Introduction
Colorectal cancer, is a common malignant tumour
worldwide, with >1 million new cases each year, accounting for
~10% of new cancer cases, and a mortality rate accounting for 9.4%
of all cancer deaths, making it a focus of attention in the field
of public health (1,2). Despite considerable progress in
multimodal treatment approaches, including precision surgery and
tailored chemoradiotherapy, achieving long-term survival and
effective disease management poses significant challenges for
patients with advanced or highly invasive forms of this cancer
(3). Recent advancements in medical
technology and the implementation of screening programs have
improved the early diagnosis and treatment outcomes of colorectal
cancer. Nonetheless, its prognosis is still determined by a complex
interplay of tumor biology and molecular biomarkers (4–6).
The surge in molecular biology, transcriptomics and
genomics research has spotlighted the pivotal role of long
non-coding (lnc)RNAs in cancer development, growth and metastasis
(7). lncRNAs are more complex than
small RNAs and involve multiple regulatory mechanisms, including
gene silencing, chromosomal alterations and protein translation
regulation (8,9). Specifically, ferroptosis-related
lncRNAs have emerged as key players in the pathogenesis if
colorectal cancer (10–12). Ferroptosis, identified as a unique
form of non-apoptotic and non-necrotic cell death, is considered a
potential therapeutic target in several malignancies (13). The aberrant expression and
dysfunction of iron death-related lncRNAs have been associated with
prognosis, tumor progression and metastasis in colorectal cancer,
suggesting their potential as novel biomarkers and therapeutic
targets (14–16). Evidence has shown that iron
death-related lncRNAs have specific expression patterns in
colorectal cancer tissue and are closely associated with disease
progression and prognosis (11,12).
However, despite the increasing volume of research, the exact
molecular mechanisms, biological functions and associations with
clinical features of iron death-related lncRNAs in colorectal
cancer are still controversial (10–20).
Therefore, the present study aimed to perform an
exhaustive meta-analysis through systematic integration and
analysis of existing literature to evaluate the expression
differences of iron death-related lncRNAs in colorectal cancer
tissues and further clarify their relationship with clinical
prognosis. Through this comprehensive analysis, the study aimed to
solidify the scientific foundation for considering iron
death-related lncRNAs as viable prognostic biomarkers in colorectal
cancer and to offer insightful recommendations for future
investigations and clinical practices within this burgeoning
domain.
Materials and methods
Search strategy
To thoroughly assess studies on the expression of
ferroptosis-related lncRNAs in colorectal cancer tissues and their
prognostic significance, the present study performed a systematic
search across several databases, including PubMed (https://pubmed.ncbi.nlm.nih.gov/), Web of Science
(https://www.webofscience.com/), Embase
(https://www.embase.com/), Cochrane Library
(https://www.cochranelibrary.com/), CNKI
(https://www.cnki.net/), Wanfang (https://www.wanfangdata.com.cn/) and VIP
(http://www.cqvip.com/) databases. The retrieval
time was limited from the establishment of each database to October
2023. The search strategy used a combination of keywords and
phrases such as ‘ferroptosis’ OR ‘iron death’ AND ‘LncRNA’ OR ‘long
non-coding RNA’ AND ‘colorectal cancer’ OR ‘colon cancer’ AND
‘prognosis’. To ensure the search was exhaustive, the reference
lists of relevant articles were also reviewed to ensure important
studies were not missed. The search strategy was as follows: (‘Iron
death’ OR ‘Ferroptosis’) AND (‘LncRNA’ OR ‘Long non-coding RNA’)
AND (‘Colorectal cancer’ OR ‘Colon cancer’) AND (‘Prognosis’).
Furthermore, the references of pertinent articles were also
manually reviewed to identify any studies that were potentially
overlooked. The detailed search methodology of the present study
was aligned with the Preferred Reporting Items for Systematic
Reviews and Meta-Analyses checklist, with the aim to guarantee the
thoroughness and precision of the search.
Inclusion and exclusion criteria
The inclusion criteria were as follows: i) Research
studies on patients with colorectal cancer, specifically reporting
on the expression levels of ferroptosis-related lncRNAs within
colorectal cancer tissues; and ii) studies providing relevant data
on the association between lncRNA expression and prognosis in
patients with colorectal cancer, such as survival rates and
recurrence rates, with ≥2 outcome metrics.
The exclusion criteria were as follows: i) Studies
that were not relevant to the topic or did not include the
expression and prognosis of iron death-related lncRNAs in
colorectal cancer; ii) studies with incomplete data, duplicate
publications, or those only in abstract form; and iii) review
articles, overviews, case reports, conference abstracts and other
non-original research publications.
Data extraction
The following key information was methodically
extracted: First author, publication year, sample size, patient
demographics (male/female ratio), follow-up outcomes, and
prognostic markers (tumor stage, T stage, lymph node metastasis,
distant metastasis and risk score). Statistical outcomes, such as
hazard ratios (HRs) and 95% confidence intervals (CIs) were also
extracted. This process was performed independently by two
researchers to ensure accuracy, with any discrepancies resolved
through consultation with a third researcher.
Literature quality evaluation
The quality of the included literature was assessed
using the Newcastle-Ottawa Scale (NOS) (21), a tool for evaluating observational
studies, especially cohort and case-control studies, by scoring ≤9
points based on three dimensions, namely selection criteria,
comparability and outcome measures. Studies scoring ≥7 were deemed
high-quality; scores of 4–6 indicated moderate quality; and scores
of ≤3 were considered low-quality.
Statistical analysis
In the present meta-analysis, R software version
4.3.1 (The R Foundation) was used to statistically analyze the
collated data. The association between iron death-related lncRNAs
and prognosis in colorectal cancer were quantitatively evaluated
using HRs and 95% CIs. The I2 test and Q test were used
to assess heterogeneity among studies in the meta-analysis.
Heterogeneity between studies was not considered significant when
I2<50% or P>0.05, and conversely, significant
heterogeneity was considered to exist. As the studies included in
the present study are from different groups and geographical
locations and may be subject to heterogeneity, the results of the
meta-analyses were thus interpreted using a random-effects model to
accommodate the inherent heterogeneity between the different
studies. Publication bias was evaluated through funnel plot
analyses, Begg's and Egger's test. Sensitivity analysis was
performed to assess the robustness and reliability of the findings.
