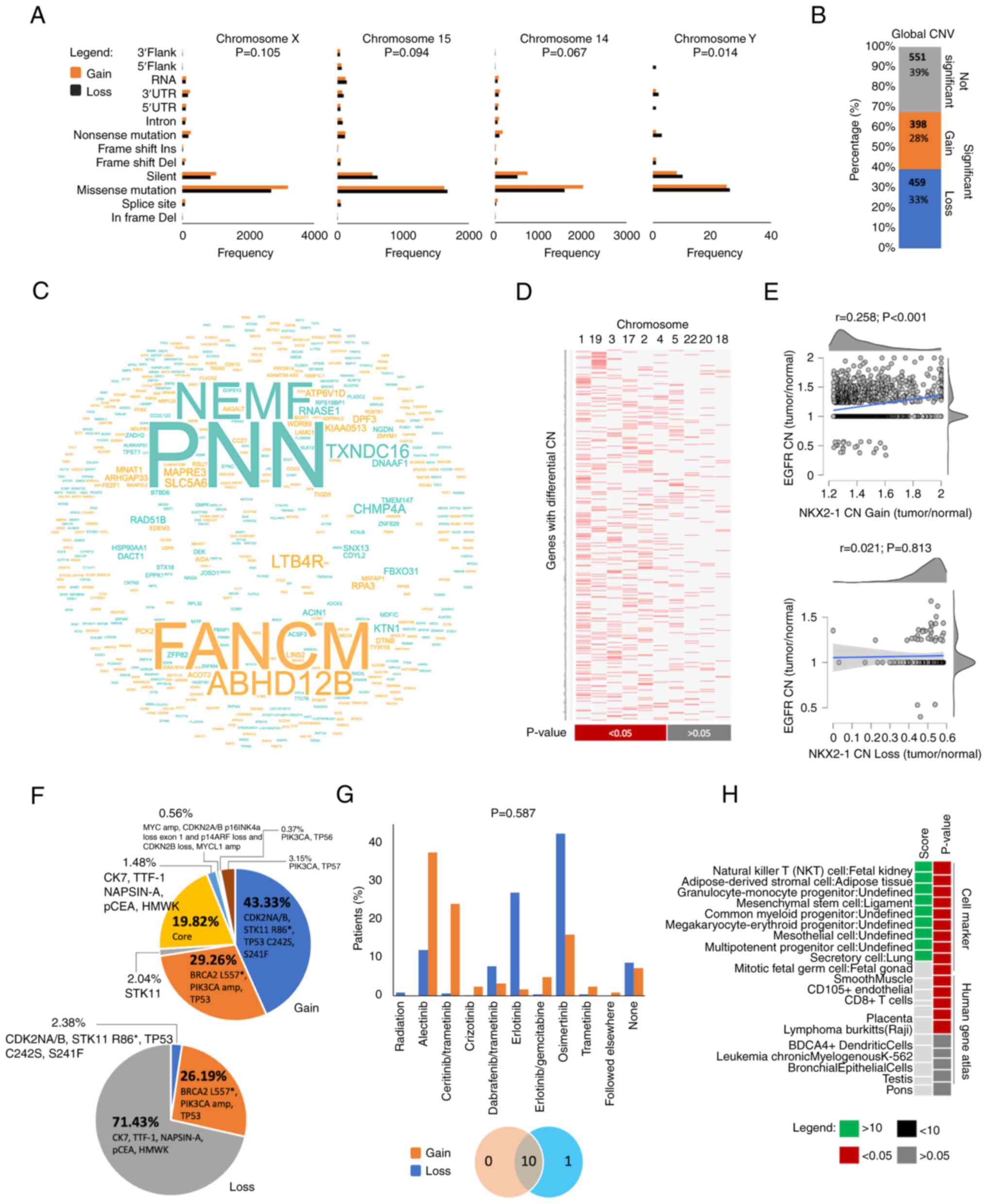
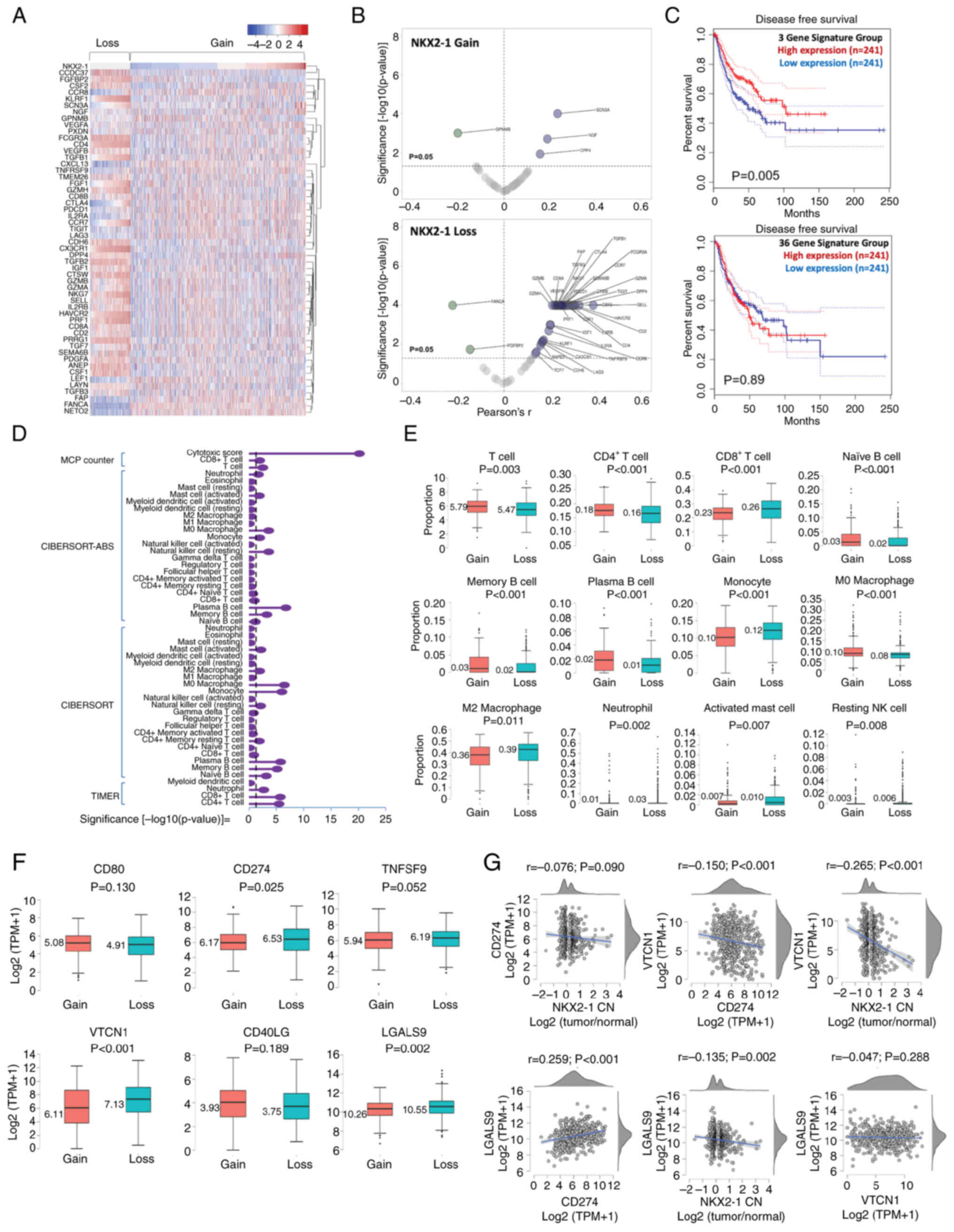
|
1
|
Feuk L, Carson AR and Scherer SW:
Structural variation in the human genome. Nat Rev Genet. 7:85–97.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Steele CD, Abbasi A, Islam SMA, Bowes AL,
Khandekar A, Haase K, Hames-Fathi S, Ajayi D, Verfaillie A, Dhami
P, et al: Signatures of copy number alterations in human cancer.
