Introduction
Glioma, a primary central nervous system tumor,
comprises 45.2% of all intracranial neoplasms, with an annual
incidence between 0.003 and 0.005% (1). Gliomas range in grade from I to IV:
Higher-grade gliomas (III and IV) are characterized by aggressive,
poorly differentiated cells, while low-grade gliomas (LGG, I and
II) consist of slowly proliferating, well-differentiated cells
(2). The cornerstone of glioma
management involves a multimodal approach encompassing
chemotherapy, surgical resection and radiation therapy (3,4).
Although surgical assistance and molecular pathology have developed
rapidly in recent times, the efficacy in patients with glioma
remains poor due to its high recurrence rate, short survival time,
aggressive growth and strong invasiveness (5,6). Thus,
the continued exploration of glioma mechanisms is critical to
identifying prospective clinical indicators.
RNA-binding proteins (RBPs), ubiquitously present in
all living organisms, were first discovered in yeast and mammalian
cell extracts due to their ability to interact with RNA molecules
(7). In human embryonic kidney
cells, more than a thousand proteins have been identified to be
able to bind with RNAs. These proteins participate in numerous
biological functions beyond dictating the fate of RNA molecules
(8). A study by Muleya and
Marondedze (9) suggested that
unallocated RBP may have a key role in adjusting metabolic changes
in response to multiple stressful stimuli. Furthermore, the
lifecycle of RNA transcription and processing from the nucleus to
translation and degradation in the cytoplasm is influenced by
several related proteins, including numerous RBPs (10). In all kinds of organisms, from yeast
to humans, cap structures are co-transcriptionally affixed to the
5′ end of RNA. When coupled with polymerase II transcripts, this
cap structure becomes an essential signal for binding these
transcripts to downstream proteins (11). Despite these insights, further
investigation is necessary to clarify the mechanisms of RBPs in
human disease pathology.
The present study delved into the role and
mechanisms of RBPs in gliomas to identify key RBPs with clinical
value and to explore the potential impact of the central gene G
protein nucleolar 2 (GNL2) on glioma progression. It was
hypothesized that aberrant expression of RBPs is a driver of glioma
progression and the identification of key RBPs among them (e.g.,
GNL2) may render them potential prognostic biomarkers. As a pivotal
gene, GNL2 has a crucial role in the pathogenesis of gliomas by
influencing key cellular processes, particularly enhanced protein
synthesis, which ultimately contributes to the development of
gliomas. These findings provide insight into targeted therapies for
gliomas based on regulatory RNA-binding proteins and lay the
foundation for future identification of new biomarkers and
development of therapeutic strategies. The present study provides
important scientific support to deepen the understanding of the
molecular mechanisms of gliomas and develop more individualized
therapeutic options for patients.
Materials and methods
Identification of differentially
expressed RBPs in glioma samples
The identification analysis from the The Cancer
Genome Atlas (TCGA) dataset was conducted using R software (version
3.3.1; http://www.r-project.org/) and the edgeR
package. The criteria for identifying differentially expressed
genes (DEGs) were set as follows: Fold change (FC) <0.5 for
downregulated genes and FC >2 for upregulated genes, and
P<0.05 for each. The heatmap and volcano map were plotted by the
gplots package of R software. A list of genes encoding RBPs was
referenced from the reference ‘a census of human RNA-binding
proteins’ (12). The overlapping
genes between the RBP-encoding genes and the identified DEGs were
determined by the Venn package of R software.
Functional enrichment and prognostic
analysis of key overlapping genes
Interactions between proteins are crucial for
exploring tumor mechanisms. In the present study, protein-protein
interaction (PPI) analyses of the overlapping genes were built
using the Search Tool for the Retrieval of Interacting Genes and
proteins (STRING; http://string-db.org). Next, ‘Molecular Complex
Detection’ (MCODE) function in Cytoscape software (version 3.7.2;
http://cytoscape.org), a clustering algorithm
based on a node-weighted algorithm, was used to identify highly
interacting gene clusters. Next, Gene Ontology (GO) analysis was
performed on the genes screened by MCODE based on the Database for
Annotation, Visualization and Integrated Discovery (DAVID;
http://david.ncifcrf.gov/) database.
Subsequently, overlapping genes were analyzed for overall survival
(OS) prognosis by univariate Cox regression, and hazard ratios
(HR), P-values and confidence intervals (CI) for hazard
coefficients were shown in a forest plot.
Construction of prognostic risk
model
A prognostic gene signature was constructed using
the least absolute shrinkage and selection operator (LASSO)
regression method, implemented through the ‘glmnet’ package in R
software, to predict glioma prognosis. Ten-fold cross-validation
was used to extract the ideal value from the least partial
likelihood deviation in order to increase objectivity and
dependability. The average risk score was applied to classify
patients into high-risk and low-risk groups. Kaplan-Meier (KM)
survival curves were generated using the ‘survival’ package in R
software to evaluate OS probabilities in two groups, with the
log-rank test used to determine statistical significance. A
receiver operating characteristic (ROC) curve was drawn to assess
the prediction accuracy of the risk model. To gauge how well the
model predicted patient survival, the area under the curve (AUC)
value was calculated.
Predictive nomogram construction with
key genes related to glioma prognosis
Next, a univariate/multivariate Cox regression
analysis was performed on the top 10 prognosis-related genes using
the ‘survival’ package of R software, and forest plots were
generated with the ‘forest plot’ package of R software to display
the variables available as nomograms. A total of 3 key genes were
selected due to their high association with glioma prognosis. The
‘rms’ program (https://CRAN.R-project.org/package=rms) in R software
was then employed to create a nomogram to forecast the 1-, 3- and
5-year OS rates. The closeness of the Nomogram model to the
calibration curve resembles the model's prediction ability.
Expression level and survival analyses
on the hub gene
Based on the above analysis, one hub gene (GNL2) was
selected in the present study for the next analysis. First, the
levels of GNL2 in LGG and glioblastoma (GBM) samples were compared
using GEPIA (http://gepia.cancer-pku.cn). Following this, the
association between GNL2 levels and various survival probabilities,
including disease-specific survival (DSS), OS and progression-free
survival (PFS), was systematically investigated utilizing the KM
plotter online database (https://kmplot.com). The log-rank P-values and HRs
were calculated with 95% CIs to measure the statistical
significance of the observed differences in survival outcomes.
Cell culture and transfection
Glioma cell lines (U251MG, SW1783 and U373) and
normal human astrocyte cells (NHA) were obtained from the Chinese
Academy of Sciences Cell Bank (Shanghai, China). They were kept in
DMEM with 10% FBS (Absin) at 37°C with 5% CO2. For
transfection, cells were seeded in appropriate culture vessels and
transfected with siRNAs targeting GNL2 (si-GNL2 #1, si-GNL2 #2 and
si-GNL2 #3) or control small interfering RNA (siRNA) following the
manufacturer's instructions for Lipofectamine 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). The relevant sequence
information was as follows: si-GNL2#1,
5′-CACGTGTGATTAAGCAGTCATCATT-3′; si-GNL2#2,
5′-CCATACAAAGTTGTCATGAAGCAAA-3′; si-GNL2#3,
5′-GGGGTTCTCCACTTTAGGTTAA-3′; and si-negative control (si-NC),
5′-UUCUCCGAACGUGUCACGUTT-3′.
Western blot (WB) analysis
Cells were lysed in RIPA buffer with a protease
inhibitor cocktail (Sigma-Aldrich; Merck KGaA) on ice for 30 min.
