Introduction
Renal cell carcinoma (RCC) is one of the most common
cancers in the world, ranking sixth and tenth in men and women,
respectively (1). Clear cell
(cc)RCC accounts for 75–80% of all instances of RCC, with the
remaining % comprised of several non-cc cancer subtypes (2,3). The
incidence of ccRCC, also known as kidney renal clear cell carcinoma
(KIRC), has progressively increased in recent years, accounting for
~5% of adult malignant tumors (4,5).
Therefore, its treatment and development warrant increased
attention. Early ccRCC is often overlooked by patients due to its
mild symptoms, such as fever and fatigue, with ~30% of patients
already presenting with metastasis at the time of diagnosis
(6–8). Although surgery can be used for tumor
removal, ~25% of patients still experience relapse and metastasis
(9). The 5-year survival rate is
notably high at 90% for early ccRCC but drops to ~33.3% for
advanced cases (10).
In recent years, the widespread use of immune
checkpoint inhibitors (ICIs) in tumor therapy, especially for
ccRCC, has marked revolutionary progress. Immunotherapy
demonstrates a significant advantage over conventional
chemotherapy, radiation therapy and standard tyrosine kinase
inhibitor therapy, either alone or combined with other agents such
as nivolumab plus ipilimumab or cabozantinib, pembrolizumab plus
axitinib or lenvatinib, and avelumab plus axitinib (11). Additionally, several biological
target molecules associated with the diagnosis and prognosis of RCC
immunotherapy have been elucidated, including programmed cell death
ligand 1 (PD-L1), tumor mutation burden and specific single gene
mutations, such as Von Hippel-Lindau, polybromo 1, SET domain
containing 2, breast cancer gene 1-associated protein 1 and lysine
demethylase 5C. Moreover, DNA-damage repair gene alterations,
including common mutations like checkpoint kinase 2, ataxia
telangiectasia mutated kinase, MutS homolog 6 and MutY DNA
glycosylase, are associated with immunotherapy (12). There are highly infiltrating immune
cells in ccRCC, and it was one of the first cancers used in
immunotherapy (13); however, the
molecular mechanism of ccRCC development is still unclear.
Furthermore, the results of a meta-analysis by Santoni et al
(14) demonstrated that sex-related
differences in the efficacy of ICIs remain an important but
undiscussed aspect in cancer immunotherapy trials. In addition,
clinical trials should emphasize assessment such as quality of life
(15) and European Cancer
Organization performance status (16) when evaluating the treatment effect
of ICIs in RCC.
The signaling lymphocyte activation molecule family
(SLAMF) is a group of receptors that are closely associated with
the immune system. They are mainly expressed in hematopoietic cells
and immune cells, and they have also been reported in cancer cells
(17,18). The family has nine members,
including SLAMF1 (CD150), SLAMF2 (CD48), SLAMF3 (CD299; Ly-9),
SLAMF4 (CD244, 2B4), SLAMF5 (CD84), SLAMF6 (CD352; Ly108; NTB-A),
SLAMF7 (CD319; CRACC; CS1), SLAMF8 (CD353; BLAME) and SLAMF9
(CD84H1; CD2F10; SF2001) (19).
Increasing research results indicate that SLAMFs serve a crucial
role in tumor immune regulation (18,20).
SLAMF1 and SLAMF3 have been reported to be the key genes regulating
immune infiltration in ovarian cancer (21). The activation of SLAMF5 upregulates
PD-L1 expression and myeloid-derived suppressor cells formation,
inhibiting T cell function (22).
Reduced SLAMF7 in mouse models leads to decreased programmed cell
death protein 1 (PD-1) in CD8+ T cells, reducing T cell failure and
tumor progression (23).
Additionally, SLAMF4 and SLAMF6 are associated with immune cell
exhaustion (24,25). However, the molecular mechanism of
SLAMFs in ccRCC remains insufficiently investigated.