Nature. 606:984–991. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bjaanæs MM, Nilsen G, Halvorsen AR,
Russnes HG, Solberg S, Jørgensen L, Brustugun OT, Lingjærde OC and
Helland Å: Whole genome copy number analyses reveal a highly
aberrant genome in TP53 mutant lung adenocarcinoma tumors. BMC
Cancer. 21:10892021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shao X, Lv N, Liao J, Long J, Xue R, Ai N,
Xu D and Fan X: Copy number variation is highly correlated with
differential gene expression: A pan-cancer study. BMC Med Genet.
20:1752019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Heo Y, Heo J, Han SS, Kim WJ, Cheong HS
and Hong Y: Difference of copy number variation in blood of
patients with lung cancer. Int J Biol Markers. 36:3–9. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qiu ZW, Bi JH, Gazdar AF and Song K:
Genome-wide copy number variation pattern analysis and a
classification signature for non-small cell lung cancer. Genes
Chromosomes Cancer. 56:559–569. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen S, Lu L, Xian J, Shi C, Chen J, Rao
B, Qiu F, Lu J and Yang L: Prognostic value of germline copy number
variants and environmental exposures in Non-small cell lung cancer.
Front Genet. 12:681852021.
|
|
8
|
Aujla S, Aloe C, Vannitamby A, Hendry S,
Rangamuwa K, Wang H, Vlahos R, Selemidis S, Leong T, Steinfort D
and Bozinovski S: Programmed Death-Ligand 1 Copy number loss in
NSCLC associates with reduced programmed Death-Ligand 1 tumor
staining and a cold immunophenotype. J Thorac Oncol. 17:675–687.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alden S, Ricciuti B, Spurr L, Gupta H,
Lamberti G, Li Y, Sholl L, Cherniack A and Awad A: P33.04
programmed death-ligand 1 (PD-L1) changes in non-small-cell lung
cancer (NSCLC): Clinical, pathologic, and genomic correlates. J
Thorac Oncol. 16:S406–S407. 2021. View Article : Google Scholar
|
|
10
|
Inoue Y, Yoshimura K, Mori K, Kurabe N,
Kahyo T, Mori H, Kawase A, Tanahashi M, Ogawa H, Inui N, et al:
Clinical significance of PD-L1 and PD-L2 copy number gains in
non-small-cell lung cancer. Oncotarget. 7:32113–32128. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei J, Meng P, Terpstra MM, van Rijk A,
Tamminga M, Scherpen F, Ter Elst A, Alimohamed MZ, Johansson LF,
Stigt J, et al: Clinical Value of EGFR Copy number gain determined
by amplicon-based targeted next generation sequencing in patients
with EGFR-Mutated NSCLC. Target Oncol. 16:215–226. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peng D, Liang P, Zhong C, Xu P, He Y, Luo
Y, Wang X, Liu A and Zeng Z: Effect of EGFR amplification on the
prognosis of EGFR-mutated advanced non-small-cell lung cancer
patients: A prospective observational study. BMC Cancer.