All statistical tests were two-sided, and P<0.05 was considered
to indicate a statistically significance difference.
Results
Literature screening and
characteristics
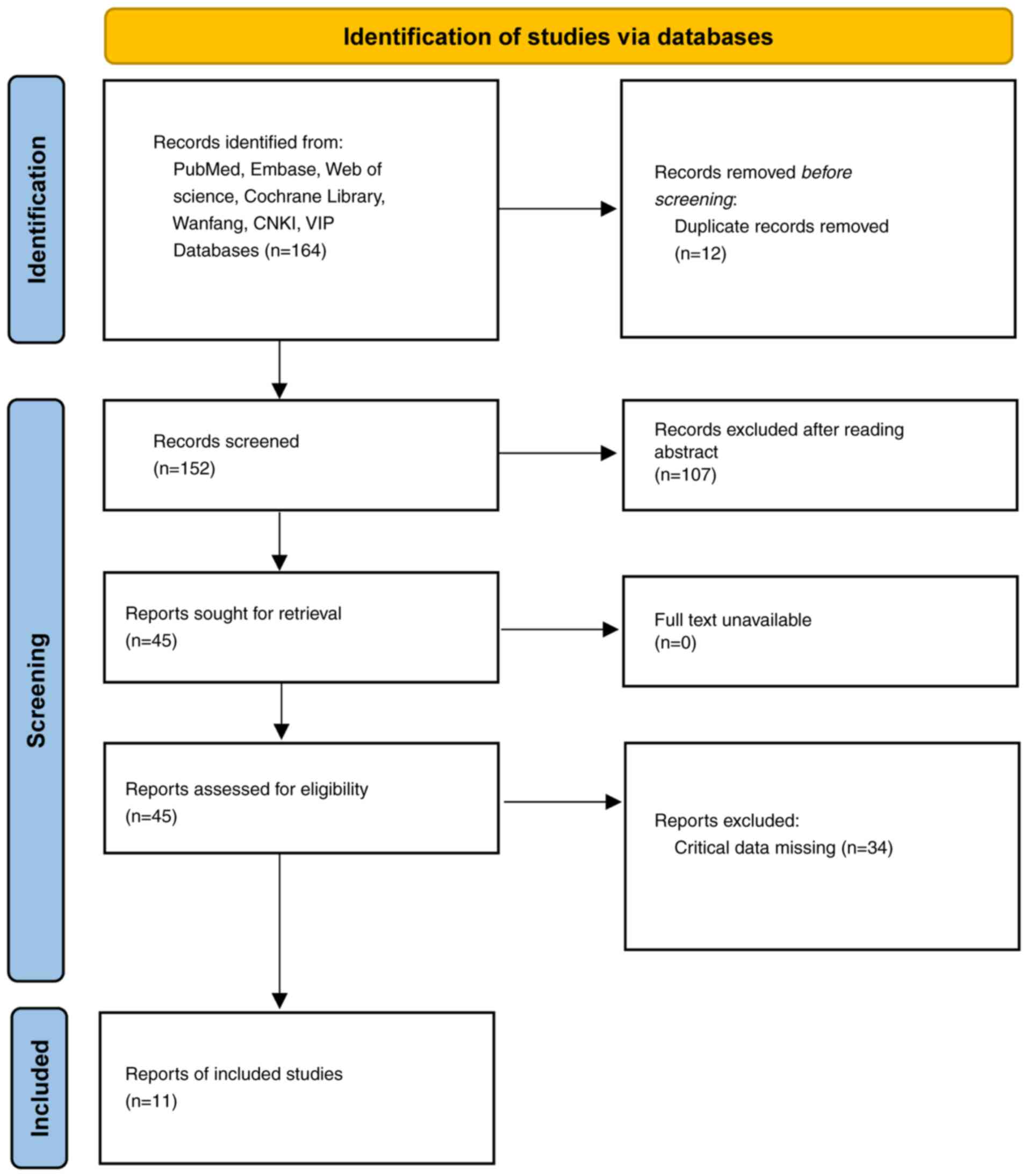
Following the outlined search strategy, 164 relevant
articles were initially retrieved, from which 152 were selected for
preliminary screening after removing duplicates. After the
screening of titles and abstracts, 107 articles were excluded due
to obvious irrelevance to the topic. The remaining 45 articles were
then read in full, and 34 articles were further excluded due to
missing key data. Finally, 11 articles were included in the
meta-analysis (Fig. 1). The
included literature mainly dealt with the expression levels of iron
death-related lncRNAs and their association with prognosis in
patients with colorectal cancer. The main characteristics of the
included studies are presented in Table
I. The results of risk-of-bias assessments of the included
literature demonstrated that all the included studies had NOS
scores ≥6, which met the quality requirements of the present paper
(Table II).
 | Table I.Characteristics of included
studies. |
Table I.
Characteristics of included
studies.
|
|
|
| Age, years |
|
|
|
|
|
|---|
|
|
|
|
|
|
| Follow-up results
(survival/death) |
|
|
|---|
| First author/s,
year | Country | Sample size, n | <65, n ≥65, n | Sex
(male/female) | TNM staging
(I–II/III–IV/unknown) | Outcome index | (Refs.) |
|---|
| Cai et al,
2021 | China | 208 | 89 | 119 | 114/94 | None | 173/35 | 1, 2, 3, 4, 5 | (13) |
| Chen et al,
2022 | China | 92 | 56 | 36 | 47/45 | 48/41/3 | None | 1, 2, 3, 4, 5 | (17) |
| Guo et al,
2023 | China | 478 | None | None | None | None | None | 1, 5 | (20) |
| Li et al,
2021 | China | 459 | 126 | 333 | 216/243 | 253/194/11 | None | 2, 3, 4, 5 | (10) |
| Li et al,
2022 | China | 626 | 373 | 253 | 333/293 | 336/269/20 | None | 1, 5 | (14) |
| Lu et al,
2022 | China | 185 | 107 | 78 | 86/99 | 37/148 | None | 1, 3, 4, 5 | (15) |
| Qiu et al,
2022 | China | 355 | 196 | 159 | 162/193 | 204/151 | None | 1, 2, 3, 4, 5 | (16) |
| Wang et al,
2022 | China | 428 | None | None | None | None | None | 1, 2, 3, 4, 5 | (18) |
| Wu et al,
2021 | China | 170 | 45 | 125 | 94/76 | 111/69/0 | None | 1, 3, 4, 5 | (11) |
| Zhang et al,
2021 | China | 408 | 118 | 290 | 192/216 | 219/175/14 | None | 1, 2, 5 | (12) |
| Li et al,
2023 | China | 405 | 176 | 229 | 188/217 | 212/193 | 350/55 | 1, 2, 3, 4, 5 | (19) |
 | Table II.Literature quality assessment
(Newcastle-Ottawa Scale). |
Table II.