In the present study, the expression of SLAMFs in
ccRCC was analyzed on the basis of The Cancer Genome Atlas (TCGA)
and Genotype-Tissue Expression (GTEx) data. The relationship
between SLAMFs expression and prognosis in ccRCC was assessed using
the Kaplan-Meier database. Finally, the Tumor Immune Estimation
Resource (TIMER) was used to assess the correlation of SLAMFs with
immune infiltration. The findings of the present study contribute
to the understanding of the molecular mechanisms of SLAMFs in ccRCC
development.
Materials and methods
mRNA expression levels of SLAMFs in
cancers
TCGA (https://portal.gdc.cancer.gov/) is a comprehensive
cancer analysis database that contains data, such as mRNA
expression, micro mi(RNA) expression, patient clinical information
and methylation. The GTEx portal (https://www.genome.gov/Funded-Programs-Projects/Genotype-Tissue-Expression-Project)
serves as a database for transcriptome sequencing of human tissues
(26). Data from the TCGA and GTEx
databases were downloaded and processed for analysis in the present
study (27). The mRNA levels of
SLAMFs in 33 cancer and paracancerous tissues were assessed using R
software (version 4.2.0; The R Foundation). Additionally, the
transcription levels of SALMF1/4/7/8 in ccRCC cell lines were
evaluated based on GSE20491 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20491).
Analysis of SLAMFs with clinical
characteristics
Gene Expression Profiling Interactive Analysis
(GEPIA)2 (http://gepia2.cancer-pku.cn/#index) is a comprehensive
web server designed for large-scale expression profiling and
interactive analysis. It encompasses modules such as gene
expression modules, prognostic modules and clinical features
modules (28). The University of
Alabama at Birmingham Cancer data analysis Portal (UALCAN;
http://ualcan.path.uab.edu/index.html) is a dedicated
portal for facilitating tumor subgroup gene expression and survival
analyses (29). The correlations of
SLAMFs with clinical features of ccRCC, such as cancer stage,
ethnicity, sex, age, tumor grade and lymph node metastasis, were
assessed using the GEPIA2 and UALCAN databases.
Analysis of diagnostic value and
survival rate
Receiver operating characteristic (ROC) analysis, a
widely accepted method for analyzing and comparing diagnostic
accuracy (30), was used to assess
the diagnostic value of SLAMFs in ccRCC. The area under the curve
(AUC) from the ROC curves, indicative of the diagnostic value, was
calculated using data from the TCGA database. To evaluate the
impact of SLAMFs on survival in cancers, especially ccRCC, the
Kaplan-Meier Plotter (http://kmplot.com/analysis) was used. This platform
incorporates data on 30,000 genes (mRNA, miRNA and protein) and
covers 21 types of tumors (31).
The overall survival (OS) of SLAMFs in ccRCC was analyzed using the
Kaplan-Meier Plotter with default settings, and the data set of
kidney renal clear cell carcinoma (n=530) was used for this
analysis.
Analysis of gene mutations and
correlation
The cBioPortal website (https://www.cbioportal.org/) serves as a comprehensive
database for assessing the genomic features of tumors. Currently,
it stores genomic information, such as DNA copy number data, mRNA
and miRNA expression data, non-synonymous mutations and protein
level and phosphoprotein level data. Gene alterations of SLAMFs
were evaluated using the cBioPortal portal (32,33).
Furthermore, an analysis of the correlations and co-expression of
genes with SLAMFs in ccRCC was performed using the TCGA database.
Genes with a correlation coefficient >0.6 and P<0.001 were
screened. The intersection of co-expressed genes of SLAMFs was
depicted using an online Venn diagram tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Subsequently, the top 20 co-expressed genes were selected, and
their correlations to SLAMFs were plotted.
Gene enrichment analysis
The Gene Ontology (GO) database (http://geneontology.org/) categorizes protein function
into three components based on its biological information:
Biological process (BP), cellular component (CC) and molecular
function (MF). Additionally, the Kyoto Encyclopedia of Genes and
Genomes (KEGG) database (https://www.genome.jp/kegg/) was used to assess the
pathways associated with the genes. To evaluate the molecular
mechanism of ccRCC, GO/KEGG analyses of the co-expressed genes
related to SLAMFs were performed. The threshold for GO/KEGG
analysis was P.adj<0.05, and the threshold for gene count with
the default setting was >1. Furthermore, gene set enrichment
analysis (GSEA) of the co-expression genes of SLAMFs was performed
(34). The gene sets used for GSEA
were obtained from the Molecular Signatures Database (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp).
