|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ychou M, Boige V, Pignon JP, Conroy T,
Bouché O, Lebreton G, Ducourtieux M, Bedenne L, Fabre JM,
Saint-Aubert B, et al: Perioperative chemotherapy compared with
surgery alone for resectable gastroesophageal adenocarcinoma: An
FNCLCC and FFCD multicenter phase III trial. J Clin Oncol.
29:1715–1721. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Longley DB, Harkin DP and Johnston PG:
5-Fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang N, Yin Y, Xu SJ and Chen WS:
5-Fluorouracil: Mechanisms of resistance and reversal strategies.
Molecules. 13:1551–1569. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sethy C and Kundu CN: 5-Fluorouracil
(5-FU) resistance and the new strategy to enhance the sensitivity
against cancer: Implication of DNA repair inhibition. Biomed
Pharmacother. 137:1112852021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reyes ME, de La Fuente M, Hermoso M, Ili
CG and Brebi P: Role of CC chemokines subfamily in the platinum
drugs resistance promotion in cancer. Front Immunol. 11:9012020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zajkowska M and Mroczko B: Eotaxins and
their receptor in colorectal cancer-a literature review. Cancers
(Basel). 12:13832020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mora-Lagos B, Cartas-Espinel I, Riquelme
I, Parker AC, Piccolo SR, Viscarra T, Reyes ME, Zanella L,
Buchegger K, Ili C and Brebi P: Functional and transcriptomic
characterization of cisplatin-resistant AGS and MKN-28 gastric
cancer cell lines. PLoS One. 15:e02283312020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jöhrer K, Zelle-Rieser C, Perathoner A,
Moser P, Hager M, Ramoner R, Gander H, Höltl L, Bartsch G, Greil R
and Thurnher M: Up-regulation of functional chemokine receptor CCR3
in human renal cell carcinoma. Clin Cancer Res. 11:2459–2465. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee YS, Kim SY, Song SJ, Hong HK, Lee Y,
Oh BY, Lee WY and Cho YB: Crosstalk between CCL7 and CCR3 promotes
metastasis of colon cancer cells via ERK-JNK signaling pathways.
Oncotarget. 7:36842–36853. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang WY, Lin YS, Lin YC, Nieh S, Chang