Literature quality assessment
(Newcastle-Ottawa Scale).
| First author/s,
year | Selection (maximum,
4 points) | Comparability
(maximum, 2 points) | Outcome exposure
measure (maximum, 3 points) | Total score
(maximum, 9 points) | (Refs.) |
|---|
| Cai et al,
2021 | 3 | 2 | 2 | 7 | (13) |
| Chen et al,
2022 | 2 | 2 | 2 | 6 | (17) |
| Guo et al,
2023 | 4 | 1 | 3 | 8 | (20) |
| Li et al,
2021 | 3 | 2 | 2 | 7 | (10) |
| Li et al,
2022 | 3 | 2 | 2 | 7 | (14) |
| Lu et al,
2022 | 2 | 1 | 3 | 6 | (15) |
| Qiu et al,
2022 | 3 | 2 | 3 | 8 | (16) |
| Wang et al,
2022 | 3 | 2 | 2 | 8 | (18) |
| Wu et al,
2021 | 2 | 2 | 2 | 6 | (11) |
| Zhang et al,
2021 | 3 | 1 | 3 | 7 | (12) |
| Li et al,
2023 | 4 | 2 | 2 | 8 | (19) |
Meta-analysis results
Tumor stage classification
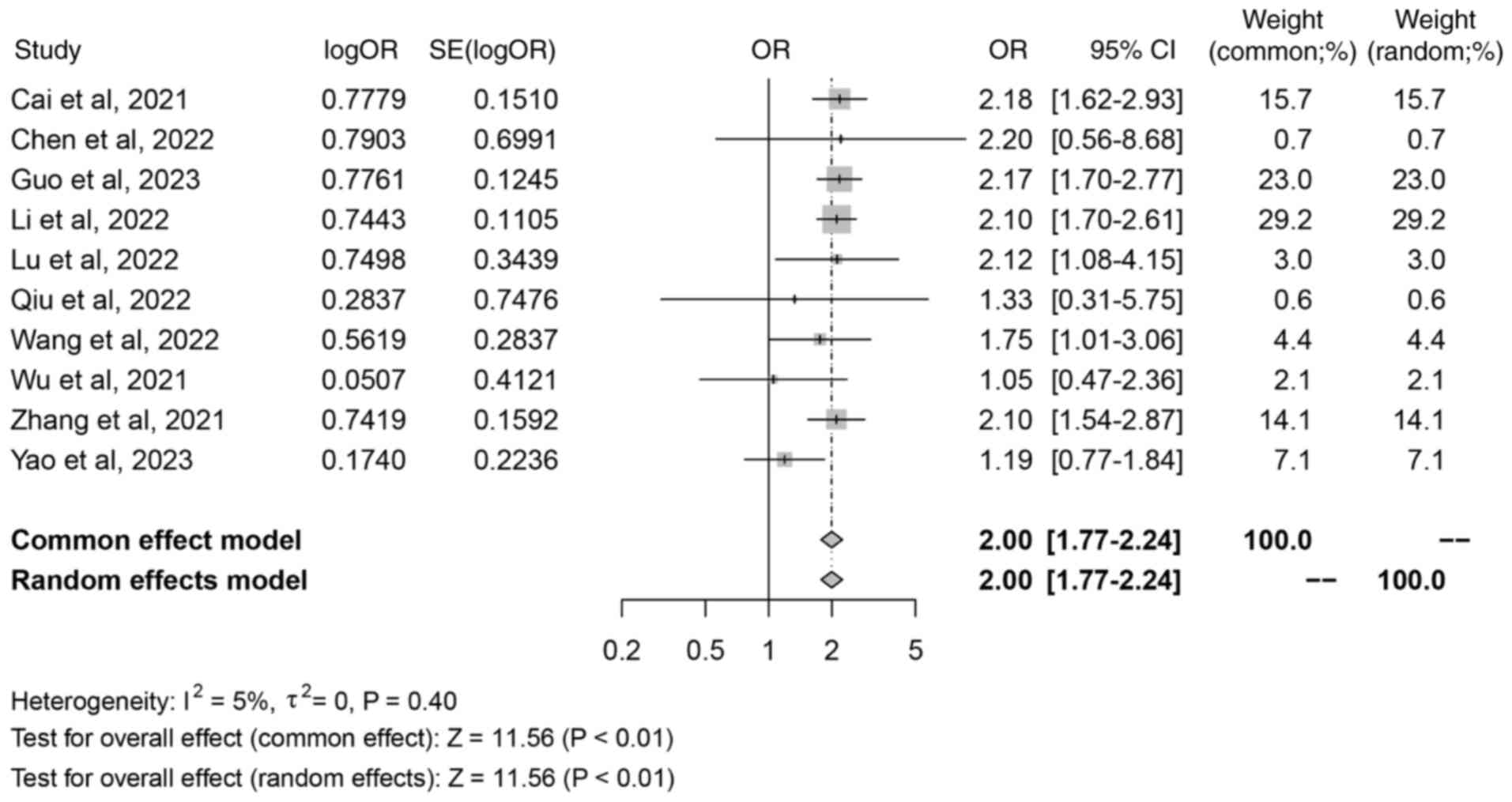
A total of 10 studies were included in the analysis,
all of which assessed the association between iron death-related
lncRNAs and tumor stage classification. There were 5 main outcome
measures in the present study, and if there were >2 outcome
measures included in the literature, the study met the criteria of
the present study. Therefore, certain outcome measures did not
include all included in the literature, so <11 studies were
included in this result. The meta-analysis demonstrated a no
significant heterogeneity (I2=5%; P=0.40) and was
analyzed using a fixed-effects model. The random effects model
analysis revealed a combined odds ratio (OR) of 2.00 (95% CI,
1.77–2.24; Z=11.56; P<0.01; Fig.
