Clin Cancer Res. 26:5543–5545. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Xiang J, Tang L, Li J, Lu Q, Tian
G, He BS and Yang J: Identifying breast cancer-related genes based
on a novel computational framework involving KEGG pathways and ppi
network modularity. Front Genet. 12:5967942021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
D'Souza-Schorey C and Chavrier P: ARF
proteins: Roles in membrane traffic and beyond. Nat Rev Mol Cell
Biol. 7:347–358. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Donaldson JG: Multiple roles for Arf6:
Sorting, structuring, and signaling at the plasma membrane. J Biol
Chem. 278:41573–41576. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Volpicelli-Daley LA, Li Y, Zhang CJ and
Kahn RA: Isoform-selective effects of the depletion of
ADP-ribosylation factors 1–5 on membrane traffic. Mol Biol Cell.
16:4495–4508. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grossmann AH, Zhao H, Jenkins N, Zhu W,
Richards JR, Yoo JH, Winter JM, Rich B, Mleynek TM, Li DY and
Odelberg SJ: The small GTPase ARF6 regulates protein trafficking to
control cellular function during development and in disease. Small
GTPases. 10:1–12. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pallen MJ: Microbial bioinformatics 2020.
Microb Biotechnol. 9:681–686. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ernst B and Anderson KS: Immunotherapy for
the treatment of breast cancer. Curr Oncol Rep. 17:52015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Laplagne C, Domagala M, Le Naour A,
Quemerais C, Hamel D, Fournié JJ, Couderc B, Bousquet C, Ferrand A
and Poupot M: Latest advances in targeting the tumor
microenvironment for tumor suppression. Int J Mol Sci. 20:47192019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim CG, Hong MH, Kim KH, Seo IH, Ahn BC,
Pyo KH, Synn CB, Yoon HI, Shim HS, Lee YI, et al: Dynamic changes
in circulating PD-1+ CD8+ T lymphocytes for predicting treatment
response to PD-1 blockade in patients with non-small-cell lung
cancer. Eur J Cancer. 143:113–126. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lewis-Saravalli S, Campbell S and Claing
A: ARF1 controls Rac1 signaling to regulate migration of MDA-MB-231
invasive breast cancer cells. Cell Signal. 25:1813–1819. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smith SA, Holik PR, Stevens J, Melis R,
White R and Albertsen H: Isolation and mapping of a gene encoding a
novel human ADP-ribosylation factor on chromosome 17q12-q21.
Genomics. 28:113–115. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sztul E, Chen PW, Casanova JE, Cherfils J,
Dacks JB, Lambright DG, Lee FS, Randazzo PA, Santy LC, Schürmann A,
et al: ARF GTPases and their GEFs and GAPs: Concepts and
challenges. Mol Biol Cell. 30:1249–1271. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang W, Ma L, Lin L, Gu L, Liu X, Cai H,
Yu Y, Tan X, Zhai Y, Xu X, et al: Identification of novel hub genes
associated with liver metastasis of gastric cancer. Int J Cancer.
125:2844–2853. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Z, Li H, Zhao Y, Guo Q, Yu Y, Zhu S,
Zhang S, Min L and Li P: Asporin promotes cell proliferation via
interacting with PSMD2 in gastric cancer. Front Biosci (Landmark
Ed). 24:1178–1189. 2019. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang D, Pei Y, Dai C, Huang Y, Chen H,
Chen X, Zhang X, Lin C, Wang H, Zhang R, et al: Up-regulated
ADP-Ribosylation factor 3 promotes breast cancer cell proliferation
through the participation of FOXO1. Exp Cell Res. 384:1116242019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shome K, Vasudevan C and Romero G: ARF
proteins mediate insulin-dependent activation of phospholipase D.
Curr Biol. 7:387–396. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim SW, Hayashi M, Lo JF, Yang Y, Yoo JS
and Lee JD: ADP-ribosylation factor 4 small GTPase mediates
epidermal growth factor receptor-dependent phospholipase D2
activation. J Biol Chem. 278:2661–2668. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Woo IS, Eun SY, Jang HS, Kang ES, Kim GH,
Kim HJ, Lee JH, Chang KC, Kim JH, Han CW and Seo HG: Identification
of ADP-ribosylation factor 4 as a suppressor of N-(4-hydroxyphenyl)
retinamide-induced cell death. Cancer Lett. 276:53–60. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Honda A, Nogami M, Yokozeki T, Yamazaki M,
Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman
MA and Kanaho Y: Phosphatidylinositol 4-phosphate 5-kinase α is a
downstream effector of the small G protein ARF6 in membrane ruffle
formation. Cell. 99:521–532. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jaworski J: ARF6 in the nervous system.
Eur J Cell Biol. 86:513–524. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sabe H: Requirement for Arf6 in cell
adhesion, migration, and cancer cell invasion. J Biochem.
134:485–489. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Onodera Y, Hashimoto S, Hashimoto A,
Morishige M, Mazaki Y, Yamada A, Ogawa E, Adachi M, Sakurai T,
Manabe T, et al: Expression of AMAP1, an ArfGAP, provides novel
targets to inhibit breast cancer invasive activities. EMBO J.
24:963–973. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Morishige M, Hashimoto S, Ogawa E, Toda Y,
Kotani H, Hirose M, Wei S, Hashimoto A, Yamada A, Yano H, et al:
GEP100 links epidermal growth factor receptor signalling to Arf6
activation to induce breast cancer invasion. Nat Cell Biol.
10:85–92. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Menju T, Hashimoto S, Hashimoto A, Otsuka
Y, Handa H, Ogawa E, Toda Y, Wada H, Date H and Sabe H: Engagement
of overexpressed Her2 with GEP100 induces autonomous invasive
activities and provides a biomarker for metastases of lung
adenocarcinoma. PLoS One. 6:e253012011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sato H, Hatanaka KC, Hatanaka Y,
Hatakeyama H, Hashimoto A, Matsuno Y, Fukuda S and Sabe H: High
level expression of AMAP1 protein correlates with poor prognosis
and survival after surgery of head and neck squamous cell carcinoma
patients. Cell Commun Signal. 12:172014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang K, Lin Y, Wang K, Shen J and Wei D:
ARFIP2 regulates EMT and autophagy in hepatocellular carcinoma in
part through the PI3K/Akt signalling pathway. J Hepatocell
Carcinoma. 9:1323–1339. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jeong AL, Han S, Lee S, Su Park J, Lu Y,
Yu S, Li J, Chun KH, Mills GB and Yang Y: Patient derived mutation
W257G of PPP2R1A enhances cancer cell migration through
SRC-JNK-c-Jun pathway. Sci Rep. 6:273912016. View Article : Google Scholar : PubMed/NCBI
|