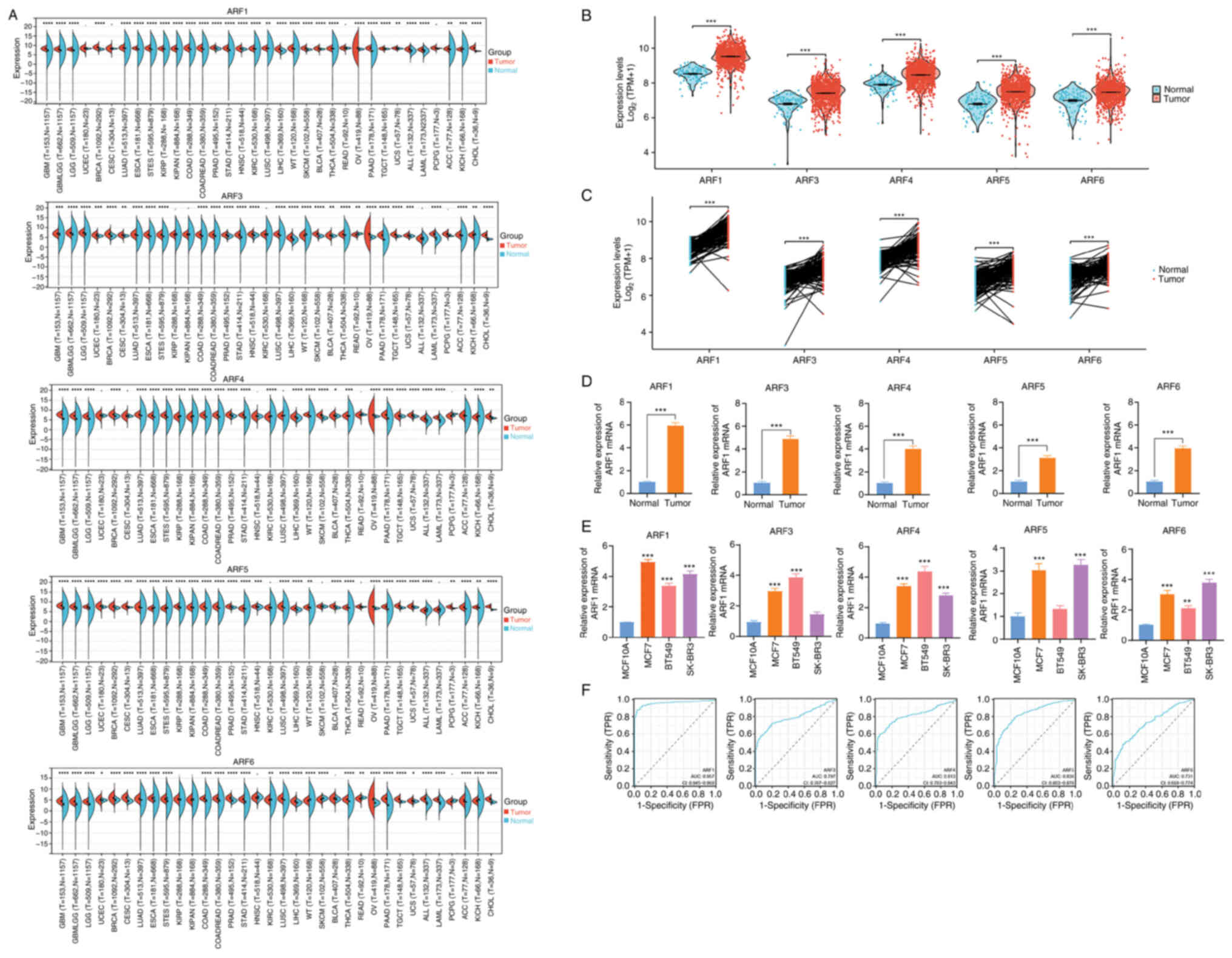
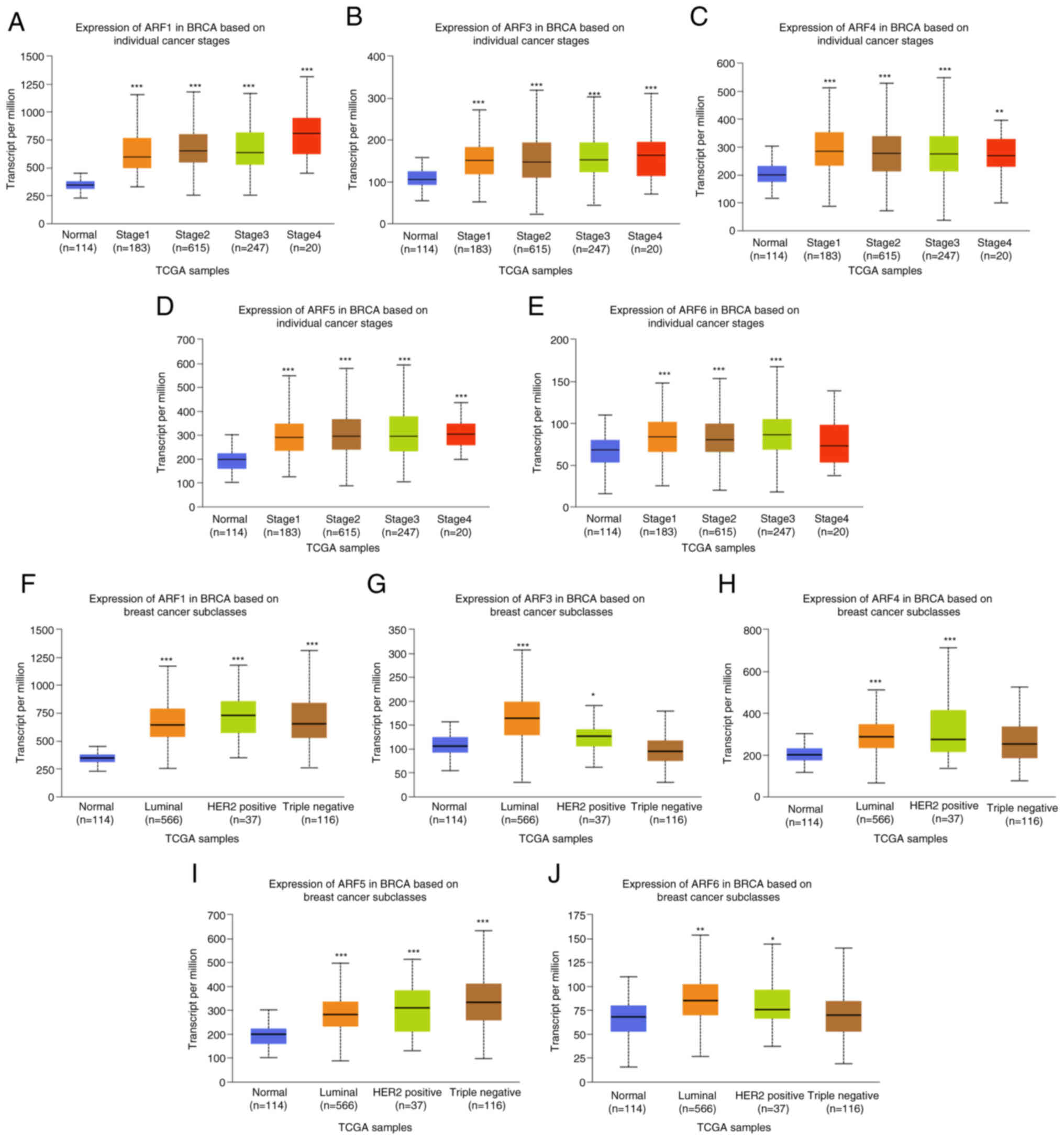
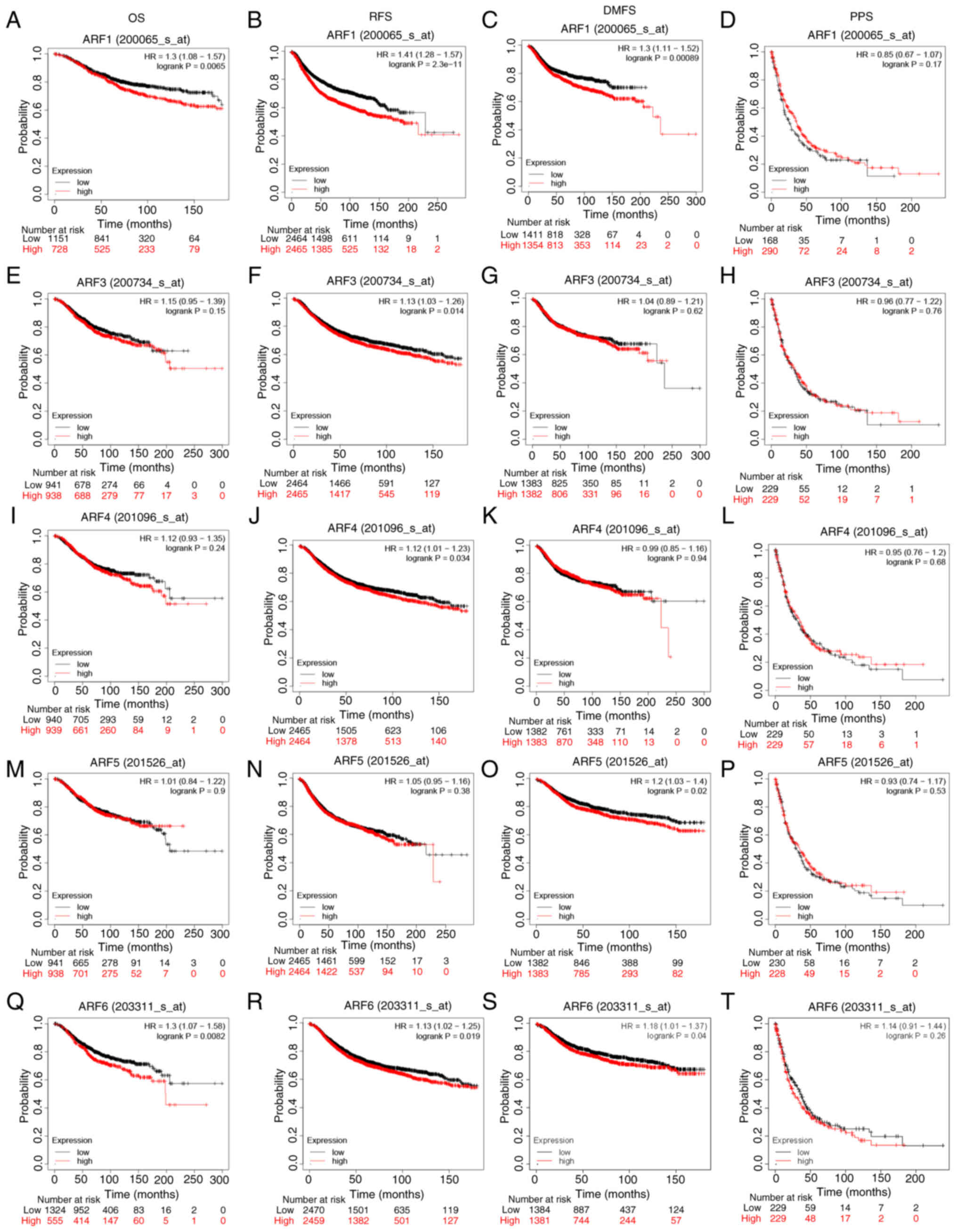
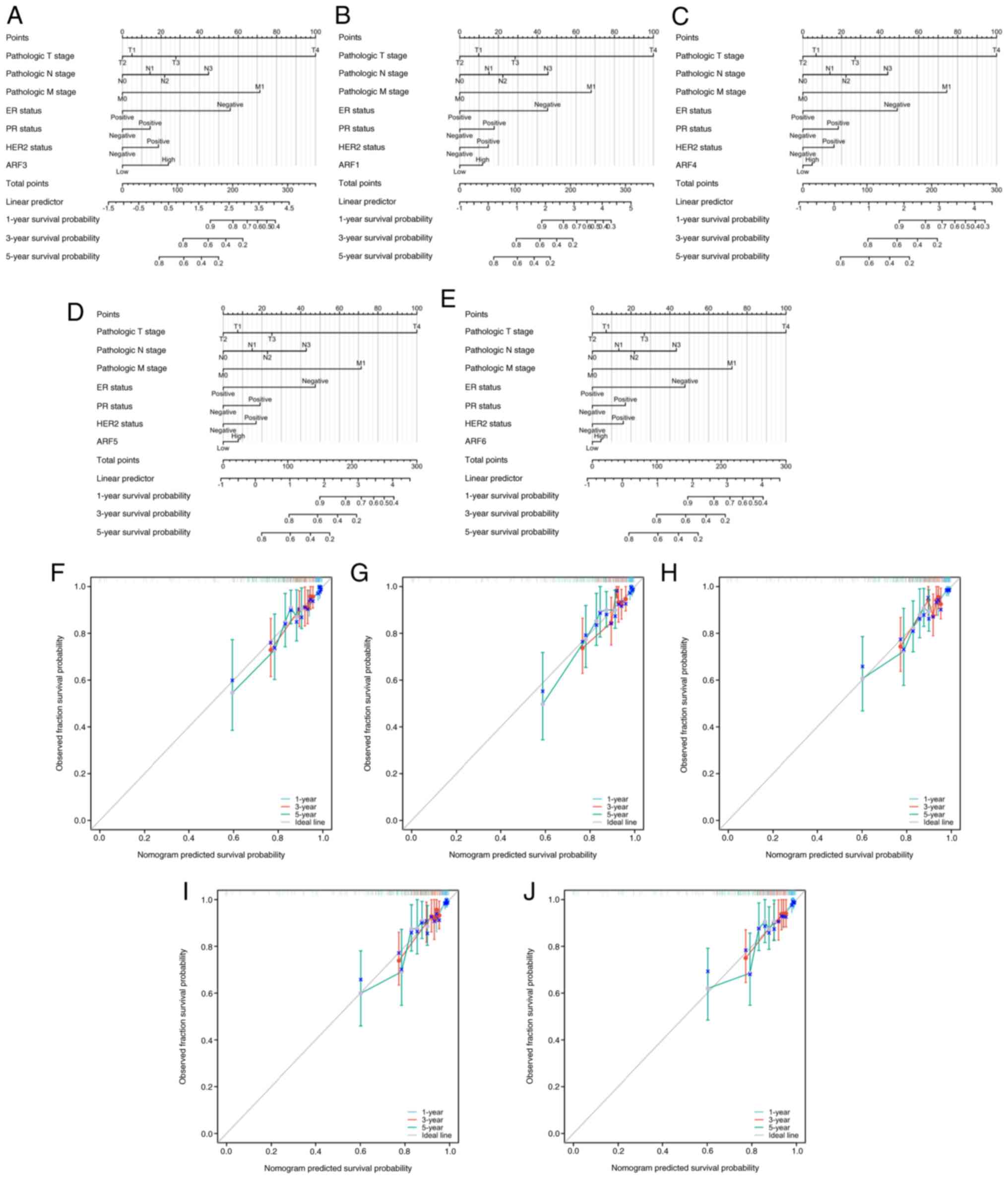
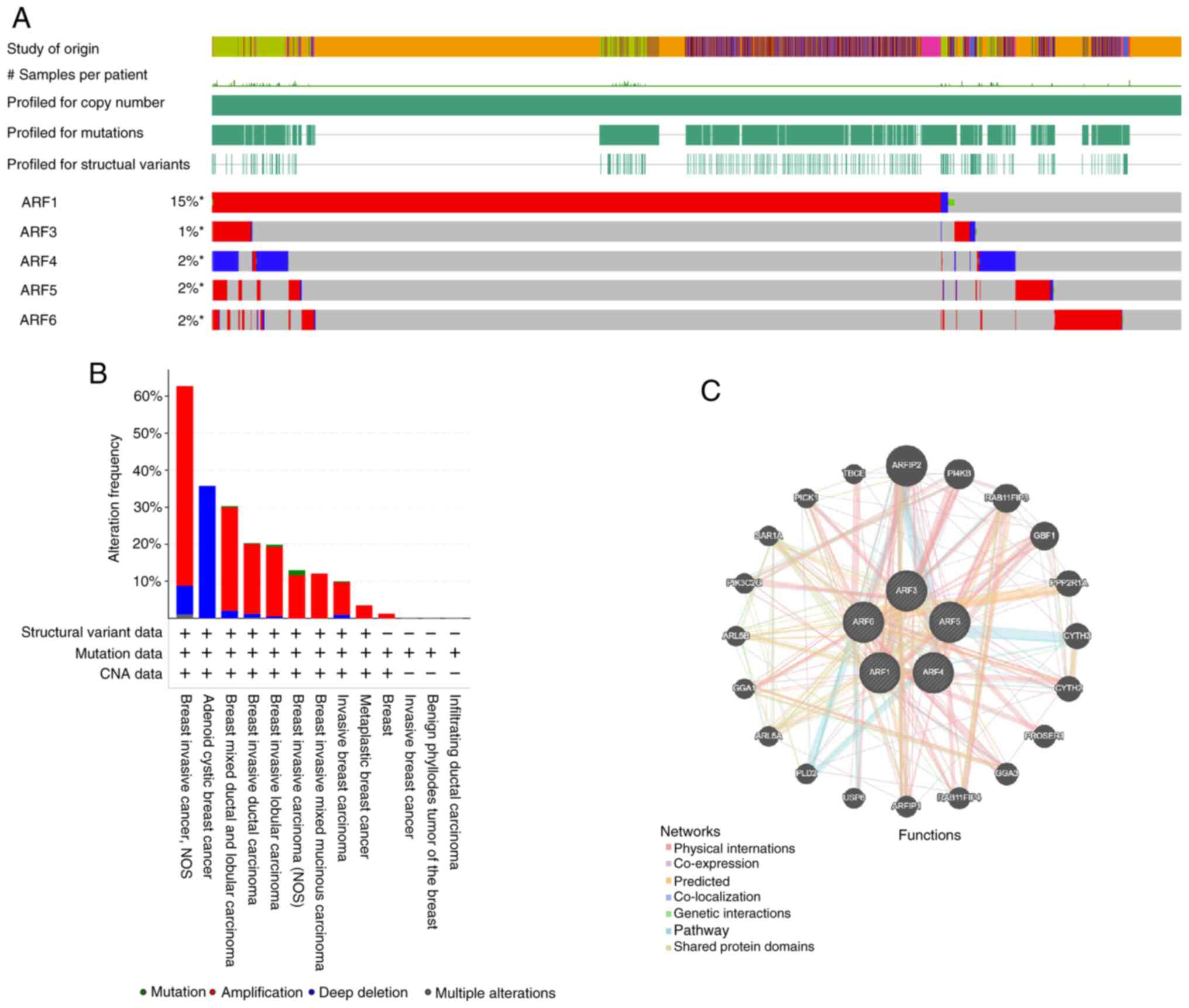
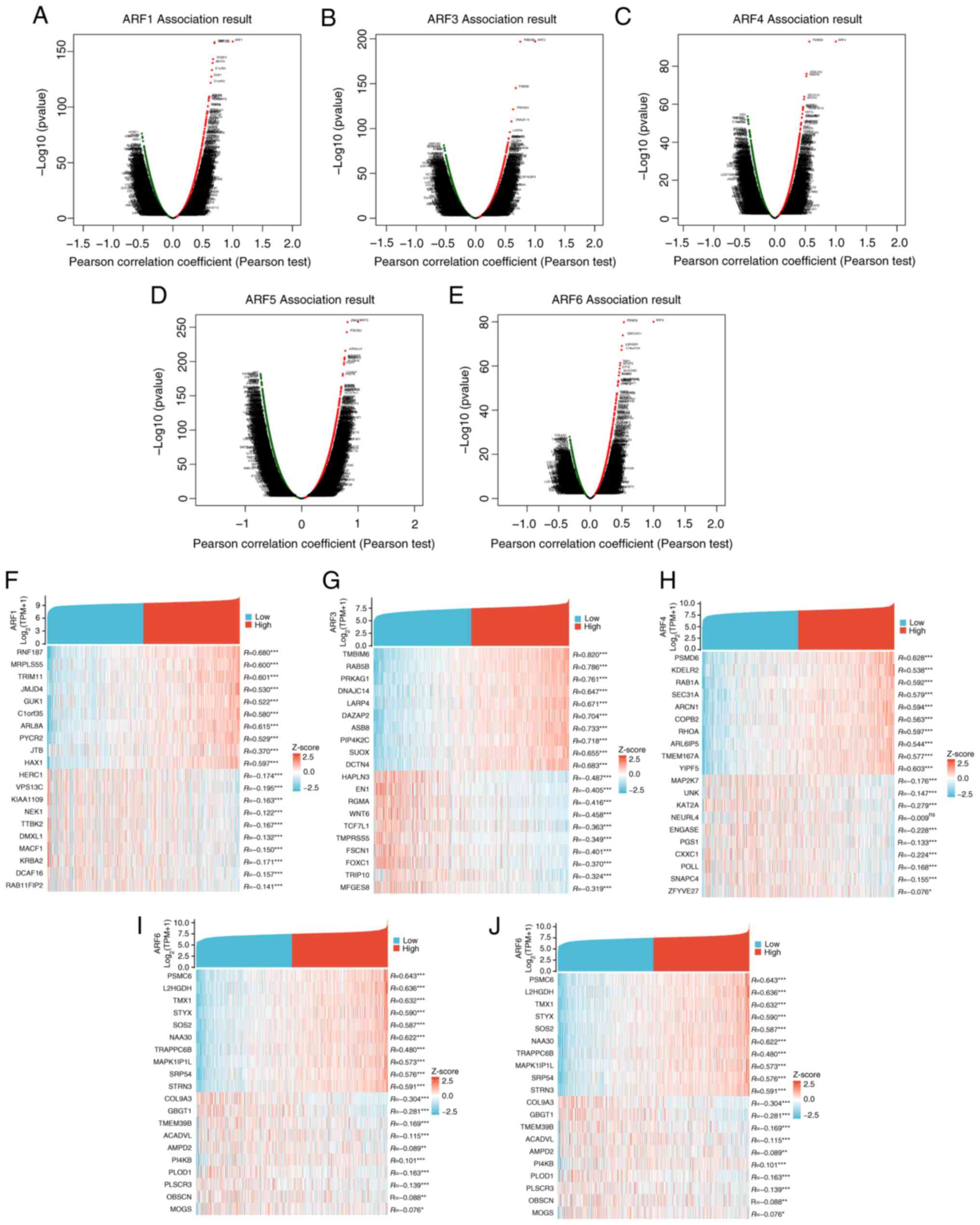
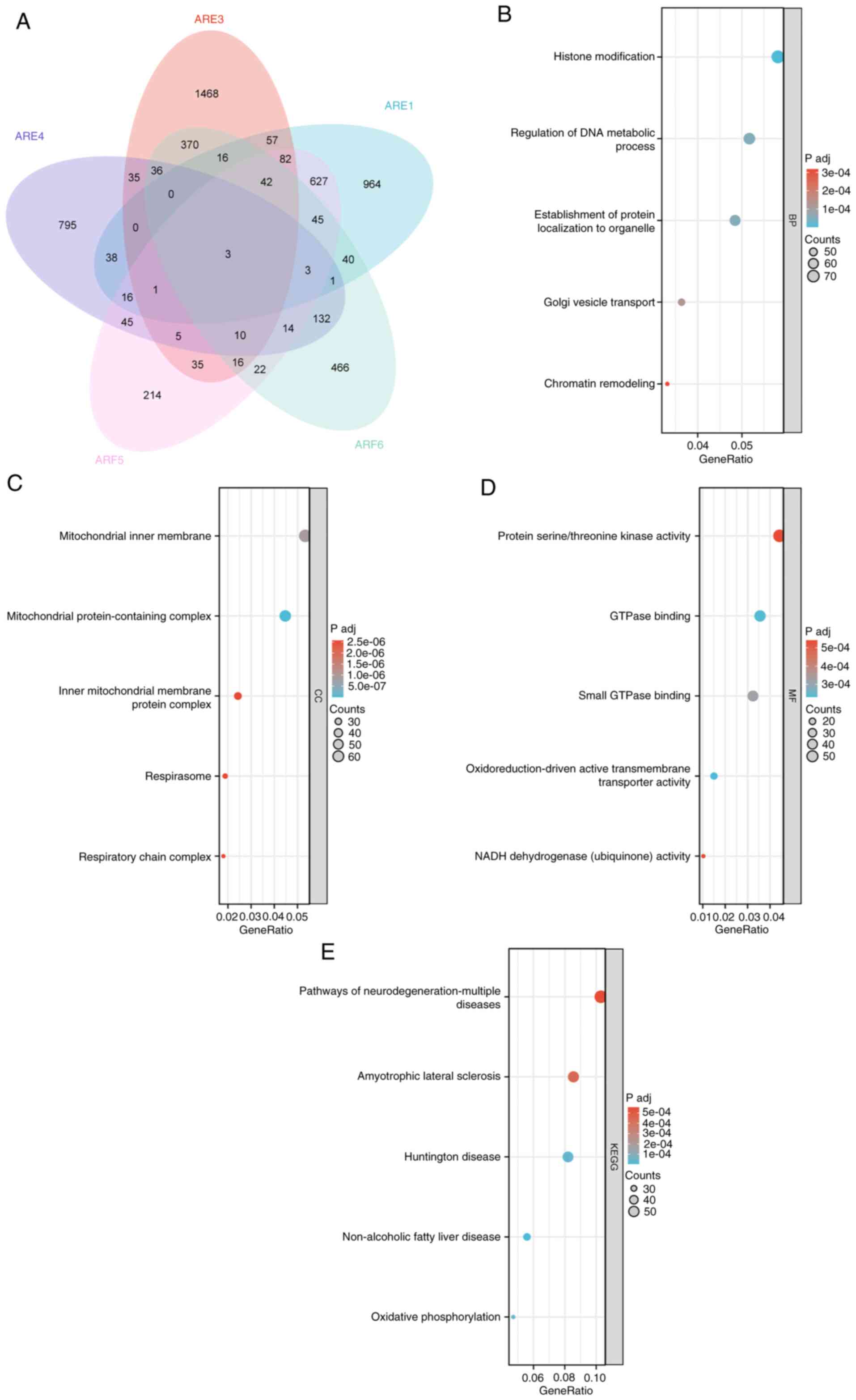
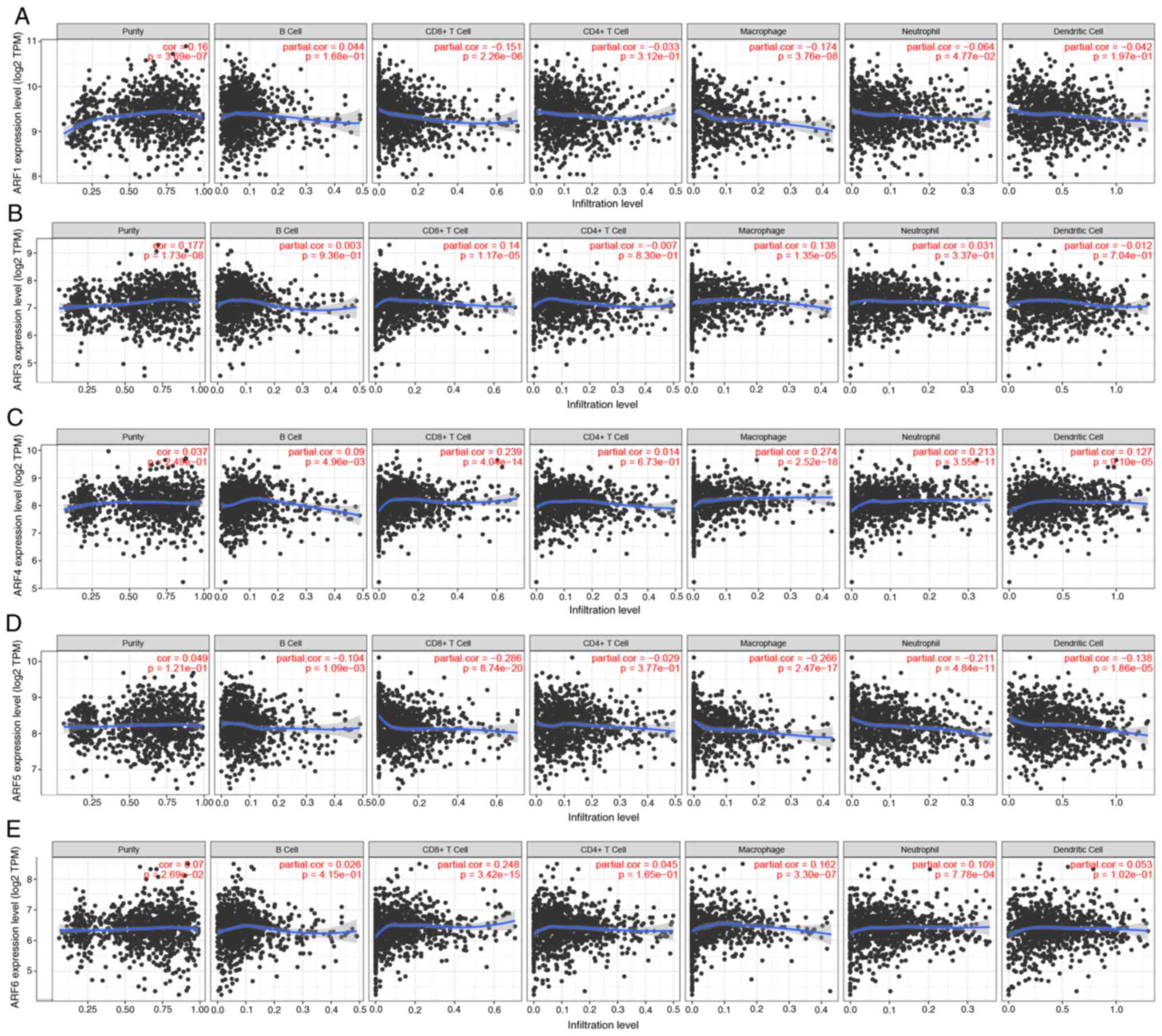
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu H, Qiu C, Wang B, Bing P, Tian G,
Zhang X, Ma J, He B and Yang J: Evaluating DNA methylation, gene