To obtain nuclear and cytosolic extracts, the cell lysates were
centrifuged at 800 × g for 5 min at 4°C to pellet the nuclei. The
supernatant with the cytosolic fraction was transferred to a fresh
tube, and the nuclear pellet was resuspended in a nuclear
extraction buffer. After centrifuging lysates at 14,000 × g for 15
min at 4°C, the supernatant was collected. Protein concentrations
were determined using the BCA protein assay kit from Thermo Fisher
Scientific, Inc. 10% SDS-PAGE was used to separate identical
quantities of protein (30–50 g) before they were transferred to
PVDF membranes (EMD Millipore). The membranes were first treated
with 5% non-fat milk in Tris-buffered saline containing Tween-20
(TBST) for 1 h at room temperature (~25°C) to block them. Following
this, the membranes were incubated overnight with primary
antibodies at 4°C at the following dilutions: Anti-GNL2 (1:1,000
dilution; Abcam), anti-Lamin A/C (1:1,000 dilution; Cell Signaling
Technology, Inc.), anti-ribosomal protein L11 (RPL11; 1:1,000
dilution; Abcam) and anti-Tubulin (1:2,000 dilution; Cell Signaling
Technology, Inc.). TBST was used to wash the membranes and then
horseradish peroxidase-conjugated secondary antibody (1:2,000
dilution; Cell Signaling Technology, Inc.) was applied for 1 h at
room temperature. Enhanced chemiluminescence substrate (Beyotime
Institute of Biotechnology) was employed to detect signals and a
chemiluminescence imaging system (Bio-Rad ChemiDoc; Bio-Rad
Laboratories, Inc.) was used for visualization. Finally, protein
bands were quantified using ImageJ software (version 1.51; National
Institutes of Health).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A NanoDrop
2000 spectrophotometer (Thermo Fisher Scientific, Inc.) was
employed to determine the purity and concentration of the RNA. cDNA
was generated from 1 g of total RNA by using a High-capacity cDNA
RT kit (Applied Biosystems; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. qPCR was performed on
an Applied Biosystems 7500 Fast real-time PCR machine (Thermo
Fisher Scientific, Inc.) using SYBR Green Master Mix (Sangon
Biotech Co., Ltd.) according to the manufacturer's instructions.
For the qPCR, the thermocycling conditions were as follows: 10 min
of initial denaturation at 95°C and 40 cycles of denaturation at
95°C for 15 sec and 1 min of annealing and extension at 60°C. The
primer sequences used were as follows: GNL2 forward,
5′-ATCCAAATGTTGGCAAGAGC-3′ and reverse,
5′-ACACCTGGACAGTCAATCAGG-3′; GAPDH forward,
5′-TGCACCACCAACTGCTTAGC-3′ and reverse,
5′-GGCATGGACTGTGGTCATGAG-3′. The relative gene expression was
calculated using the 2−ΔΔCq method (13), with GAPDH as the internal
control.
Cell Counting Kit-8 (CCK-8) assay
The impact of si-GNL2 #2 on SW1783 and U373 cells
was determined using a CCK-8 (Beyotime Institute of Biotechnology).
Cellular samples were seeded in 96-well plates and subjected to
transfection with si-GNL2 #2 or control siRNA. Following
transfection, the cells were incubated for 0, 1, 2, 3 or 4 days. To
evaluate cell proliferation, 10 µl of CCK-8 solution was added to
each well and allowed to incubate at 37°C for 2 h. Subsequently, a
microplate reader was used to measure the optical density values at
450 nm.
Transwell assay
The impact of si-GNL2 #2 on the invasion and
migration of SW1783 and U373 cells was assessed using Transwell
assays. For the invasion assay, the upper chamber of a 24-well
Transwell plate (8 µm pore size; Nanjing KeyGen Biotech Co., Ltd.)
was coated with Matrigel® (Beijing Solarbio Science
& Technology Co., Ltd.). Following transfection with either
si-GNL2 #2 or control siRNA, the cells were suspended in serum-free
medium and then seeded in the upper chamber. Simultaneously, medium
supplemented with 10% FBS was added to the lower chamber. After 24
h of incubation at 37°C, the non-migrated cells on the upper
surface of the membrane were gently removed with a cotton swab.
Cells that migrated to the lower surface were then fixed with 4%
paraformaldehyde for 20 min at room temperature and stained with
DAPI for 15 min. After staining, cells were washed, dried and
counted under a light microscope. The migration assay was performed
in a similar manner to the invasion assay, with the key difference
being the omission of Matrigel® in the Transwell plate
to allow for the assessment of cell migration without extracellular
matrix components.
Labeling and detection of newly
synthesized proteins
Based on a previous study (14), 100 µCi of
35S-methionine/35S-cysteine
(EXPRE35S35S Protein Labeling Mix; Perkin
Elmer) was added to each ml of cell culture half an hour before the
end of cell treatment to label newly generated proteins. Following
treatment, cells were lysed in a modified RIPA solution containing
protease and phosphatase inhibitors (Beijing Solarbio Science &
Technology Co., Ltd.) after being rinsed with PBS. Using a Bradford
Protein Assay Kit (Bio-Rad Laboratories, Inc.), the protein
concentrations were measured, and 50 µg of the extract was placed
on Whatman paper following the protocol described in Tailler et
al (14). Using a cold 5%
trichloroacetic acid (TCA) and methionine solution, polypeptides
were precipitated. This was followed by boiling in 5% TCA for 15
min and washing with ethanol. Subsequently, 30 µg of radiolabeled
proteins were separated by 10% SDS-PAGE, transferred to a PVDF
membrane and scintillation was measured with a Typhoon FLA 7000
machine (GE Healthcare).
Statistical analysis
All experimental results were expressed as the mean
± standard deviation from a minimum of three separate experiments.
Student's t-test was used to compare differences between two
groups. One-way ANOVA was performed for comparisons between
multiple groups, followed by Tukey's post-hoc test to adjust for
multiple comparisons. P<0.05 was considered to indicate a
statistically significant difference. All statistical analyses were
conducted using the SPSS 25.0 software package (IBM Corp.). Graphs
were generated using GraphPad Prism 8.0 (GraphPad Software;
Dotmatics).
Results
Identification of 1,063 overlapping
genes from TCGA-DEGs and RBPs
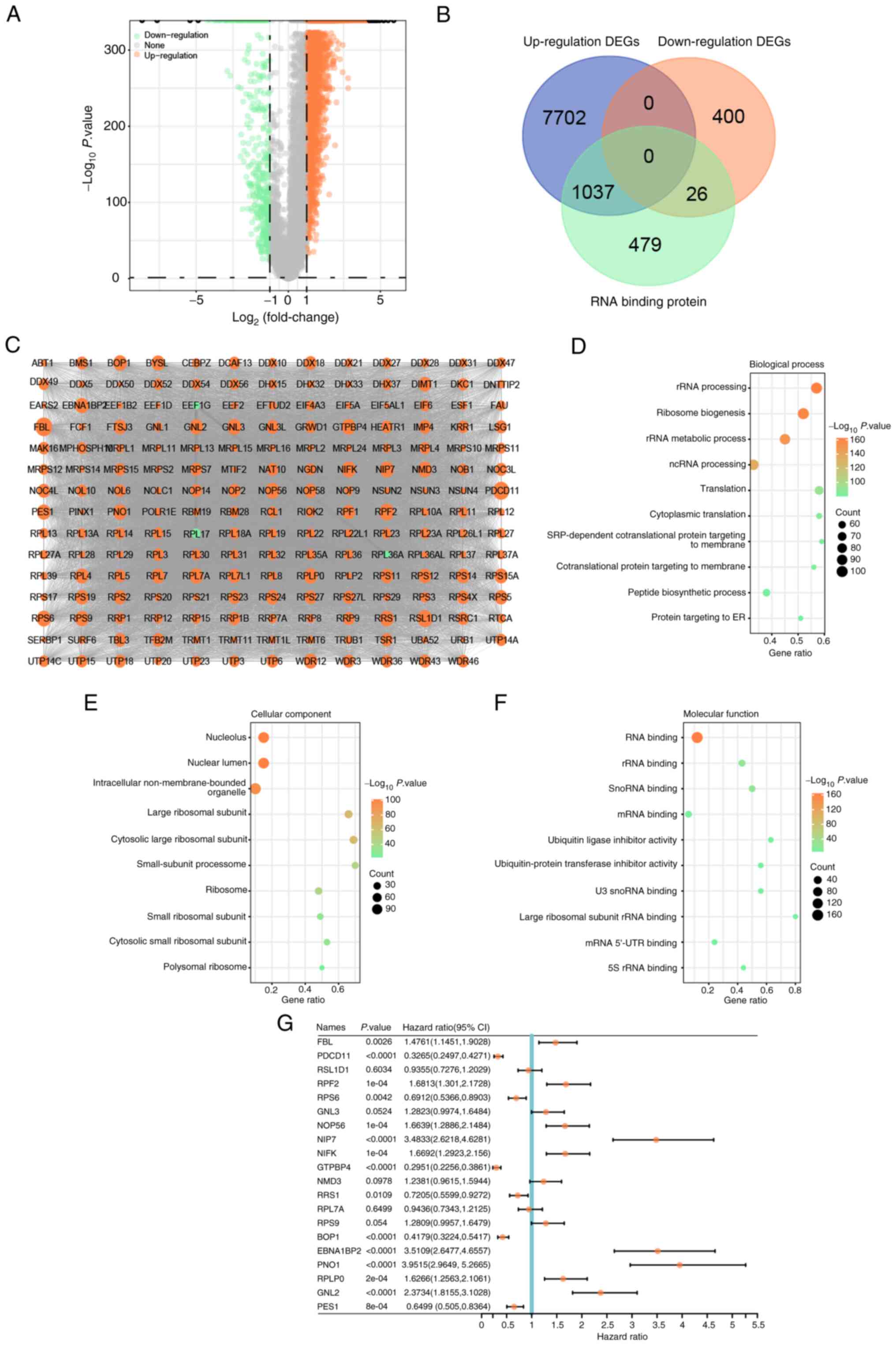
The volcano plot (Fig.