YM, Lee TY, Chen SF and Yang KD: Cancer-associated fibroblasts
promote tumor aggressiveness in head and neck cancer through
chemokine ligand 11 and C-C motif chemokine receptor 3 signaling
circuit. Cancers (Basel). 14:31412022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Andalib A, Doulabi H, Hasheminia S, Maracy
M and Rezaei A: CCR3, CCR4, CCR5, and CXCR3 expression in
peripheral blood CD4+ lymphocytes in gastric cancer patients. Adv
Biomed Res. 2:312013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
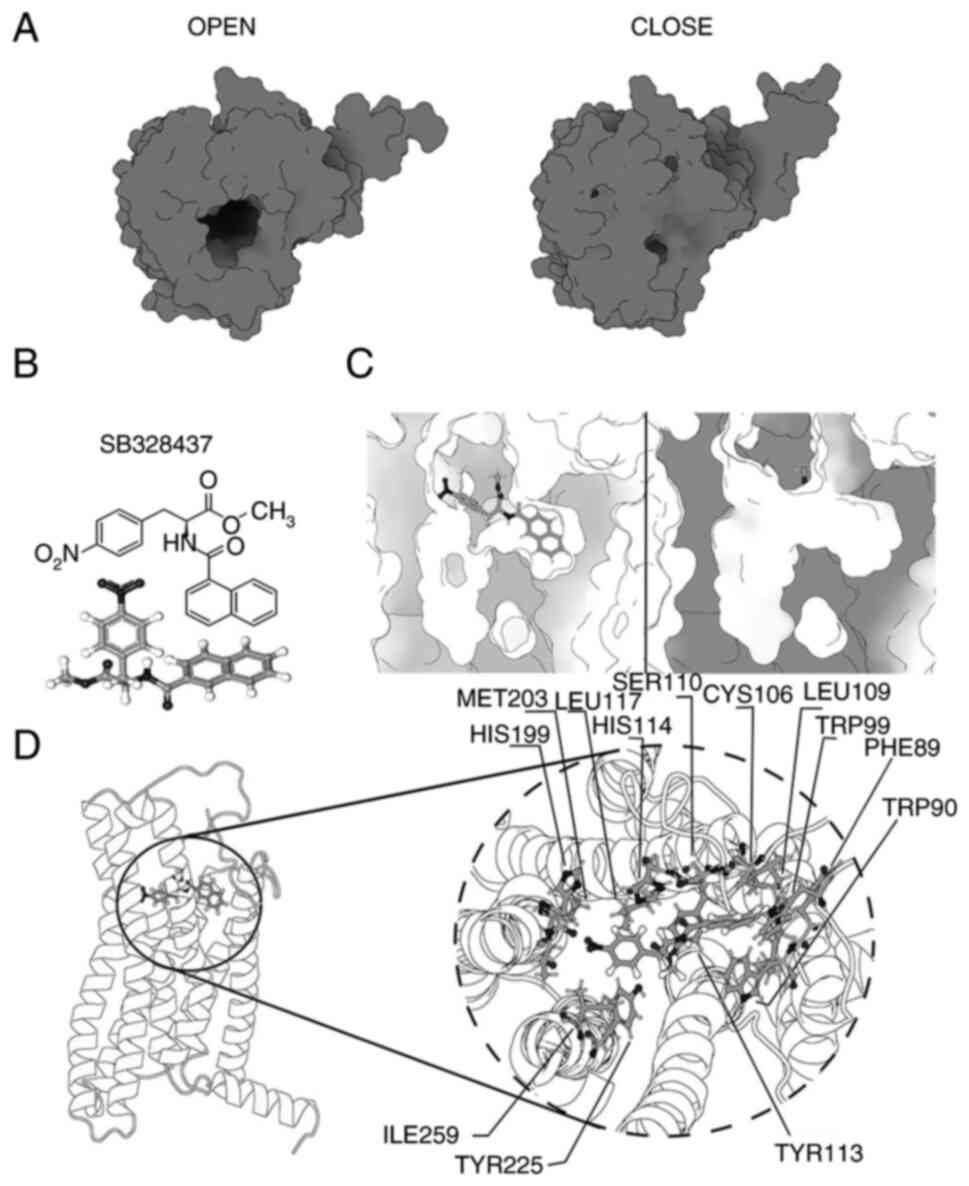
Filippone RT, Dargahi N, Eri R, Uranga JA,
Bornstein JC, Apostolopoulos V and Nurgali K: Potent CCR3 receptor
antagonist, SB328437, suppresses colonic eosinophil chemotaxis and
inflammation in the winnie murine model of spontaneous chronic
colitis. Int J Mol Sci. 23:77802022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang C, Wang Y, Hong T, Cheng B, Gan S,
Chen L, Zhang J, Zuo L, Li J and Cui X: Blocking the autocrine
regulatory loop of Gankyrin/STAT3/CCL24/CCR3 impairs the
progression and pazopanib resistance of clear cell renal cell
carcinoma. Cell Death Dis. 11:1172020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shao Z, Tan Y, Shen Q, Hou L, Yao B, Qin
J, Xu P, Mao C, Chen LN, Zhang H, et al: Molecular insights into
ligand recognition and activation of chemokine receptors CCR2 and
CCR3. Cell Discov. 8:442022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
White JR, Lee JM, Dede K, Imburgia CS,
Jurewicz AJ, Chan G, Fornwald JA, Dhanak D, Christmann LT, Darcy
MG, et al: Identification of potent, selective non-peptide CC
chemokine receptor-3 antagonist that inhibits eotaxin-, eotaxin-2-,
and monocyte chemotactic protein-4-induced eosinophil migration. J
Biol Chem. 275:36626–36631. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Coley HM: Development of drug-resistant
models. Methods Mol Med. 88:267–274. 2004.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Berman HM, Westbrook J, Feng Z, Gilliland
G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE: The protein
data bank. Nucleic Acids Res. 28:235–242. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jo S, Kim T, Iyer VG and Im W: CHARMM-GUI:
A web-based graphical user interface for CHARMM. J Comput Chem.