Analysis of immune cell infiltration
and somatic copy number
TIMER (https://cistrome.shinyapps.io/timer) is a web server
designed for the comprehensive analysis of tumor-infiltrating
immune cells (TIICs) in several cancer types based on TCGA
(35). The abundance of TIICs,
including B cells, CD4+ T cells, CD8+ T
cells, neutrophils, macrophages and dendritic cells, was evaluated
using the TIMER portal. The estimate algorithm was used to analyze
the immune score in the overall internal environment of ccRCC.
Furthermore, the somatic copy number alterations (SCNA) module in
TIMER was used to assess the relationship between SLAMFs SCNA and
tumor immune-related cell infiltration levels (35). Additionally, the Tumor Immune
Dysfunction and Exclusion (TIDE) score was used to assess the
efficacy of immune checkpoint blockade (ICB) in ccRCC with high and
low expression of SLAMF (4,9,17,21,36).
Correlation analysis of SLAMFs with
immunoinhibitory checkpoints
TIMER2.0 (http://timer.cistrome.org/) offers three modules,
immunity, exploration and estimation, to assess the associations
between immune infiltrates and genetic or clinical features based
on the TCGA cohorts (37). In
comparison with TIMER, TIMER 2.0 provides more detailed information
on immune infiltrates cells. The present study used TIMER 2.0 to
evaluate the correlation between three tumor-related immune cells
[(M2, Treg and cancer-associated fibroblasts (CAFs)] and the
expression of SLAMFs by three algorithms (EPIC, QUANTISEQ and
CIBERSORT-ABS). Additionally, using R software (version 4.2.0), the
correlations between PD-1, PD-L1, PD-L2 and SLAMF were
assessed.
Correlation analysis of SLAMFs with
immune-related molecules
Chemokine/chemokine receptors serve a crucial role
in the development and homeostasis of the immune system,
influencing several immune and inflammatory responses (38). Furthermore, chemokine/chemokine
receptors are implicated in several essential steps of tumor
metastasis, such as tumor cell adhesion, vascular extravasations,
metastatic colonization, angiogenesis and proliferation (39). In addition, major histocompatibility
complex (MHC) molecules are key players in antigen presentation and
the induction of immune responses (40). Therefore, an analysis was performed
to assess the correlations between SLAMFs expression and
chemokines/chemokine receptors, as well as with MHC molecules in
ccRCC.
Effects of immune cells combined with
SLAMFs on the prognosis of ccRCC
The Kaplan-Meier plotter website was used to analyze
the impact of SLAMFs on the prognosis of ccRCC considering changes
in the content of immune cells. Using the four types of immune
cells available in the TIMER database (https://cistrome.shinyapps.io/timer) (35), namely B cells, CD4+ T
cells, CD8+ T cells and macrophages, their effects on
the prognosis of patients with ccRCC in relation to SLAMFs
expression was assessed.
Cell culture and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
The cell lines were purchased from Pricella (Procell
Life Science & Technology Co., Ltd.), and the reverse
transcription kit was purchased from Thermo Fisher Scientific, Inc.
Human ccRCC 786-O and 769-P cell lines were cultured in an
incubator (37°C; 5% CO2) in RPMI 1640 medium containing
10% fetal bovine serum (cat. no. C04001; ViaCell Inc.) and 1%
penicillin/streptomycin (cat. no. C0222; Beyotime Institute of
Biotechnology) for 2 days. To assess the expression levels of
SLAMF1/4/7/8 in the ccRCC cell lines, cell clusters from 786-O and
769-P were extracted. Total RNA was extracted with RNAiso PLUS
(cat. no. 9108; Takara Bio, Inc.), and then cDNA was synthesized
using the RevertAid First Strand cDNA Synthesis Kit (cat. no.