1A) revealed 8,739 upregulated and 426 downregulated DEGs
related to glioma in the TCGA dataset. The Venn package was applied
to analyze the intersecting genes between up- and down-regulated
DEGs and genes encoding RBPs, respectively. As presented in
Fig. 1B, the overlapping genes
included 1,037 upregulated and 26 downregulated genes. These
overlapping genes were specifically selected for further
investigation in the present study, as they may shed light on the
molecular mechanisms underlying glioma and potentially represent
valuable therapeutic targets.
PPI network construction and module
analysis
To further explore the relationship between glioma
and the intersecting genes, a PPI network was generated to reveal
the interaction between these genes. The genes were imported into
Cytoscape and analyzed using the plugin MCODE, resulting in a PPI
network with 194 nodes and 11,364 edges (Fig. 1C). GO term enrichment analysis was
then performed on 194 genes. In the GO enrichment results, the main
enrichment items of these genes in cellular component, biological
process and molecular function included ‘ribosomal RNA (rRNA)
processing’, ‘ribosome biogenesis’, ‘large ribosomal subunit’, ‘RNA
binding’ and ‘large ribosomal subunit rRNA binding’ (Fig. 1D-F). After performing OS prognostic
analysis on 194 genes, the results of the top 20 signature genes
(P<0.05) were selected for visualization in a forest plot
(Fig. 1G). These 20 genes may be
used as prognostic genes for glioma and progressed to the
subsequent analysis.
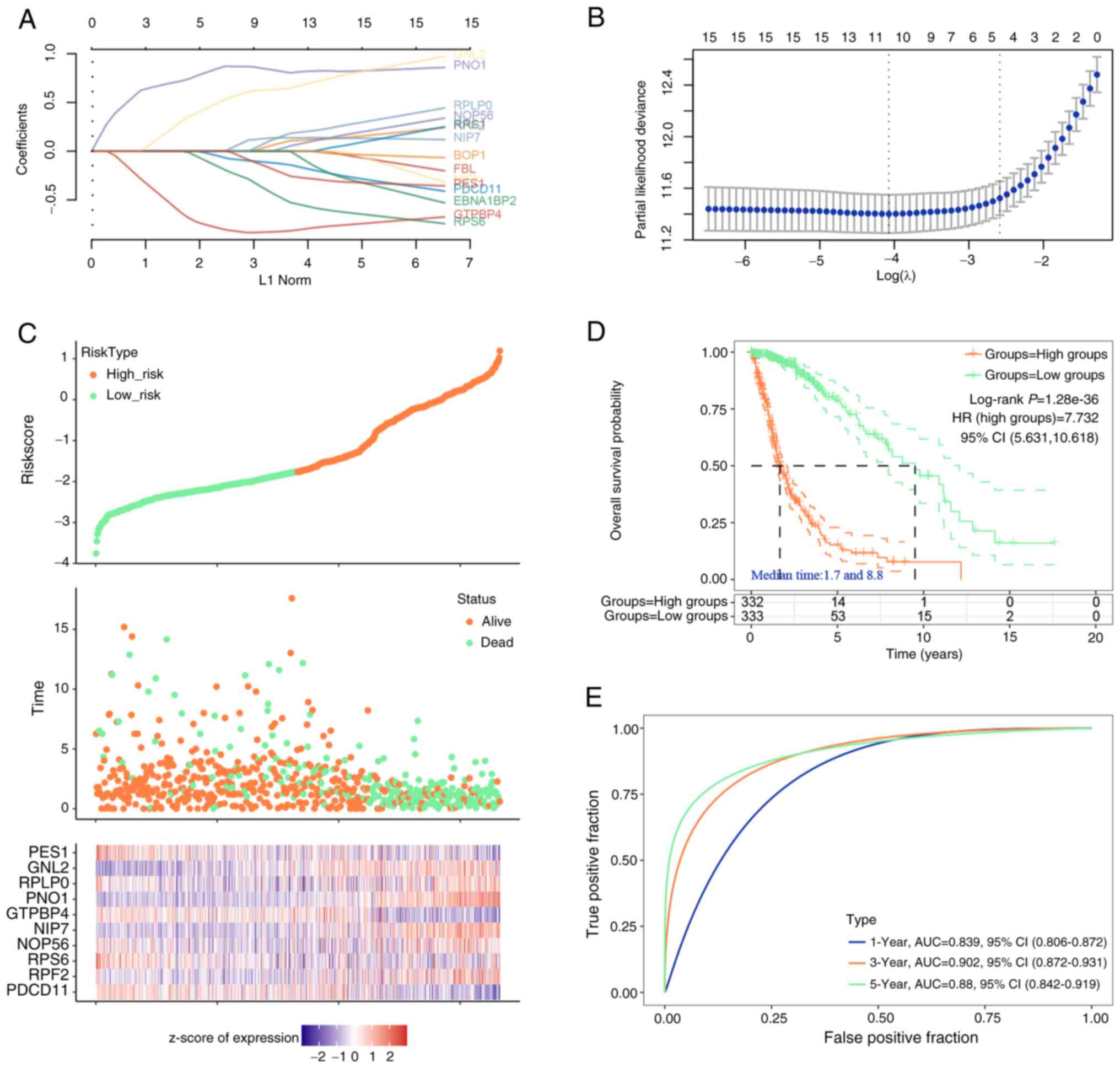
Prognostic risk model of the top 20
genes
The top 20 significant genes were used for Lasso
proportional hazards regression and 10-fold cross-validation to
construct the optimal gene signature, and 10 candidate genes were
finally determined (Fig. 2A and B).
The risk scores, survival duration and status of the glioma
patients, along with a heatmap detailing z-score normalized
expression of prognostic genes derived from the TCGA-glioma
dataset, were illustrated in Fig.