2), indicating a significant association between
ferroptosis-related lncRNAs and tumor stage prediction.
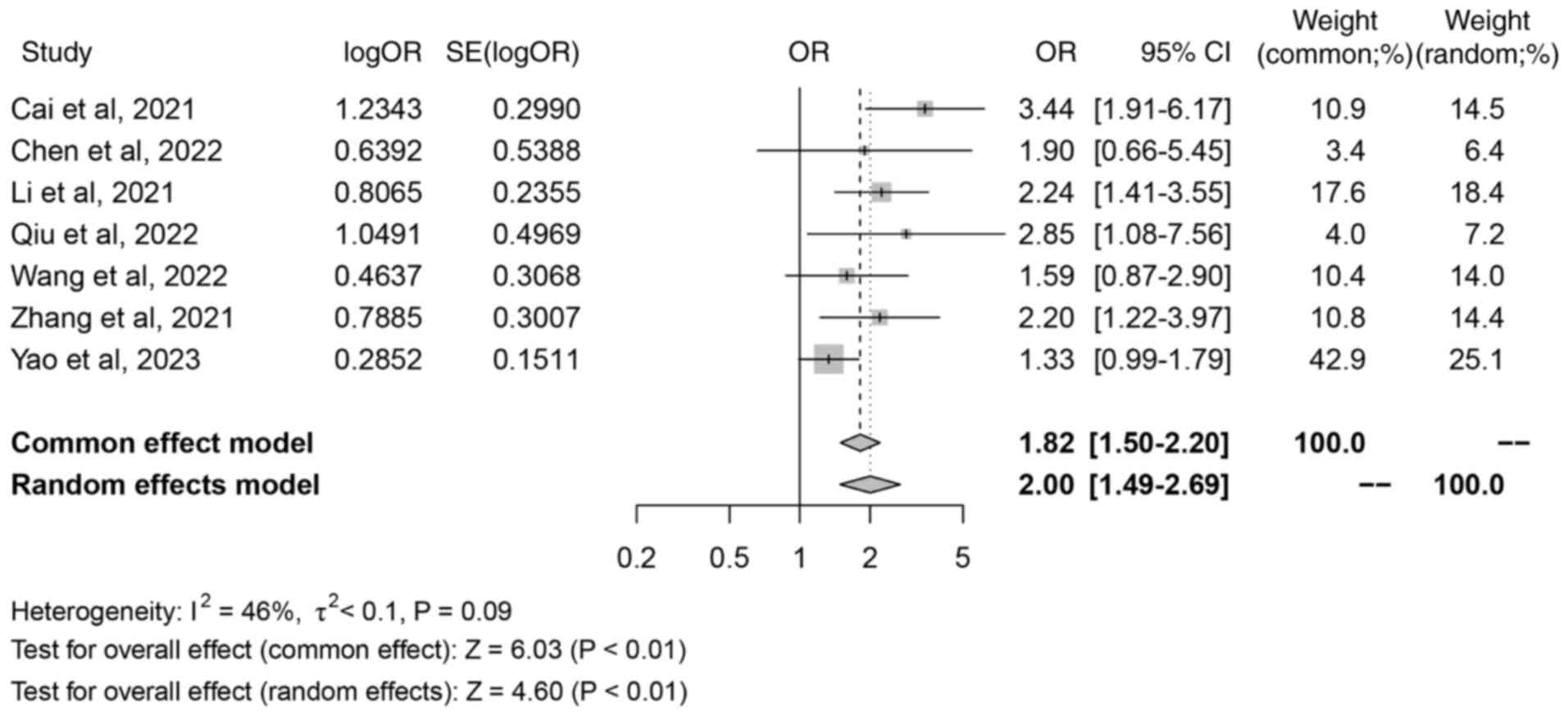
T stage
A total of 7 studies were included in the analysis
of the relationship between iron death-related lncRNAs and the T
stage. There were 5 main outcome measures in the present study, and
if there were >2 outcome measures included in the literature,
the study met the criteria of the present study. Therefore, certain
outcome measures did not include all included in the literature, so
<11 studies were included in this result. The meta-analysis of
the combined results showed that the included literature presented
moderate, but not significant, heterogeneity (I2=46%,
P=0.09), and a fixed-effect model was used for analysis. The random
effects model analysis demonstrated a combined OR of 2.00 (95% CI,
1.49–2.69;, Z=4.60; P<0.01; Fig.
3), which indicated that there was a significant association
between iron death-related lncRNAs and T stage.