29:1859–1865. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brooks BR, Brooks CL III, Mackerell AD Jr,
Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C,
Boresch S, et al: CHARMM: The biomolecular simulation program. J
Comput Chem. 30:1545–1614. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu EL, Cheng X, Jo S, Rui H, Song KC,
Dávila-Contreras EM, Qi Y, Lee J, Monje-Galvan V, Venable RM, et
al: CHARMM-GUI membrane builder toward realistic biological
membrane simulations. J Comput Chem. 35:1997–2004. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Morris GM, Huey R, Lindstrom W, Sanner MF,
Belew RK, Goodsell DS and Olson AJ: AutoDock4 and AutoDockTools4:
Automated docking with selective receptor flexibility. J Comput
Chem. 30:2785–2791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tan Q, Zhu Y, Li J, Chen Z, Han GW,
Kufareva I, Li T, Ma L, Fenalti G, Li J, et al: Structure of the
CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex.
Science. 341:1387–1390. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Berendsen HJC, van der Spoel D and van
Drunen R: GROMACS: A message-passing parallel molecular dynamics
implementation. Comput Phys Commun. 91:43–56. 1995. View Article : Google Scholar
|
|
26
|
Amadei A, Linssen AB and Berendsen HJ:
Essential dynamics of proteins. Proteins. 17:412–425. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tomasello G, Armenia I and Molla G: The
protein imager: A full-featured online molecular viewer interface
with server-side HQ-rendering capabilities. Bioinformatics.
36:2909–2911. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu ZY, Tang JN, Xie HX, Du YA, Huang L, Yu
PF and Cheng XD: 5-Fluorouracil chemotherapy of gastric cancer
generates residual cells with properties of cancer stem cells. Int
J Biol Sci. 11:284–294. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hembruff SL and Cheng N: Chemokine
signaling in cancer: Implications on the tumor microenvironment and
therapeutic targeting. Cancer Ther. 7:254–267. 2009.PubMed/NCBI
|
|
30
|
Oppermann M: Chemokine receptor CCR5:
Insights into structure, function, and regulation. Cell Signal.
16:1201–1210. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Salmas RE, Yurtsever M and Durdagi S:
Investigation of inhibition mechanism of chemokine receptor CCR5 by
micro-second molecular dynamics simulations. Sci Rep. 5:131802015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wasilko DJ, Johnson ZL, Ammirati M, Che Y,
Griffor MC, Han S and Wu H: Structural basis for chemokine receptor
CCR6 activation by the endogenous protein ligand CCL20. Nat Commun.
11:30312020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Duchesnes CE, Murphy PM, Williams TJ and
Pease JE: Alanine scanning mutagenesis of the chemokine receptor
CCR3 reveals distinct extracellular residues involved in
recognition of the eotaxin family of chemokines. Mol Immunol.
43:1221–2131. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
van Aalst EJ, McDonald CJ and Wylie BJ:
Cholesterol biases the conformational landscape of the chemokine
receptor CCR3: A MAS SSNMR-filtered molecular dynamics study. J
Chem Inf Model. 63:3068–3085. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Malgija MB, Rajendran HAD, Darvin SS,
Nachimuthu S and Priyakumari J: In silico exploration of HIV entry
co-receptor antagonists: A combination of molecular modeling,
docking and molecular dynamics simulations. Acta Sci Pharm Sci.
3:60–67. 2019.
|
|
36
|
Manna M, Niemelä M, Tynkkynen J,
Javanainen M, Kulig W, Müller DJ, Rog T and Vattulainen I:
Mechanism of allosteric regulation of β2-adrenergic
receptor by cholesterol. Elife. 5:e184322016. View Article : Google Scholar : PubMed/NCBI
|