K1622; Thermo Fisher Scientific, Inc.) according to the
instructions provided by the manufacturer. qPCR was performed with
UltraSYBR Mixture containing high ROX (cat. no. CW2602M; CoWin
Biosciences) following the manufacturer's instructions. The mRNA
expression of SLAMF members in ccRCC cell lines was assessed using
the human kidney HK-2 cell line as control cells and 18S rRNA as an
internal reference. The primers for RT-qPCR were designed by
PrimerBank (The Massachusetts General Hospital; http://pga.mgh.harvard.edu/primerbank/index.html)
(41) and the sequences are as
follows: SLAMF1 (NM_003037), sense: 5′-GGAGAACAGTGTCGAGAACAAA-3′
and antisense: 5′-CGTATCCCCAGGGTGAGATTC-3′; SLAMF4 (also known as
CD244, NM_001166663), sense: 5′-TCGTGATTCTAAGCGCACTGT-3′ and
antisense: 5′-CAGGTTCTTGTGACGTGGGAG-3′; SLAMF7 (NM_021181), sense:
5′-GGCAGCTCACAGGGTCAG-3′ and antisense:
5′-GGGTTGTGTTGAAGGTCCAGA-3′; SLAMF8 (NM_020125), sense:
5′-CTTCTCTGGGAAGATGCAGTG-3′ and antisense:
5′-TTTCGCTGATGTTGGGGGC-3′; 18S rRNA, sense:
5′-GTAACCCGTTGAACCCCATT-3′ and antisense:
5′-CCATCCAATCGGTAGTAGCG-3′ (42).
Relative mRNA expression was calculated using the comparative
2−ΔΔCq method (43).
Statistical analysis
All analyses were performed using R software
(version 4.2.0; The R Foundation; http://www.r-project.org/) (44), with graph making and statistical
analysis using the Xiantao Academic Platform (www.xiantaozi.com) and GraphPad (version 9.5.1). The
Wilcoxon rank-sum test was used to analyze differences in
expression in Figs. 1A, S1, 5E,
8B and 13. The paired t-test was used to analyze
differences in Fig. 1C. One-way
ANOVA was performed on the GEPIA website to analyze the differences
in gene expression between pathological stage in Fig. 2A-I. One-way ANOVA with Tukey's
honestly significant difference test was used for analysis of
Fig. 5A-D and F. Spearman's rank
was used for correlation analysis. pROC package (version 1.17.0.1)
(https://cran.r-project.org/web/packages/pROC/index.html)
was used for the ROC curve analysis, whilst the clusterprofiler
package (version 3.14.3) was used for enrichment analysis and
visualization (45). The estimate
package (version 1.0.13) (https://rdrr.io/rforge/estimate/) was used to analyze
the immune score (46). P<0.05
was considered to indicate a statistically significant
difference.