2C. Each column in the heat map represents a gene expression
profile for an individual patient and corresponds to the patient
data shown in the risk and survival plots above. To identify RBP
features suitable for survival prediction, patients with glioma,
based on the average risk score, were classified into low-risk
(n=333) and high-risk groups (n=332). KM curve analysis indicated
that compared to low-risk patients, high-risk individuals had worse
OS (Fig. 2D). Furthermore, ROC
curves demonstrated that the AUC of the risk model at 1, 3 and 5
years was 0.839, 0.902 and 0.88 in the training set, respectively
(Fig. 2E), which indicated the good
predictive ability of this model.
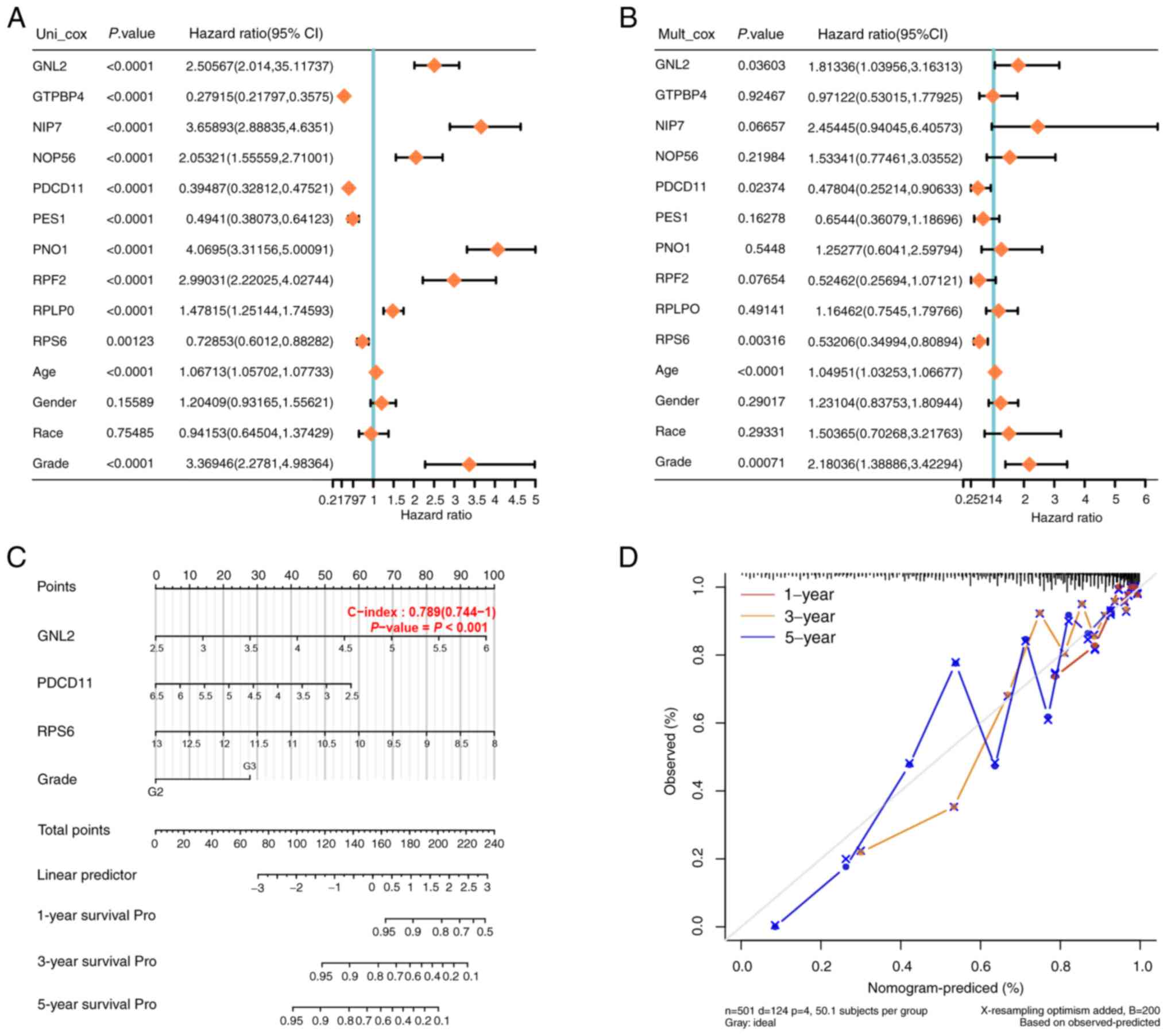
Construction and verification of the
nomogram of glioma prognosis
Univariate/multivariate Cox regression analyses were
performed on the 10 genes and clinical variables included in the
prognostic risk model. GNL2, programmed cell death 11 (PDCD11),
ribosomal protein S6 (RPS6) and Grade (P<0.05) were suggested to
be independent prognostic indicators for OS (Fig. 3A and B). To establish a more
reliable clinical prediction method, a comprehensive nomogram
containing GNL2, PDCD11, RPS6 and Grade was constructed to predict
1-, 3- and 5-year OS for patients with glioma (Fig. 3C). The calibration plot of patient
survival prediction in the TCGA-glioma cohort showed that the
predicted results of the prognostic nomogram were similar to the
actual results (Fig. 3D). The GNL2,
PDCD11 and RPS6 genes may serve as new biomarkers in glioma
prognosis. In the present study, GNL2 was chosen to be the target
gene for further analysis.
Expression and prognostic analysis of
GNL2 in glioma
Next, the levels of GNL2 were analyzed in LGG and
GBM, and it was found that the levels of GNL2 in LGG and GBM were
higher than those in normal tissues (Fig. 4A). In the OS, PFS and DSS analyses,
the survival rate of patients with high GNL2 expression was lower
(Fig. 4B-D). Collectively, these
findings indicate that GNL2 functions as an oncogene in glioma.