22:13232022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gloriane C Luna H, Severino Imasa M, Juat
N, Hernandez KV, May Sayo T, Cristal-Luna G, Marie Asur-Galang S,
Bellengan M, John Duga K, Brian Buenaobra B, et al: Expression
landscapes in non-small cell lung cancer shaped by the thyroid
transcription factor 1. Lung Cancer. 176:121–131. 2023. View Article : Google Scholar
|
|
14
|
Yoshimura K, Inoue Y, Mori K, Iwashita Y,
Kahyo T, Kawase A, Tanahashi M, Ogawa H, Inui N, Funai K, et al:
Distinct prognostic roles and heterogeneity of TTF1 copy number and
TTF1 protein expression in non-small cell lung cancer. Genes
Chromosomes Cancer. 56:570–581. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Wan L, Shen H, Geng J, Nie J, Wang
G, Jia N, Dai M and Bai X: Thyroid transcription Factor-1
amplification and expressions in lung adenocarcinoma tissues and
pleural effusions predict patient survival and prognosis. J Thorac
Oncol. 7:76–84. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Clarke N, Biscocho J, Kwei KA, Davidson
JM, Sridhar S, Gong X and Pollack JR: Integrative genomics
implicates EGFR as a downstream mediator in NKX2-1 amplified
non-small cell lung cancer. PLoS One. 10:e01420612015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Luna HG, Prieto E, Dimayacyac-Esleta BR,
Imasa MS, Juat N, Hernandez KV, Sayo TM, Cristal-Luna GR,
Asur-Galang SM, Bellengan M, et al: 342P Prognostic implications of
PD-L1 co-expression among Filipino EGFR MT mNSCLC. Ann Oncol. 33
(Suppl):S15762022. View Article : Google Scholar
|
|
18
|
Luna HGC, Imasa MS, Juat N, Hernandez KV,
Sayo TM, Cristal-Luna G, Asur-Galang SM, Bellengan M and Duga KJ:
The differential prognostic implications of PD-L1 expression in the
outcomes of Filipinos with EGFR-mutant NSCLC treated with tyrosine
kinase inhibitors. Transl Lung Cancer Res. 12:1896–1911. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cancer Genome Atlas Research Network, .
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Maynard A, McCoach CE, Rotow JK, Harris L,
Haderk F, Kerr DL, Yu EA, Schenk EL, Tan W, Zee A, et al:
Therapy-Induced evolution of human lung cancer revealed by
single-cell RNA sequencing. Cell. 182:1232–1251.e22. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Grossman RL, Heath AP, Ferretti V, Varmus
HE, Lowy DR, Kibbe WA and Staudt LM: Toward a shared vision for
cancer genomic data. N Engl J Med. 375:1109–1112. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Luna HGC, Imasa MS, Juat N, Hernandez V,
Sayo MT, Cristal-Luna G, Asur-Galang SM, Bellengan M, Duga KJ,
Buenaobra BB, et al: Prognostic Value of PD-L1 in Metastatic NSCLC
with EGFR-Sensitizing Mutations: A Benchmark Filipino Cohort Study.
SSRN. 2022.doi: 10.2139/ssrn.4286198. View Article : Google Scholar
|
|
24
|
Torous VF, Rangachari D, Gallant BP, Shea
M, Costa DB and VanderLaan PA: PD-L1 testing using the clone 22C3
pharmDx kit for selection of patients with non-small cell lung
cancer to receive immune checkpoint inhibitor therapy: Are cytology
cell blocks a viable option? J Am Soc Cytopathol. 7:133–141. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gu K, Shah V, Ma C, Zhang L and Yang M:
Cytoplasmic immunoreactivity of thyroid transcription Factor-1
(Clone 8G7G3/1) in hepatocytestrue positivity or cross-reaction? Am
J Clin Pathol. 128:382–388. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bardou P, Mariette J, Escudié F, Djemiel C
and Klopp C: Jvenn: An interactive Venn diagram viewer. BMC
Bioinformatics. 15:2932014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kuleshov MV, Jones MR, Rouillard AD,
Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM,
Lachmann A, et al: Enrichr: A comprehensive gene set enrichment
analysis web server 2016 update. Nucleic Acids Res. 44:W90–W97.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xie Z, Bailey A, Kuleshov M V, Clarke DJB,
Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML,
Kropiwnicki E, Jagodnik KM, et al: Gene set knowledge discovery
with enrichr. Curr Protoc. 1:e902021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang R, Grishagin I, Wang Y, Zhao T,
Greene J, Obenauer JC, Ngan D, Nguyen DT, Guha R, Jadhav A, et al:
The NCATS BioPlanet-An integrated platform for exploring the
universe of cellular signaling pathways for toxicology, systems
biology, and chemical genomics. Front Pharmacol. 10:4452019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanehisa M, Furumichi M, Sato Y,
Ishiguro-Watanabe M and Tanabe M: KEGG: Integrating viruses and
cellular organisms. Nucleic Acids Res. 49:D545–D551. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gene Ontology Consortium: The Gene
Ontology resource: Enriching a GOld mine. Nucleic Acids Res.