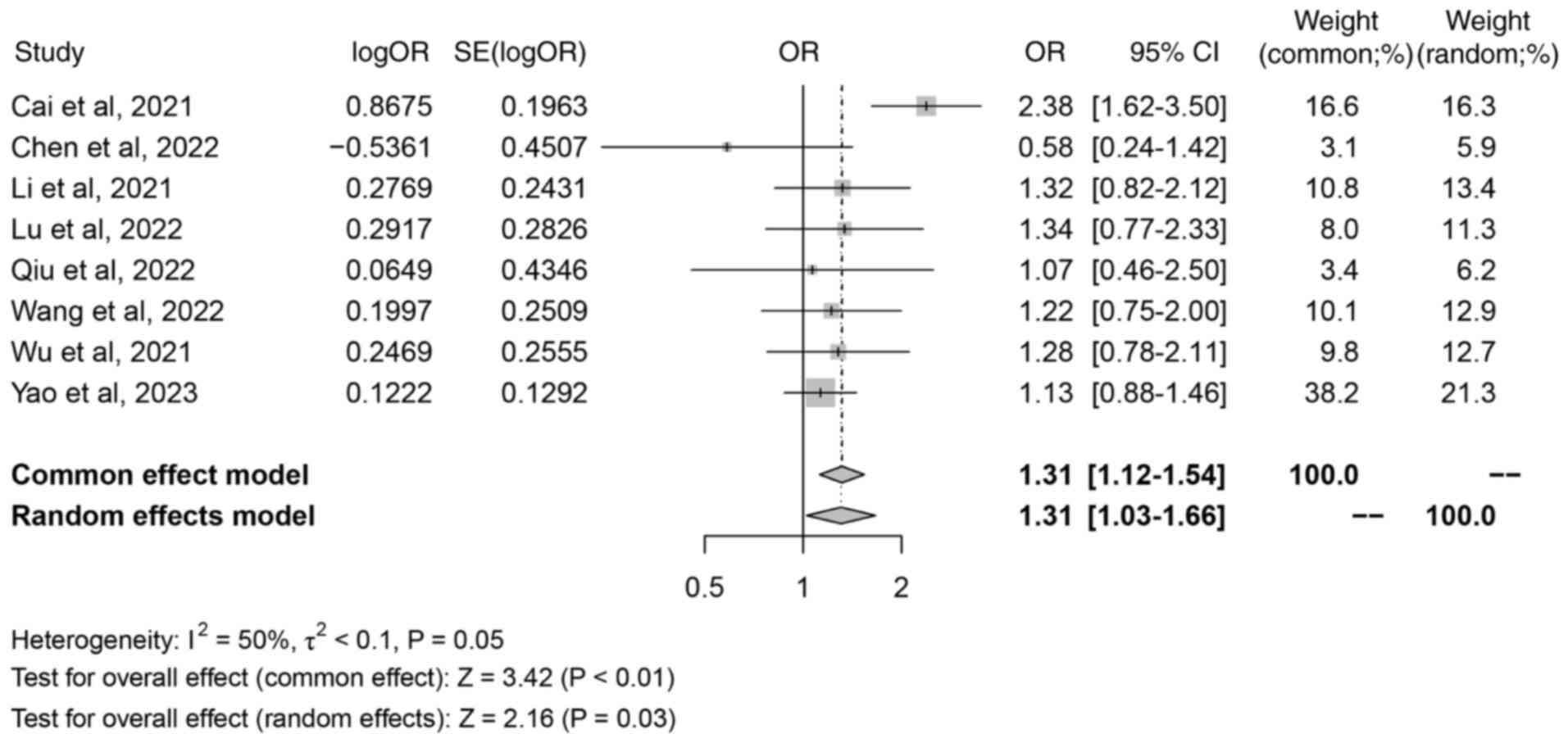
Lymphatic metastasis of colorectal
cancer
A total of 8 studies reported the association
between iron death-related lncRNAs and lymphatic metastasis of
colorectal cancer. There were 5 main outcome measures in the
present study, and if there were >2 outcome measures included in
the literature, the study met the criteria of the present study.
Therefore, certain outcome measures did not include all included in
the literature, so <11 studies were included in this result. The
meta-analysis showed moderate heterogeneity in the included
literature (I2=50%; P=0.05), which was analyzed by a
random effect model. The random effects model analysis revealed the
combined OR was 1.31 (95% CI, 1.03–1.66; Z=2.16; P=0.03; Fig. 4), which indicated that there was a
significant association between iron death-related lncRNAs and
lymph node metastasis of colorectal cancer.
Distant metastasis of colorectal
cancer
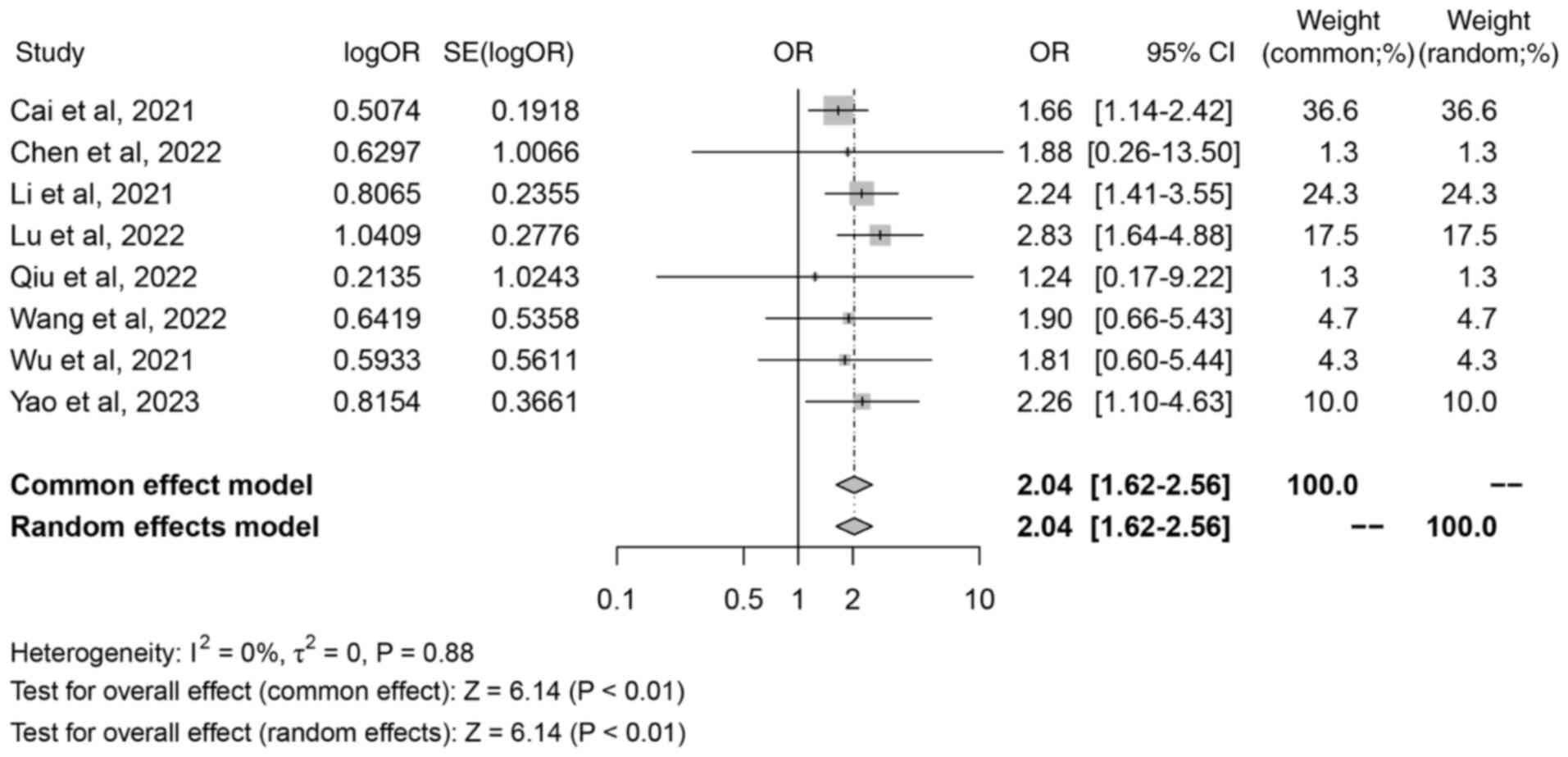
A total of 8 studies reported the association
between iron death-related lncRNAs and distant metastasis. There
were 5 main outcome measures in the present study, and if there
were >2 outcome measures included in the literature, the study
met the criteria of the present study. Therefore, certain outcome
measures did not include all included in the literature, so <11
studies were included in this result. The meta-analysis revealed no
significant heterogeneity (I2=0%; P=0.88). The random
effects model was selected to interpret the results regardless of
heterogeneity, and the results were consistent between the random
and fixed effects models. The random effects model analysis
demonstrated a combined OR of 2.04 (95% CI, 1.62–2.56; Z=6.14;
P<0.01; Fig. 5), which indicated
that there was a significant association between iron death-related
lncRNAs and distant metastasis of colorectal cancer.