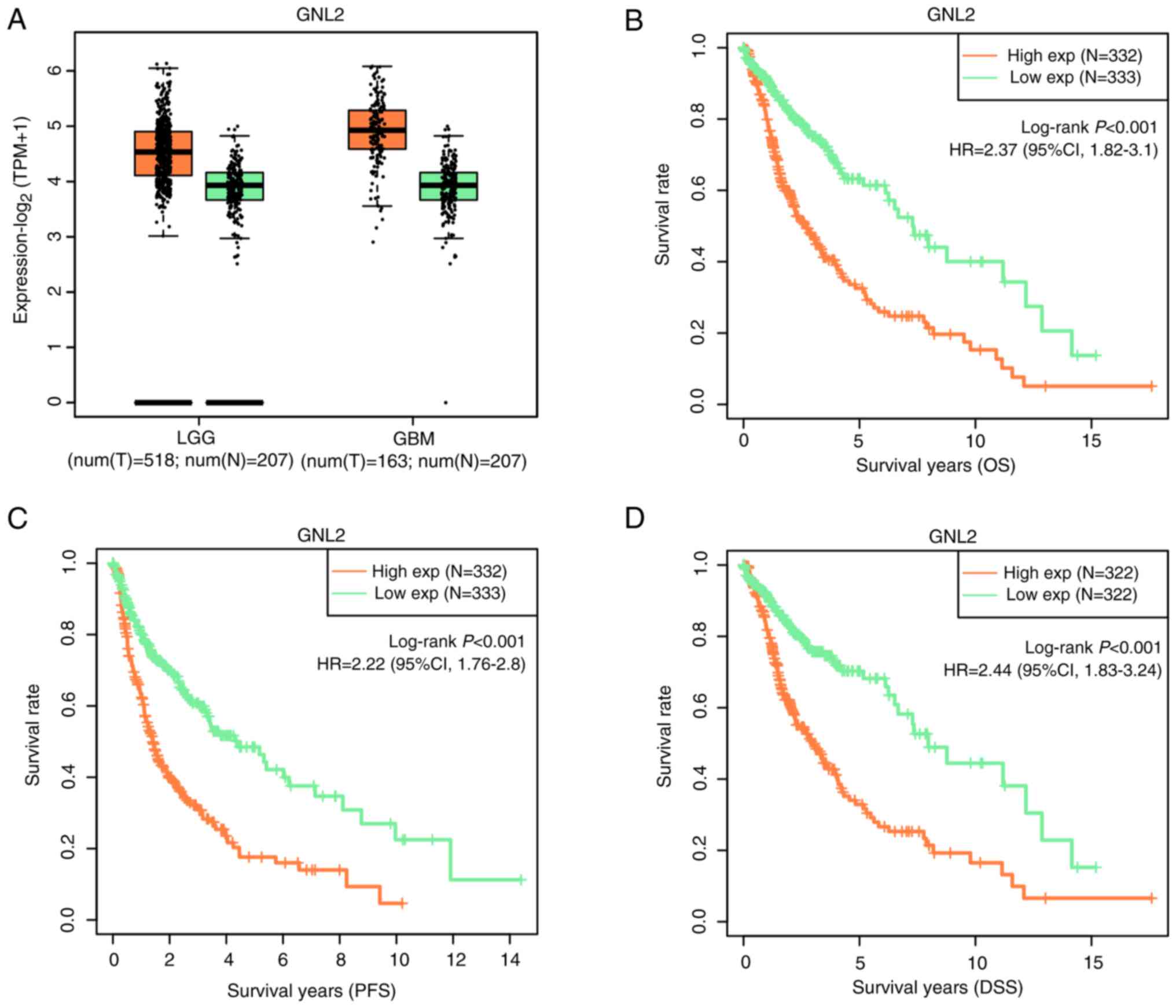 | Figure 4.GNL2 expression and its association
with survival in patients with glioma. (A) GEPIA database detects
the expression level of GNL2 in LGG and GBM; the orange box line
represents tumor samples and the green box line represents normal
samples. Each box plot shows the median expression level (central
line), the 25 to 75th percentiles (box) and the standard error of
the mean (Whisker lines). (B-D) KM survival curve analysis of the
effect of GNL2 expression on (B) OS, (C) PFS and (D) DSS in
patients with glioma. The horizontal axis displays the survival
time, while the vertical axis shows the survival probability. GNL2,
G protein nucleolar 2; LGG, low-grade glioma; GBM, glioblastoma;
OS, overall survival; PFS, progression-free survival; DSS,
disease-specific survival; exp, expression; T, tumor; N, normal
tissue sample; HR, hazard ratio. |
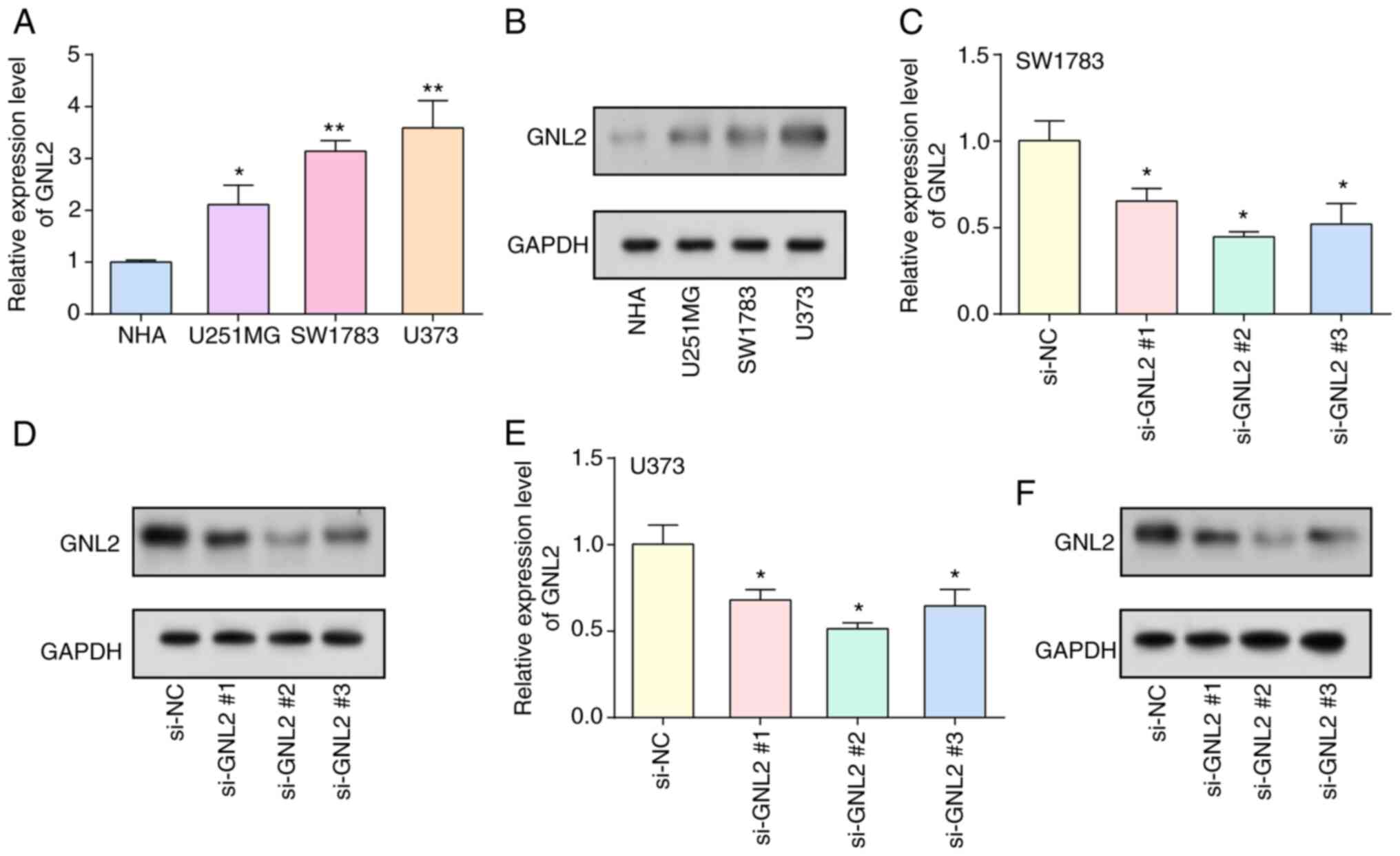
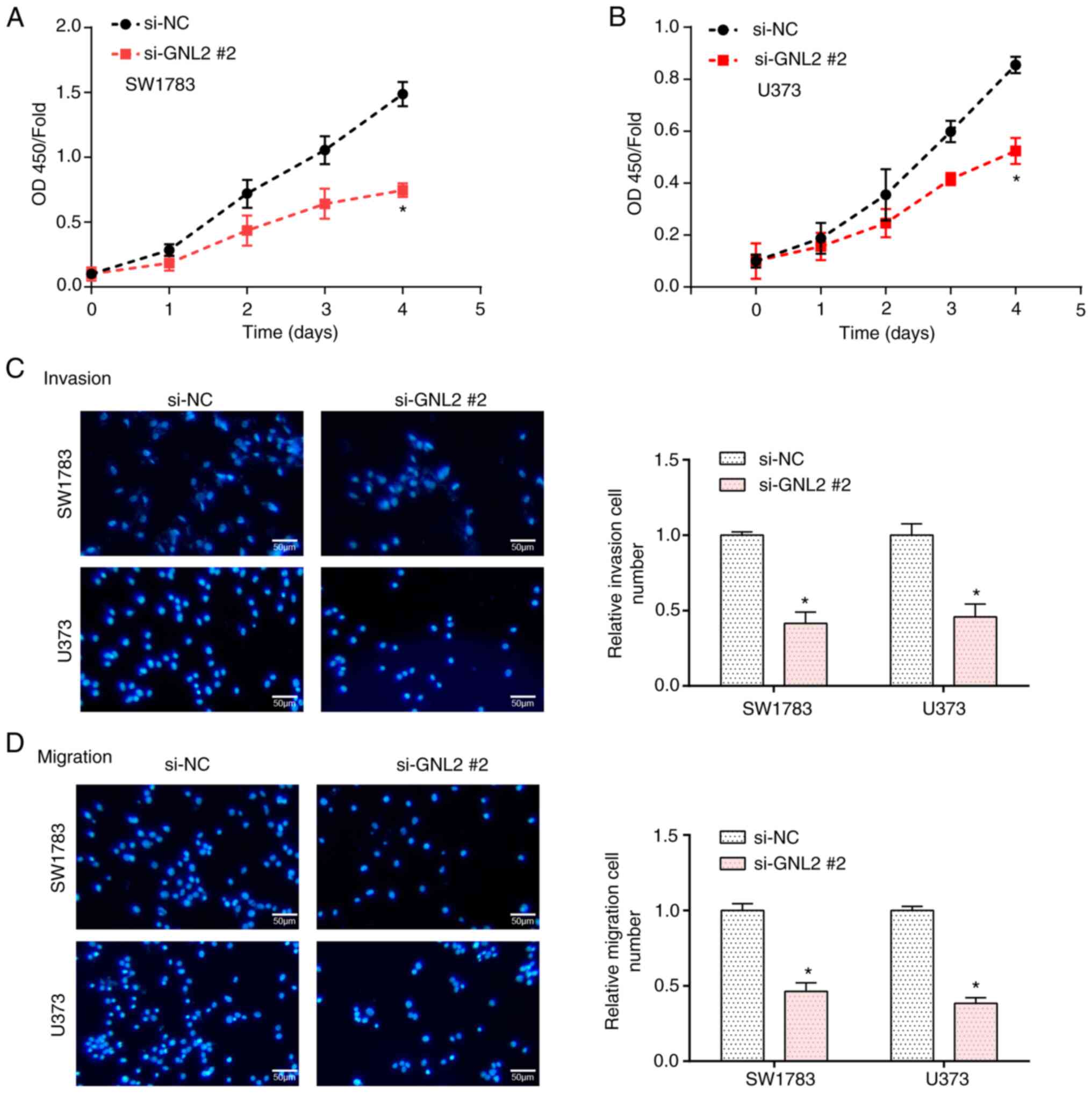
Knockdown of GNL2 suppresses the
proliferation, invasion and migration of glioma cells
By RT-qPCR and WB analyses, the levels of GNL2 were
examined in glioma cells and normal cells. The results demonstrated
enhanced levels of GNL2 in glioma cells, with particularly
pronounced increases observed in the SW1783 and U373 cell lines
(Fig. 5A and B). Subsequently, GNL2
was silenced within glioma cell lines using three distinct siRNAs:
si-GNL2 #1, si-GNL2 #2 and si-GNL2 #3. Among these, si-GNL2 #2
demonstrated the most optimal knockdown efficiency, as validated by
both WB and RT-qPCR (Fig. 5C-F).
After further investigation using CCK-8 and Transwell assays, in
GNL2-knockdown glioma cell lines, a significant reduction in cell
proliferation, invasion and migration was detected in comparison to
the control group (Fig. 6A-D). In
summary, the present results suggest that GNL2 is a potential
oncogene in glioma and its inhibition may impair key oncogenic
characteristics of glioma cells, highlighting its prospective value
as a therapeutic target.
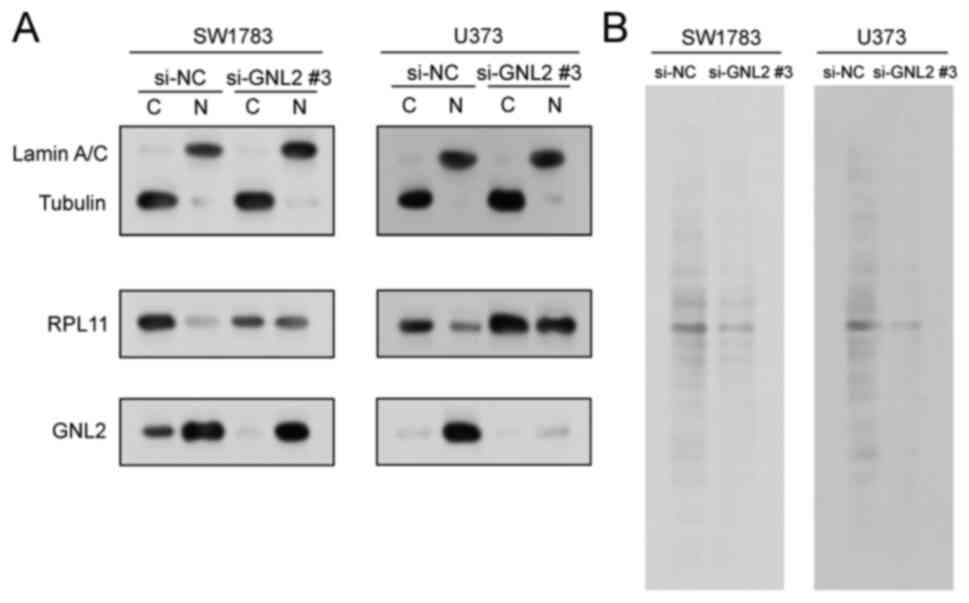
GNL2 silencing alters RPL11
localization and suppresses protein synthesis in glioma cells
GNL2 is involved in various processes related to
rRNA synthesis and processing, and assembly of ribosomal subunits
(15,16). According to earlier research, the
cytoplasmic/nuclear ratio of 60S ribosomal protein RPL11 was
decreased in ovarian cancer cells when GNL2 was silenced (17). In the present study, the protein
lysates from SW1783 and U373 cells transfected with si-GNL2 or