49:D325–D334. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee BT, Barber GP, Benet-Pagès A, Casper
J, Clawson H, Diekhans M, Fischer C, Gonzalez JN, Hinrichs AS, Lee
CM, et al: The UCSC Genome Browser database: 2022 update. Nucleic
Acids Res. 50:D1115–D1122. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Binder JX, Pletscher-Frankild S, Tsafou K,
Stolte C, O'Donoghue SI, Schneider R and Jensen LJ: COMPARTMENTS:
Unification and visualization of protein subcellular localization
evidence. Database (Oxford). 2014:bau0122014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang X, Lan Y, Xu J, Quan F, Zhao E, Deng
C, Luo T, Xu L, Liao G, Yan M, et al: CellMarker: A manually
curated resource of cell markers in human and mouse. Nucleic Acids
Res. 47:D721–D728. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu C, MacLeod I and Su AI: BioGPS and
MyGene.info: Organizing online, gene-centric information. Nucleic
Acids Res. 41:D561–D565. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu M, Mei F, Liu W and Jiang J:
Comprehensive characterization of tumor infiltrating natural killer
cells and clinical significance in hepatocellular carcinoma based
on gene expression profiles. Biomed Pharmacother. 121:1096372020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Goedhart J and Luijsterburg MS: VolcaNoseR
is a web app for creating, exploring, labeling and sharing volcano
plots. Sci Reports. 10:205602020.PubMed/NCBI
|
|
40
|
Babicki S, Arndt D, Marcu A, Liang Y,
Grant JR, Maciejewski A and Wishart DS: Heatmapper: Web-enabled
heat mapping for all. Nucleic Acids Res. 44:W147–W153. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Becht E, Giraldo NA, Lacroix L, Buttard B,
Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautès-Fridman
C, Fridman WH and de Reyniès A: Estimating the population abundance
of tissue-infiltrating immune and stromal cell populations using
gene expression. Genome Biol. 17:2182016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Galland L, Le Page AL, Lecuelle J, Bibeau
F, Oulkhouir Y, Derangère V, Truntzer C and Ghiringhelli F:
Prognostic value of thyroid transcription Factor-1 expression in
lung adenocarcinoma in patients treated with anti PD-1/PD-L1.
Oncoimmunology. 10:19576032021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lamberti G, Spurr LF, Li Y, Ricciuti B,
Recondo G, Umeton R, Nishino M, Sholl LM, Meyerson ML, Cherniack AD
and Awad MM: Clinicopathological and genomic correlates of
programmed cell death ligand 1 (PD-L1) expression in nonsquamous
non-small-cell lung cancer. Ann Oncol. 31:807–814. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yamaguchi T, Yanagisawa K, Sugiyama R,
Hosono Y, Shimada Y, Arima C, Kato S, Tomida S, Suzuki M, Osada H
and Takahashi T: NKX2-1/TITF1/TTF-1-Induced ROR1 is required to
sustain EGFR survival signaling in lung adenocarcinoma. Cancer
Cell. 21:348–361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li Y, Liu Z, Zhao Y, Yang J, Xiao TS,
Conlon RA and Wang Z: PD-L1 expression is regulated by ATP-binding
of the ERBB3 pseudokinase domain. Genes Dis. 10:1702–1713. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hu X, Li J, Fu M, Zhao X and Wang W: The
JAK/STAT signaling pathway: From bench to clinic. Signal Transduct
Target Ther. 6:4022021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jahangiri A, Dadmanesh M and Ghorban K:
STAT3 inhibition reduced PD-L1 expression and enhanced antitumor