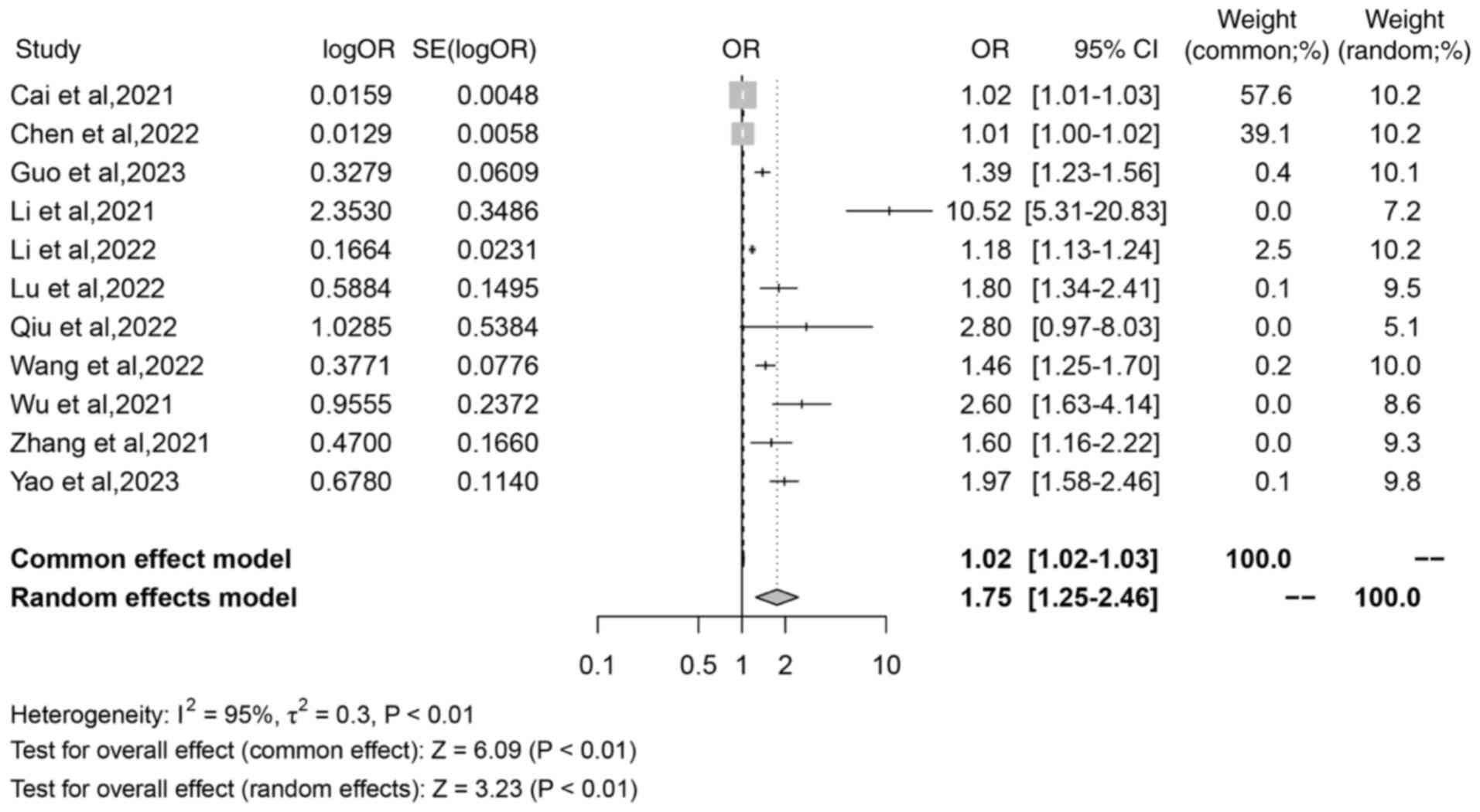
Colorectal cancer risk score
A total of 11 studies reported the association
between iron death-related lncRNAs and distant metastasis. The
meta-analysis revealed significant and considerable heterogeneity
(I2=95%; P<0.01), which was assessed using a random
effect model. The random effects model analysis demonstrated a
combined OR of 1.75 (95% CI, 1.25–2.46; Z=3.23; P<0.01; Fig. 6), which indicated that there was a
significant association between iron death-related lncRNAs and
colorectal cancer risk score.