si-NC we separated into nuclear and cytoplasmic fractions and the
localization of RPL11 was assessed using WB. Following the
knockdown of GNL2, the expression of RPL11 increased in the nuclei
of both SW1783 and U373 cells (Fig.
7A), suggesting that the export of 60S ribosomal subunits was
impaired, and GNL2 knockdown decreased the cytoplasmic/nuclear
ratio of RPL11. Furthermore, by examining newly synthesized
S35-tagged total protein, it was found that knockdown of
GNL2 inhibited overall protein synthesis in glioma cells (Fig. 7B). These findings underscore a vital
function for GNL2 in the modulation of RPL11 localization and
protein synthesis in glioma cells. Silencing of GNL2 appears to
disrupt the nuclear export of RPL11, affecting the normal
distribution of RPL11 and overall protein synthesis within the
cell. These disruptions may potentially influence the oncogenic
behavior of glioma cells. The present study thereby suggests that
GNL2 may be a potential therapeutic target in glioma.
Discussion
Gliomas account for ~45% of all intracranial tumors
(18). The prognosis of patients
with glioma patients often deteriorates with increasing degree of
malignancy, leading to gliomas being one of the deadliest malignant
tumor types, particularly in the context of grade IV GBM (19). Alarmingly, statistics suggest that
individuals afflicted with grade IV GBM exhibit a median survival
duration of just one year and <5% survive beyond five years
(20). Given their malignant
nature, surgical intervention or radiation therapy rarely yield
curative results for malignant gliomas (21), thereby necessitating the frequent
recourse to chemotherapy. However, the effectiveness of
chemotherapy is often significantly impeded by the blood-brain
barrier, which restricts the entry of intravenously administered
anticancer drugs into the glioma region (22). While molecular-targeted therapies
have somewhat improved the survival rates of patients with glioma,
the overall therapeutic efficacy remains disappointing (23). Given these challenges, it is of
utmost importance to further our understanding of the molecular
underpinnings of gliomas.
In the current investigation, an exhaustive analysis
of glioma samples from the TCGA database was performed, leading to
the identification of 8,739 upregulated and 426 downregulated DEGs.
To further refine this analysis, the DEGs were cross-referenced
with genes encoding RBPs, employing the Venn package, and an
intersection of 1,037 up- and 26 downregulated genes was obtained.