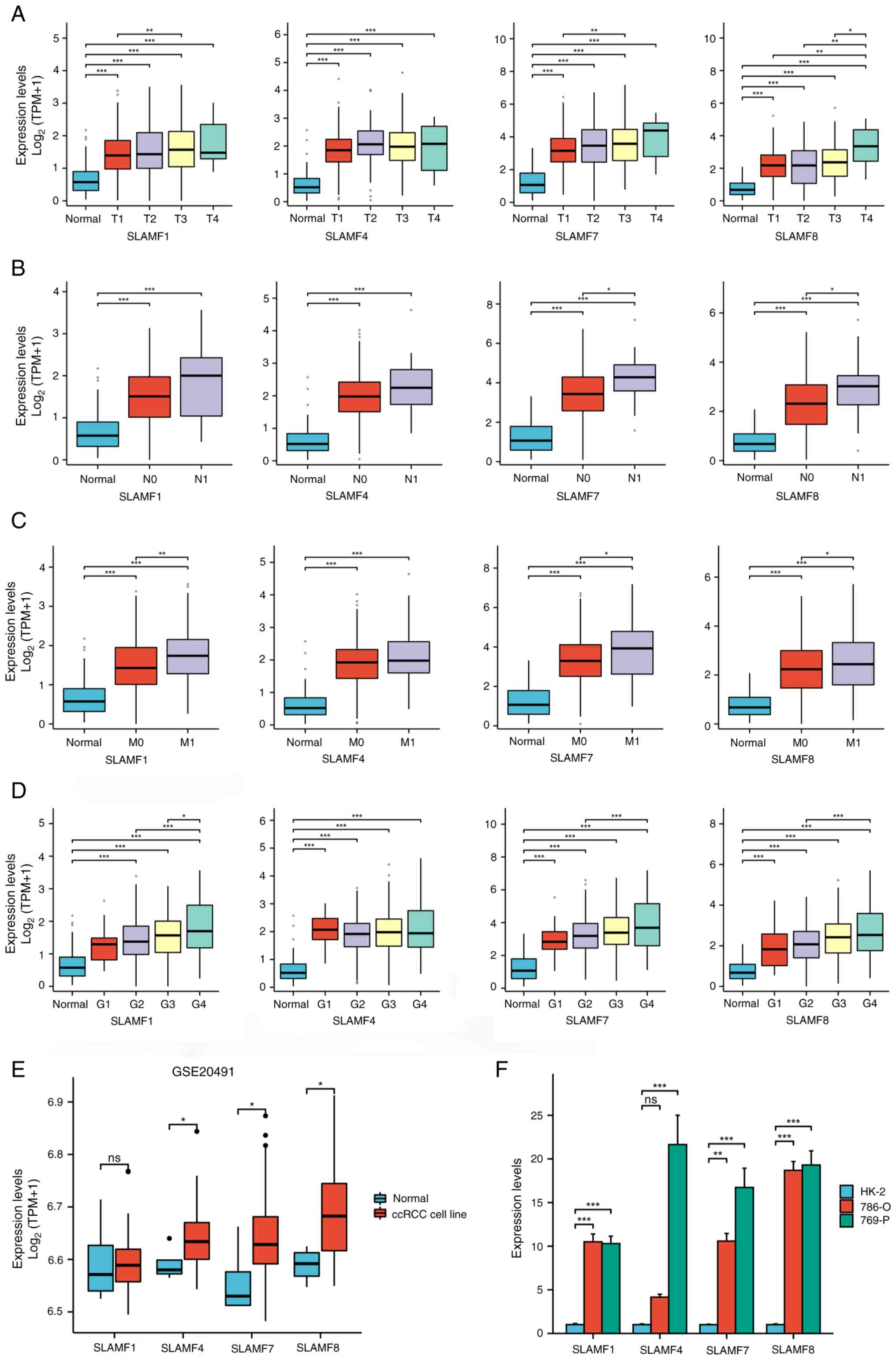 | Figure 5.Expression of SLAMF members in
different tumor stages and cell lines. Associations between SLAMF1,
4, 7 and 8 expression levels with different (A) T, (B) N and (C) M
staging, and (D) grading. Differential expression of SLAMF1, 4, 7
and 8 in (E) normal and ccRCC cell lines in GSE20491, and (F)
normal (HK-2) and ccRCC cell lines (786-O and 769-P). *P<0.05;
**P<0.01; ***P<0.001. SLAMF, signaling lymphocyte activation
molecule family; T, tumor; N, node; M, metastasis; G, grade; ccRCC,
clear cell renal cell carcinoma; TPM, transcript per million; ns,
not significant. |