expression, somatic mutation, and their combinations in inferring
tumor tissue-of-origin. Front Cell Dev Biol. 9:6193302021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hunter NB, Kilgore MR and Davidson NE: The
long and winding road for breast cancer biomarkers to reach
clinical utility. Clin Cancer Res. 26:5543–5545. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Xiang J, Tang L, Li J, Lu Q, Tian
G, He BS and Yang J: Identifying breast cancer-related genes based
on a novel computational framework involving KEGG pathways and ppi
network modularity. Front Genet. 12:5967942021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
D'Souza-Schorey C and Chavrier P: ARF
proteins: Roles in membrane traffic and beyond. Nat Rev Mol Cell
Biol. 7:347–358. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Donaldson JG: Multiple roles for Arf6:
Sorting, structuring, and signaling at the plasma membrane. J Biol
Chem. 278:41573–41576. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Volpicelli-Daley LA, Li Y, Zhang CJ and
Kahn RA: Isoform-selective effects of the depletion of
ADP-ribosylation factors 1–5 on membrane traffic. Mol Biol Cell.
16:4495–4508. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grossmann AH, Zhao H, Jenkins N, Zhu W,
Richards JR, Yoo JH, Winter JM, Rich B, Mleynek TM, Li DY and
Odelberg SJ: The small GTPase ARF6 regulates protein trafficking to