This set of overlapping genes, potentially crucial in illuminating
the molecular underpinnings of glioma, was chosen for subsequent
scrutiny in the present study, suggesting their potential as
valuable therapeutic targets. To unveil the roles of these
crossover genes in glioma, a PPI network comprising 194 genes was
constructed. This network not only revealed the interaction of
these genes, but also elucidated their potential synergy in glioma
pathogenesis. Of note, GO term enrichment analysis for these 194
genes unveiled significant enrichment in functional terms such as
‘rRNA processing’, ‘ribosome biogenesis’, ‘large ribosomal
subunit’, ‘RNA binding’ and ‘large ribosomal subunit rRNA binding’.
These processes and components are integral for the cellular
functioning and protein synthesis machinery (24,25),
highlighting the potential relevance of these genes in the context
of the aggressive cellular behavior and uncontrolled proliferation
of glioma.
Following an OS prognostic analysis conducted on 194
genes, the top 20 genes with a significant impact on outcomes were
selected. These genes were further subjected to LASSO regression,
risk model analysis and univariate/multivariate Cox regression
analyses to establish a prognostic nomogram for patients with
glioma. In the nomogram, 3 genes with clinical value in glioma
prognosis were found, namely GNL2, PDCD11 and RPS6. PDCD11, also
known as ALG4, NFBP, RRP5 and ALG-4, an NF-κB-binding protein
necessary for rRNA maturation and the production of 18S rRNA,
colocalizes with U3 RNA in the nucleolus (26,27).
To date, only a small number of studies on PDCD11 have been
published. Xing et al (28)
analyzed predictive biomarkers for triple-negative breast cancer
(TNBC) and determined that PDCD11 acts as an oncogene in TNBC. The
cytoplasmic ribosomal protein S6 (RPS6) is a subunit of the 40S
subunit (29,30). Shirakawa et al (31) demonstrated that RPS6 was present in
perinecrotic, perivascular and border niches in GBM tissues and was
markedly elevated in high-grade gliomas. To date, >88 clinical
trials of immunotherapies for GBM have been initiated and conducted
worldwide (32). In addition,
several immunotherapies have shown promising efficacy, including
dendritic cell (DC) vaccines such as DCVax-L (33) and oncolytic virus G47Δ (34). Furthermore, the swift emergence of
molecular subtypes in gliomas carries significant clinical
implications and applications. These encompass diagnostic imaging,
pathology testing prerequisites, strategic planning for clinical
trials and the implementation of targeted therapies for gliomas
(35). This consolidated evidence
amplifies the potential of these genes as vital prognostic
indicators and offers a promising avenue for therapeutic
advancements in glioma.
GNL2 enables RNA-binding activity and is predicted
to be involved in ribosome biogenesis (17). The GNL2 family encompasses two
members, namely GNL3 (nucleoprotein) and GNL3-like (GNL3L)
(36). While GNL3L functions as the
vertebrate equivalent of the nucleostemin, GNL2 has a unique
presence across both vertebrate and invertebrate species (37). The contribution of GNL2 to various
cancers has been previously highlighted in the literature. In a
compelling study by Nakamura et al (17), it was observed that healthy
fallopian tube secretory epithelial cells exhibited increased
proliferation and colony formation when GNL2 was overexpressed, but
xenograft tumor development was inhibited when GNL2 was silenced.
Furthermore, within the context of ovarian cancer, GNL2 appears to
regulate the formation of the 60S ribosomal subunit (17). Drawing from these existing
investigations, GNL2 was deemed a potential biomarker for glioma.
The subsequent analyses of the present study revealed that GNL2
levels were markedly elevated in glioma as compared with normal
tissues. Furthermore, high expression of GNL2 was associated with
diminished survival rates. Collectively, these observations led us
to conclude that GNL2 functions as an oncogene within the scope of
glioma.
The present study underscores the pivotal role of
GNL2 in glioma pathogenesis. Enhanced GNL2 expression was detected
in glioma cell lines, particularly SW1783 and U373. Following GNL2
knockdown, a significant reduction in glioma cell proliferation,
invasion and migration was observed, highlighting the potential
oncogenic and therapeutic importance of GNL2 in glioma.
Furthermore, GNL2 seems instrumental in modulating RPL11
subcellular localization and overall protein synthesis. GNL2
silencing resulted in RPL11 nuclear accumulation and a reduced
cytoplasmic/nuclear ratio, a novel observation that suggests a
potential regulatory mechanism by GNL2 in glioma pathogenesis.
Dysregulation of ribosomal proteins such as RPL11, crucial for
ribosomal biogenesis, can incite cellular stress and contribute to
diseases including cancer (38). In
gliomas, manipulation of ribosomal proteins can affect key cellular
processes and their overexpression is linked to poor prognosis
(39,40). Thus, understanding the role of RPL11
in glioma may provide important molecular insight and potential
therapeutic targets. Furthermore, GNL2 silencing inhibited overall
protein synthesis, implying GNL2 may govern protein synthesis via
RPL11 localization regulation. The current findings suggest a
mechanism through which GNL2 influences the oncogenic behavior of
glioma, potentially via RPL11, highlighting GNL2′s promise as a
therapeutic target for glioma. This combined evidence further
solidifies the key position of these genes among the prognostic
indicators of gliomas and provides a broad perspective for future
treatments. The present study revealed the critical role of these
genes in tumor development and patient prognosis, laying a
foundation for a deeper understanding of glioma biology and disease
mechanisms. In-depth study of these genes not only enables more
accurate assessment of patient prognosis, but also supports
individualized treatment. Understanding gene function and
expression patterns can help optimize therapeutic regimens and
improve targeting and effectiveness. The findings provide important
clues for the development of new therapeutic strategies and are
expected to improve the outcome of glioma treatment. The current
study lays a foundation for incorporating genetic information into
the clinical management of gliomas, facilitating the application of
personalized medicine in this field, improving patient survival and
quality of life, and pointing the way to future research and
clinical practice. Although the present study highlights the
critical role of specific genes, possible limitations in sample
size and patient heterogeneity need to be recognized, and larger
and multicenter studies are needed to address these challenges in
the future.
In conclusion, by bioinformatics analysis, 3
promising prognostic biomarkers in glioma were identified in the
present study, namely GNL2, PDCD11 and RPS6. Furthermore, GNL2 was
investigated as the hub gene in the present study, and it was found
to be upregulated in glioma tissues and closely connected with poor
prognosis. GNL2 silencing inhibits glioma cell growth and impairs
the export of 60S ribosomal subunits and promotes overall protein
synthesis in glioma cells. Collectively, GNL2 promotes the protein
synthesis of RPL11 to facilitate the development of glioma. All of
these findings provide new hints for clinical applications for
glioma.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XY and XL participated in the conception and design
of the study, as well as data collection and analysis. XY and XL
were involved in drafting the manuscript or revising it critically
for intellectual content. XY and XL confirm the authenticity of all
the raw data. Both XY and XL read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Elia-Pasquet S, Provost D, Jaffré A,
Loiseau H, Vital A, Kantor G, Maire JP, Dautheribes M, Darrouzet V,
Dartigues JF, et al: Incidence of central nervous system tumors in
Gironde, France. Neuroepidemiology. 23:110–117. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Persano L, Rampazzo E, Basso G and Viola
G: Glioblastoma cancer stem cells: Role of the microenvironment and
therapeutic targeting. Biochem Pharmacol. 85:612–622. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reddy AT and Wellons JC III: Pediatric
high-grade gliomas. Cancer J. 9:107–112. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Perkins SM, Rubin JB, Leonard JR, Smyth
MD, El Naqa I, Michalski JM, Simpson JR, Limbrick DL, Park TS and
Mansur DB: Glioblastoma in children: A single-institution
experience. Int J Radiat Oncol Biol Phys. 80:1117–1121. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bălașa A, Șerban G, Chinezu R, Hurghiș C,
Tămaș F and Manu D: The involvement of exosomes in glioblastoma
development, diagnosis, prognosis, and treatment. Brain Sci.