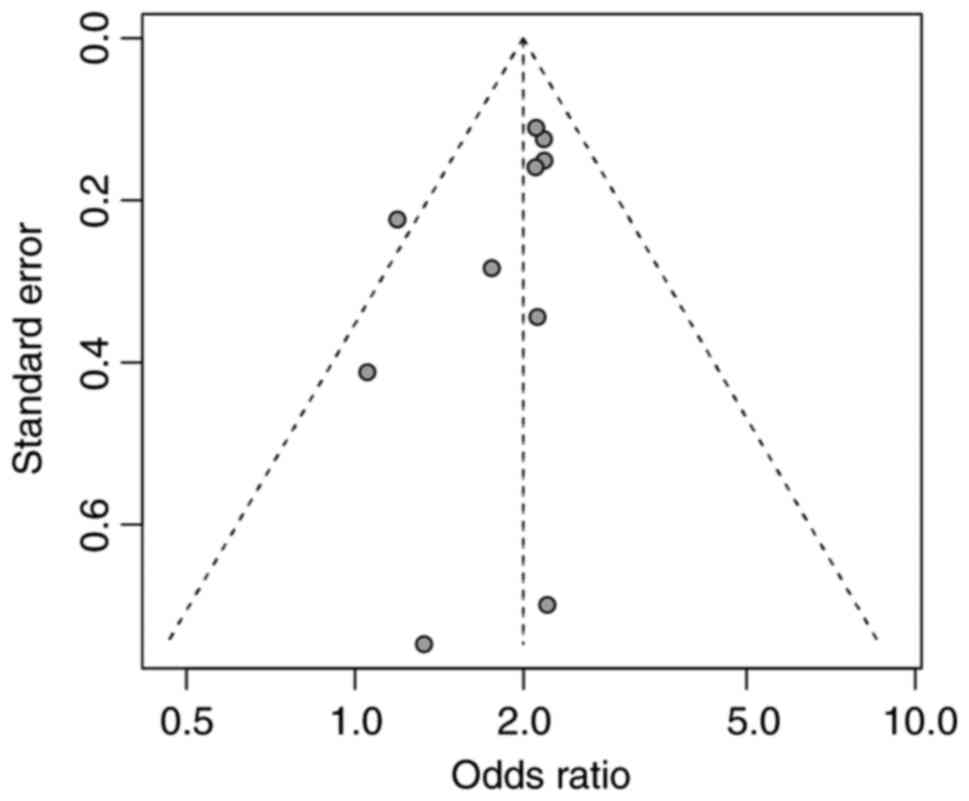
Publication bias
To assess the possible publication bias that may
exist in the research, Begg's and Egger's tests were performed. The
funnel plot results demonstrated that although there was one study
outside the funnel, there was good symmetry of the funnel plot
(Fig. 7) The results of Begg's test
(z=−2.06; P=0.04) and Egger's test (t=−1.71; P=0.12) indicate a
degree of publication bias among the studies included.
Sensitivity analysis
A sensitivity analysis, excluding each study
systematically, affirmed the stability and reliability of the
results of the meta-analyses. The direction and significance of the
overall effect remained unchanged irrespective of the study
removed, indicating robustness in the findings (data not
shown).
Discussion
With the rapid development of molecular biology and
transcriptomics technologies, lncRNAs have become a new focus in
cancer research (22–24). In colorectal cancer, lncRNAs not
only serve a role in cancer cell proliferation, metastasis and
invasion, but they are also associated with patient prognosis
(25). The present meta-analysis
assessed iron death-related lncRNAs within colorectal cancer,
shedding light on their potential clinical utility.
The results of the present study demonstrated a
significant association between the overexpression of iron
death-related lncRNAs and several clinical characteristics of
colorectal cancer, such as tumor stage, T stage, lymph node
metastasis, distant metastasis and risk scoring. These findings
highlight the central role of lncRNAs in the progression of
colorectal cancer and support the viewpoint that they serve as
important biomarkers. Particularly, given the paramount importance
of lymph node and distant metastases in prognostic evaluations, the
expression levels of iron death-related lncRNAs emerge as potent
predictors for these conditions (11,12).
Furthermore, although the present meta-analysis
results demonstrated a significant association between iron
death-related lncRNAs and colorectal cancer prognosis, the specific
biological mechanisms involved remain unclear. It is possible that
they influence the growth and metastasis of colorectal cancer by
regulating certain key genes or signaling pathways (26,27).
Future experimental studies should investigate their mechanisms of
action, which will provide insights into the development of novel
therapeutic strategies for colorectal cancer. Moreover, it is worth
noting that although the present study found a degree of
publication bias in the review, the overall quality of these trials
was acceptable. In addition, the present study assessed the
stability of the meta-analysis results through sensitivity
analysis, further strengthening the conclusions. Finally, multiple
lncRNAs associated with iron death were included, such as lncRNA
associated with poor prognosis of hepatocellular carcinoma (AWPPH),
AC004687.1, AC010973.2, AP001189.3, LINC01503, NCK1 Divergent
Transcript (NCK1−DT), GFL2-AS1, LOXL1 antisense RNA 1 (LOXL1-AS1),
Taurine Upregulated Gene 1 (TUG1) and the XIRP2-AS1 MIR31 Host Gene
(MIR31HG), all of which are upregulated lncRNAs. However, subgroup
analyses of lncRNAs were not performed, as the amount of data
available in the literature was limited, as was the analysis of the
prognostic impact of different types of lncRNAs on colon
cancer.