control cellular function during development and in disease. Small
GTPases. 10:1–12. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pallen MJ: Microbial bioinformatics 2020.
Microb Biotechnol. 9:681–686. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ernst B and Anderson KS: Immunotherapy for
the treatment of breast cancer. Curr Oncol Rep. 17:52015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Laplagne C, Domagala M, Le Naour A,
Quemerais C, Hamel D, Fournié JJ, Couderc B, Bousquet C, Ferrand A
and Poupot M: Latest advances in targeting the tumor
microenvironment for tumor suppression. Int J Mol Sci. 20:47192019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim CG, Hong MH, Kim KH, Seo IH, Ahn BC,
Pyo KH, Synn CB, Yoon HI, Shim HS, Lee YI, et al: Dynamic changes
in circulating PD-1+ CD8+ T lymphocytes for predicting treatment
response to PD-1 blockade in patients with non-small-cell lung
cancer. Eur J Cancer. 143:113–126. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lewis-Saravalli S, Campbell S and Claing
A: ARF1 controls Rac1 signaling to regulate migration of MDA-MB-231
invasive breast cancer cells. Cell Signal. 25:1813–1819. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smith SA, Holik PR, Stevens J, Melis R,
White R and Albertsen H: Isolation and mapping of a gene encoding a
novel human ADP-ribosylation factor on chromosome 17q12-q21.