10:5532020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Seymour T, Nowak A and Kakulas F:
Targeting aggressive cancer stem cells in glioblastoma. Front
Oncol. 5:1592015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Anji A and Kumari M: Guardian of genetic
messenger-RNA-binding proteins. Biomolecules. 6:42016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hentze MW, Castello A, Schwarzl T and
Preiss T: A brave new world of RNA-binding proteins. Nat Rev Mol
Cell Biol. 19:327–341. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Muleya V and Marondedze C: Functional
roles of RNA-Binding Proteins in plant signaling. Life. 10:2882020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Idler RK and Yan W: Control of messenger
RNA fate by RNA-binding proteins: An emphasis on mammalian
spermatogenesis. J Androl. 33:309–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li X, Zhang F, Ma J, Ruan X, Liu X, Zheng
J, Liu Y, Cao S, Shen S, Shao L, et al: NCBP3/SNHG6 inhibits GBX2
transcription in a histone modification manner to facilitate the
malignant biological behaviour of glioma cells. RNA Biol. 18:47–63.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gerstberger S, Hafner M and Tuschl T: A
census of human RNA-binding proteins. Nat Rev Genet. 15:829–845.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tailler M, Lindqvist LM, Gibson L and
Adams JM: By reducing global mRNA translation in several ways,
2-deoxyglucose lowers MCL-1 protein and sensitizes hemopoietic
tumor cells to BH3 mimetic ABT737. Cell Death Differ. 26:1766–1781.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Okuwaki M, Saito S, Hirawake-Mogi H and
Nagata K: The interaction between nucleophosmin/NPM1 and the large
ribosomal subunit precursors contribute to maintaining the
nucleolar structure. Biochim Biophys Acta Mol Cell Res.
1868:1188792021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yelland JN, Bravo JPK, Black JJ, Taylor DW
and Johnson AW: A single 2′-O-methylation of ribosomal RNA gates
assembly of a functional ribosome. Nat Struct Mol Biol. 30:91–98.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakamura K, Reid BM, Chen A, Chen Z, Goode
EL, Permuth JB, Teer JK, Tyrer J, Yu X, Kanetsky PA, et al:
Functional analysis of the 1p34.3 risk locus implicates GNL2 in
high-grade serous ovarian cancer. Am J Hum Genet. 109:116–135.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ostrom QT, Bauchet L, Davis FG, Deltour I,
Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh
KM, et al: The epidemiology of glioma in adults: a ‘state of the
science’ review. Neuro Oncol. 16:896–913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiao H, Ding N, Liao H, Yao Z, Cheng X,
Zhang J and Zhao M: Prediction of relapse and prognosis by
expression levels of long noncoding RNA PEG10 in glioma patients.
Medicine (Baltimore). 98:e175832019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Delgado-López P and Corrales-García E:
Survival in glioblastoma: A review on the impact of treatment
modalities. Clin Transl Oncol. 18:1062–1071. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Van Meir EG, Hadjipanayis CG, Norden AD,
Shu HK, Wen PY and Olson JJ: Exciting new advances in
neuro-oncology: The avenue to a cure for malignant glioma. CA
Cancer J Clin. 60:166–193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wohlfart S, Khalansky AS, Gelperina S,
Maksimenko O, Bernreuther C, Glatzel M and Kreuter J: Efficient
chemotherapy of rat glioblastoma using doxorubicin-loaded PLGA
nanoparticles with different stabilizers. PLoS One. 6:e191212011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chow AKM, Yau SWL and Ng L: Novel
molecular targets in hepatocellular carcinoma. World J Clin Oncol.
11:589–605. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chaillou T, Kirby TJ and McCarthy JJ:
Ribosome biogenesis: Emerging evidence for a central role in the
regulation of skeletal muscle mass. J Cell Physiol. 229:1584–1594.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Henras AK, Plisson-Chastang C, O'Donohue
MF, Chakraborty A and Gleizes PE: An overview of pre-ribosomal RNA
processing in eukaryotes. Wiley Interdiscip Rev RNA. 6:225–242.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoshida Y, Wang H, Hiwasa T, Machida T,
Kobayashi E, Mine S, Tomiyoshi G, Nakamura R, Shinmen N, Kuroda H,
et al: Elevation of autoantibody level against PDCD11 in patients
with transient ischemic attack. Oncotarget. 9:8836–8848. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eid W, Hess D, König C, Gentili C and
Ferrari S: The human exonuclease-1 interactome and phosphorylation
sites. Biochem Biophys Res Commun. 514:567–573. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xing Z, Wang R and Wang X, Liu J, Zhang M,
Feng K and Wang X: CircRNA circ-PDCD11 promotes triple-negative
breast cancer progression via enhancing aerobic glycolysis. Cell
Death Discov. 7:2182021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Williams AJ, Werner-Fraczek J, Chang IF
and Bailey-Serres J: Regulated phosphorylation of 40S ribosomal
protein S6 in root tips of maize. Plant Physiol. 132:2086–2097.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cuperjani F, Gashi L, Kurshumliu F,
Dreshaj S and Selimi F: Relationship between ribosomal protein
S6-pS240 expression and other prognostic factors in non-special
type invasive breast cancer. Breast Care (Basel). 14:171–175. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shirakawa Y, Ohta K, Miyake S, Kanemaru A,
Kuwano A, Yonemaru K, Uchino S, Yamaoka M, Ito Y, Ito N, et al:
Glioma cells acquire stem-like characters by extrinsic ribosome
stimuli. Cells. 10:29702021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mahmoud AB, Ajina R, Aref S, Darwish M,
Alsayb M, Taher M, AlSharif SA, Hashem AM and Alkayyal AA: Advances
in immunotherapy for glioblastoma multiforme. Front Immunol.
13:9444522022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liau LM, Ashkan K, Brem S, Campian JL,
Trusheim JE, Iwamoto FM, Tran DD, Ansstas G, Cobbs CS, Heth JA, et
al: Association of autologous tumor lysate-loaded dendritic cell
vaccination with extension of survival among patients with newly
diagnosed and recurrent glioblastoma: A phase 3 prospective
externally controlled cohort trial. JAMA Oncol. 9:112–121. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Todo T, Ito H, Ino Y, Ohtsu H, Ota Y,
Shibahara J and Tanaka M: Intratumoral oncolytic herpes virus G47
for residual or recurrent glioblastoma: A phase 2 trial. Nat Med.
28:1630–1639. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen R, Smith-Cohn M, Cohen AL and Colman
H: Glioma subclassifications and their clinical significance.
Neurotherapeutics. 14:284–297. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Quiroga-Artigas G, de Jong D and
Schnitzler CE: GNL3 is an evolutionarily-conserved stem cell gene
influencing cell proliferation, animal growth, and regeneration in
the hydrozoan Hydractinia. Open Biol. 12:2201202022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dong Y, Cai Q, Fu L, Liu H, Ma M and Wu X:
Study of the G protein nucleolar 2 value in liver hepatocellular
carcinoma treatment and prognosis. Biomed Res Int.
2021:48736782021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kang J, Brajanovski N, Chan KT, Xuan J,
Pearson RB and Sanij E: Ribosomal proteins and human diseases:
Molecular mechanisms and targeted therapy. Signal Transduct Target
Ther. 6:3232021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guo P, Wang Y, Dai C, Tao C, Wu F, Xie X,
Yu H, Zhu Q, Li J, Ye L, et al: Ribosomal protein S15a promotes
tumor angiogenesis via enhancing Wnt/β-catenin-induced FGF18
expression in hepatocellular carcinoma. Oncogene. 37:1220–1236.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Elhamamsy AR, Metge BJ, Alsheikh HA,
Shevde LA and Samant RS: Ribosome biogenesis: A central player in
cancer metastasis and therapeutic resistance. Cancer Res.
82:2344–2353. 2022. View Article : Google Scholar : PubMed/NCBI
|