immune responses. J Cell Physiol. 235:9457–9463. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zegeye MM, Lindkvist M, Fälker K, Kumawat
AK, Paramel G, Grenegård M, Sirsjö A and Ljungberg LU: Activation
of the JAK/STAT3 and PI3K/AKT pathways are crucial for IL-6
trans-signaling-mediated pro-inflammatory response in human
vascular endothelial cells. Cell Commun Signal. 16:552018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang N, Zeng Y, Du W, Zhu J, Shen D, Liu
Z and Huang JA: The EGFR pathway is involved in the regulation of
PDL1 expression via the IL-6/JAK/STAT3 signaling pathway in
EGFR-mutated non-small cell lung cancer. Int J Oncol. 49:1360–1368.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Federico L, McGrail DJ, Bentebibel SE,
Haymaker C, Ravelli A, Forget MA, Karpinets T, Jiang P, Reuben A,
Negrao MV, et al: Distinct tumor-infiltrating lymphocyte landscapes
are associated with clinical outcomes in localized non-small-cell
lung cancer. Ann Oncol. 33:42–56. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jamal-Hanjani M, Wilson GA, McGranahan N,
Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al: Tracking the evolution of non-small-cell lung
cancer. N Engl J Med. 376:2109–2121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Pinto JA, Vallejos CS, Raez LE, Mas LA,
Ruiz R, Torres-Roman JS, Morante Z, Araujo JM, Gómez HL, Aguilar A,
et al: Gender and outcomes in non-small cell lung cancer: An old
prognostic variable comes back for targeted therapy and
immunotherapy? ESMO Open. 3:e0003442018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
He H, Ma H, Chen Z, Chen J, Wu D, Lv X and
Zhu J: Chromosomal copy number variation predicts EGFR-TKI response
and prognosis for patients with non-small cell lung cancer.
Pharmgenomics Pers Med. 16:835–846. 2023.PubMed/NCBI
|
|
56
|
Hong L, Dibaj S, Negrao MV, Reuben A,
Roarty E, Rinsurongkawong W, Lewis J, Gibbons DL, Sepesi B,
Papadimitrakopoulou V, et al: Spatial and temporal heterogeneity of
PD-L1 and its impact on benefit from immune checkpoint blockade in
non-small cell lung cancer (NSCLC). J Clin Oncol. 37:90172019.
View Article : Google Scholar
|
|
57
|
De-Rui Huang D and Chih-Hsin Yang J:
Checkpoint inhibitor combined with tyrosine kinase inhibitor-the
end or beginning? J Thorac Oncol. 15:305–307. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Nakahama K, Kaneda H, Osawa M, Izumi M,
Yoshimoto N, Sugimoto A, Nagamine H, Ogawa K, Matsumoto Y, Sawa K,
et al: Association of thyroid transcription factor-1 with the
efficacy of immune-checkpoint inhibitors in patients with advanced
lung adenocarcinoma. Thorac Cancer. 13:2309–2317. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lee JS, Kim HR, Lee CY, Shin M and Shim
HS: EGFR and TTF-1 gene amplification in surgically resected lung
adenocarcinomas: Clinicopathologic significance and effect on
response to EGFR-tyrosine kinase inhibitors in recurred cases. Ann
Surg Oncol. 20:3015–3022. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pös O, Radvanszky J, Styk J, Pös Z, Buglyó
G, Kajsik M, Budis J and Nagy B and Nagy B: Copy number variation:
Methods and clinical applications. Appl Sci. 11:8192021. View Article : Google Scholar
|
|
61
|
Yu HA, Suzawa K, Jordan E, Zehir A, Ni A,
Kim R, Kris MG, Hellmann MD, Li BT, Somwar R, et al: Concurrent
alterations in EGFR-mutant lung cancers associated with resistance
to EGFR kinase inhibitors and characterization of MTOR as a
mediator of resistance. Clin Cancer Res. 24:3108–3118. 2018.
View Article : Google Scholar : PubMed/NCBI
|