Genomics. 28:113–115. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sztul E, Chen PW, Casanova JE, Cherfils J,
Dacks JB, Lambright DG, Lee FS, Randazzo PA, Santy LC, Schürmann A,
et al: ARF GTPases and their GEFs and GAPs: Concepts and
challenges. Mol Biol Cell. 30:1249–1271. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang W, Ma L, Lin L, Gu L, Liu X, Cai H,
Yu Y, Tan X, Zhai Y, Xu X, et al: Identification of novel hub genes
associated with liver metastasis of gastric cancer. Int J Cancer.
125:2844–2853. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Z, Li H, Zhao Y, Guo Q, Yu Y, Zhu S,
Zhang S, Min L and Li P: Asporin promotes cell proliferation via
interacting with PSMD2 in gastric cancer. Front Biosci (Landmark
Ed). 24:1178–1189. 2019. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang D, Pei Y, Dai C, Huang Y, Chen H,
Chen X, Zhang X, Lin C, Wang H, Zhang R, et al: Up-regulated
ADP-Ribosylation factor 3 promotes breast cancer cell proliferation
through the participation of FOXO1. Exp Cell Res. 384:1116242019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shome K, Vasudevan C and Romero G: ARF
proteins mediate insulin-dependent activation of phospholipase D.
Curr Biol. 7:387–396. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim SW, Hayashi M, Lo JF, Yang Y, Yoo JS
and Lee JD: ADP-ribosylation factor 4 small GTPase mediates
epidermal growth factor receptor-dependent phospholipase D2
activation. J Biol Chem. 278:2661–2668. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Woo IS, Eun SY, Jang HS, Kang ES, Kim GH,
Kim HJ, Lee JH, Chang KC, Kim JH, Han CW and Seo HG: Identification
of ADP-ribosylation factor 4 as a suppressor of N-(4-hydroxyphenyl)
retinamide-induced cell death. Cancer Lett. 276:53–60. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Honda A, Nogami M, Yokozeki T, Yamazaki M,
Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman
MA and Kanaho Y: Phosphatidylinositol 4-phosphate 5-kinase α is a
downstream effector of the small G protein ARF6 in membrane ruffle
formation. Cell. 99:521–532. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jaworski J: ARF6 in the nervous system.
Eur J Cell Biol. 86:513–524. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sabe H: Requirement for Arf6 in cell
adhesion, migration, and cancer cell invasion. J Biochem.
134:485–489. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Onodera Y, Hashimoto S, Hashimoto A,
Morishige M, Mazaki Y, Yamada A, Ogawa E, Adachi M, Sakurai T,
Manabe T, et al: Expression of AMAP1, an ArfGAP, provides novel
targets to inhibit breast cancer invasive activities. EMBO J.
24:963–973. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Morishige M, Hashimoto S, Ogawa E, Toda Y,
Kotani H, Hirose M, Wei S, Hashimoto A, Yamada A, Yano H, et al:
GEP100 links epidermal growth factor receptor signalling to Arf6
activation to induce breast cancer invasion. Nat Cell Biol.
10:85–92. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Menju T, Hashimoto S, Hashimoto A, Otsuka
Y, Handa H, Ogawa E, Toda Y, Wada H, Date H and Sabe H: Engagement
of overexpressed Her2 with GEP100 induces autonomous invasive
activities and provides a biomarker for metastases of lung
adenocarcinoma. PLoS One. 6:e253012011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sato H, Hatanaka KC, Hatanaka Y,
Hatakeyama H, Hashimoto A, Matsuno Y, Fukuda S and Sabe H: High
level expression of AMAP1 protein correlates with poor prognosis
and survival after surgery of head and neck squamous cell carcinoma
patients. Cell Commun Signal. 12:172014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang K, Lin Y, Wang K, Shen J and Wei D:
ARFIP2 regulates EMT and autophagy in hepatocellular carcinoma in
part through the PI3K/Akt signalling pathway. J Hepatocell
Carcinoma. 9:1323–1339. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jeong AL, Han S, Lee S, Su Park J, Lu Y,
Yu S, Li J, Chun KH, Mills GB and Yang Y: Patient derived mutation
W257G of PPP2R1A enhances cancer cell migration through
SRC-JNK-c-Jun pathway. Sci Rep. 6:273912016. View Article : Google Scholar : PubMed/NCBI
|