In conclusion, the present meta-analysis
demonstrates the role of iron death-related lncRNAs as innovative
prognostic biomarkers in colorectal cancer. Nevertheless, there is
a need for further prospective studies and mechanistic explorations
to corroborate the findings of the present study and pioneer new
diagnostic and therapeutic strategies for this malignancy.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XLH and JS made substantial contributions in
conceiving and drafting the manuscript. XLH and ZXD made
substantial contributions to acquisition of data. XLH and JS made
substantial contributions to the analysis and interpretation of
data. XLH and JS confirm the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xu J, Zeng W, Liu T, Wan Z, Yang X, Chen J
and Liu F: lncRNA TINCR knockdown inhibits colon cancer cells via
regulation of autophagy. Food Sci Nutr. 11:1965–1981. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu Y, Xu Z, Ni H, Jin M and Dai C:
Clinicopathological and prognostic value of long non-coding RNA
CCAT1 expression in patients with digestive system cancer. Oncol
Lett. 25:732023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qu A, Wang Q, Chang Q, Liu J, Yang Y,
Zhang X and Zhang Y, Zhang X, Wang H and Zhang Y: Prognostic and
predictive value of a lncRNA signature in patients with stage II
colon cancer. Sci Rep. 13:13502023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liang GZ, Wen XF, Song YW, Zhang ZJ, Chen
J, Chen YL, Pan WD, He XW, Hu T and Xian ZY: Construction and
validation of a novel prognosis model in colon cancer based on
cuproptosis-related long non-coding RNAs. J Clin Med. 12:15282023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liau XL, Salvamani S, Gunasekaran B,
Chellappan DK, Rhodes A, Ulaganathan V and Tiong YL: CCAT 1-a
pivotal oncogenic long non-coding RNA in colorectal cancer. Br J
Biomed Sci. 80:111032023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu S, Zhang S, Liu Y, Yang X and Zheng G:
Comprehensive analysis of cuproptosis-related long noncoding RNA
for predicting prognostic and diagnostic value and immune landscape
in colorectal adenocarcinoma. Hum Genomics. 17:222023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sepehri Z, Banerjee A, Vizeacoumar FS,
Freywald A, Vizeacoumar FJ, Dolinsky VW and Davie JR: Differential
expression of HNF1A and HNF1A-AS1 in colon cancer cells. IUBMB
Life. 74:496–507. 2022. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo J, Peng J, Xiao W, Huang S, Cao Y,
Wang T and Wang X: A novel necroptosis-related lncRNA signature for
predicting prognosis and immune response of colon cancer. Front
Genet. 13:9846962022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo Y, Huang S, Wei J, Zhou H, Wang W,
Yang J, Deng Q, Wang H and Fu Z: Long noncoding RNA LINC01606
protects colon cancer cells from ferroptotic cell death and
promotes stemness by SCD1-Wnt/β-catenin-TFE3 feedback loop
signalling. Clin Transl Med. 12:e7522022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li H, Liu L, Huang T, Jin M, Zheng Z,
Zhang H, Ye M and Liu K: Establishment of a novel
ferroptosis-related lncRNA pair prognostic model in colon
adenocarcinoma. Aging (Albany NY). 13:23072–23095. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu Z, Lu Z, Li L, Ma M, Long F, Wu R,
Huang L, Chou J, Yang K, Zhang Y, et al: Identification and
validation of ferroptosis-related LncRNA signatures as a novel
prognostic model for colon cancer. Front Immunol. 12:7833622021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang W, Fang D, Li S, Bao X, Jiang L and
Sun X: Construction and validation of a novel ferroptosis-related
lncRNA signature to predict prognosis in colorectal cancer
patients. Front Genet. 12:7093292021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cai HJ, Zhuang ZC, Wu Y, Zhang YY, Liu X,
Zhuang JF, Yang YF, Gao Y, Chen B and Guan GX: Development and
validation of a ferroptosis-related lncRNAs prognosis signature in
colon cancer. Bosn J Basic Med Sci. 21:569–576. 2021.PubMed/NCBI
|
|
14
|
Li N, Shen J, Qiao X, Gao Y, Su HB and
Zhang S: Long non-coding RNA signatures associated with ferroptosis
predict prognosis in colorectal cancer. Int J Gen Med. 15:33–43.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu J, Tan J and Yu X: A prognostic
ferroptosis-related lncRNA model associated with immune
infiltration in colon cancer. Front Genet. 13:9341962022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qiu Y, Li H, Zhang Q, Qiao X and Wu J:
Ferroptosis-related long noncoding RNAs as prognostic marker for
colon adenocarcinoma. Appl Bionics Biomech. 2022:52203682022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen W, Chen Y, Liu L, Wu Y, Fu P, Cao Y,
Xiong J, Tu Y, Li Z, Liu Y and Jie Z: Comprehensive analysis of
immune infiltrates of ferroptosis-related long noncoding RNA and
prediction of colon cancer patient prognoses. J Immunol Res.
2022:94806282022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Y, Zhang Y, Xu W and Cui X:
Construction and clinical application of colon cancer prognostic
risk model of ferroptosis-related lncRNA. Laboratory Med.
37:720–728. 2022.(In Chinese).
|
|
19
|
Li Y, Huan S and Xiang L: Construction and
validation of colorectal cancer prognosis risk model based on iron
death-related lncRNA. Zhejiang J Clin Med J. 25:167–171. 2023.(In
Chinese).
|
|
20
|
Guo S, Song B, Li L, Li H, Yang T, Cao L
and Wang J: lncRNA HCG11 promotes colorectal cancer cell malignant
behaviors via sponging mir-26b-5p. J Immunol Res. 2023:90112322023.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stang A: Critical evaluation of the
Newcastle-Ottawa scale for the assessment of the quality of
nonrandomized studies in meta-analyses. Eur J Epidemiol.
25:603–605. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hosseini FA, Rejali L, Zabihi MR, Salehi
Z, Daskar-Abkenar E, Taraz T, Fatemi N, Hashemi M,
Asadzadeh-Aghdaei H and Nazemalhosseini-Mojarad E: Long non-coding
RNA LINC00460 contributes as a potential prognostic biomarker
through its oncogenic role with ANXA2 in colorectal polyps. Mol
Biol Rep. 50:4505–4515. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao C, Gan C, Xiao Y, Liu R, Zhang L, Lan
T, Ye Y, Tong H, Huang Z, Tang C and Gao J: High expression of long
non-coding RNA Linc-A associates with poor survival in patients
with colorectal cancer. Mol Biol Rep. 47:7497–7504. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsai KW, Lo YH, Liu H, Yeh CY, Chen YZ,
Hsu CW, Chen WS and Wang JH: Linc00659, a long noncoding RNA, acts
as novel oncogene in regulating cancer cell growth in colorectal
cancer. Mol Cancer. 17:722018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gaballah HH, Gaber RA, Elrashidy MA,
Elshahat DA, Hablus MA and Ebeid AM: Expression of long non-coding
RNA CCHE1 in colorectal carcinoma: correlations with
clinicopathological features and ERK/COX-2 pathway. Mol Biol Rep.
46:657–667. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiong Z, Li W, Luo X, Lin Y, Huang W and
Zhang S: Seven bacterial response-related genes are biomarkers for
colon cancer. BMC Bioinformatics. 24:1032023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shaalan AAM, Mokhtar SH, Ahmedah HT,
Almars AI, Toraih EA, Ibrahiem AT, Fawzy MS and Salem MA:
Prognostic value of LINC-ROR (rs1942347) variant in patients with
colon cancer harboring BRAF mutation: A propensity score-matched
analysis. Biomolecules. 12:5692022. View Article : Google Scholar : PubMed/NCBI
|