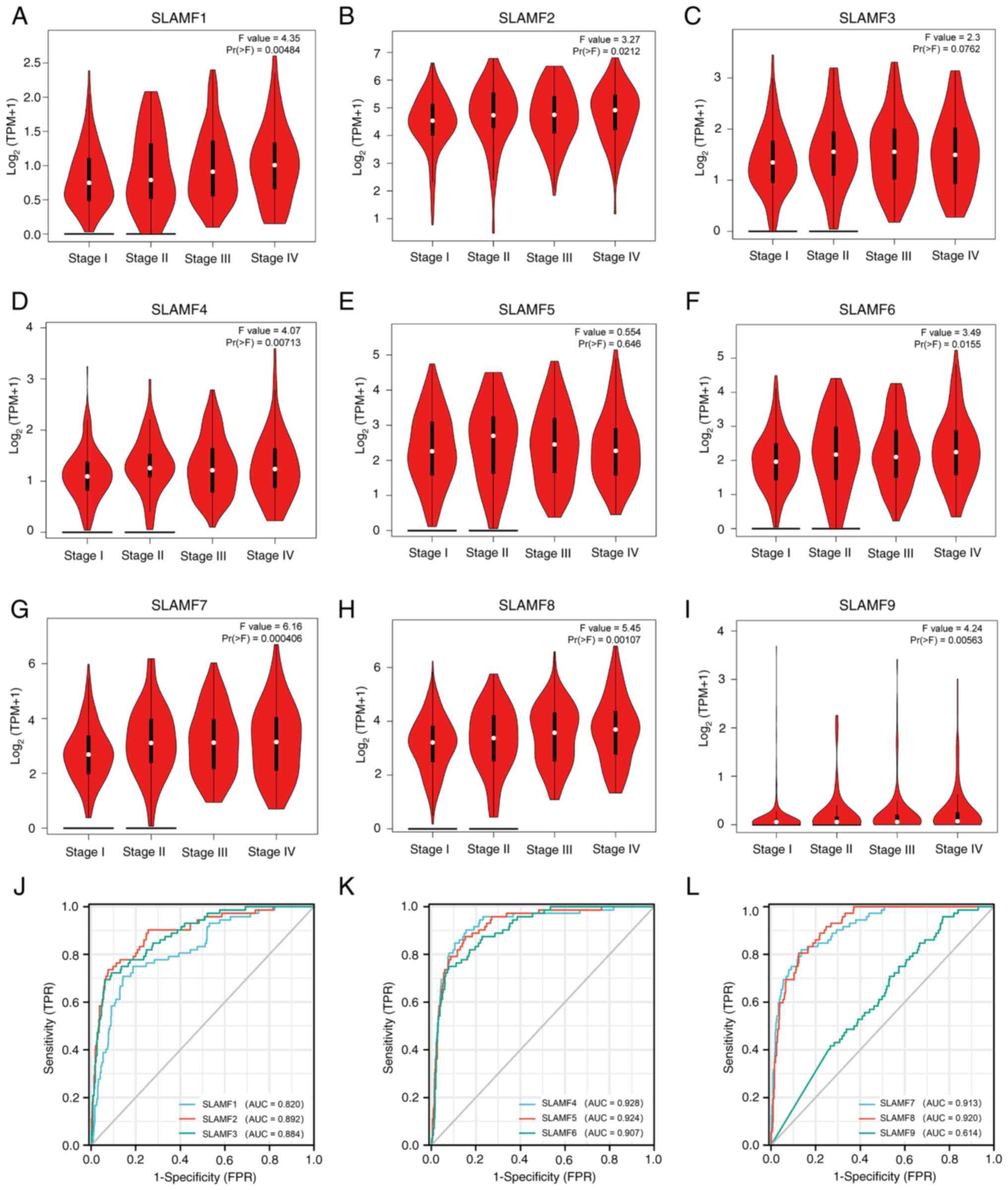 | Figure 2.SLAMF expression in different tumor
stages and the diagnostic value in ccRCC. Association between ccRCC
stage and (A) SLAMF1, (B) SLAMF2, (C) SLAMF3, (D) SLAMF4, (E)
SLAMF5, (F) SLAMF6, (G) SLAMF7, (H) SLAMF8 and (I) SLAMF9
expression, with log2(TPM+1) representing the gene transcription
level. Diagnostic value of (J) SLAMF1-3, (K) SLAMF4-6 and (L)
SLAMF7-9. SLAMF, signaling lymphocyte activation molecule family;
ccRCC, clear cell renal cell carcinoma; TPM, transcript per
million; FPR, false positive rate; TPR, true positive rate; AUC,
area under the curve. |
Results
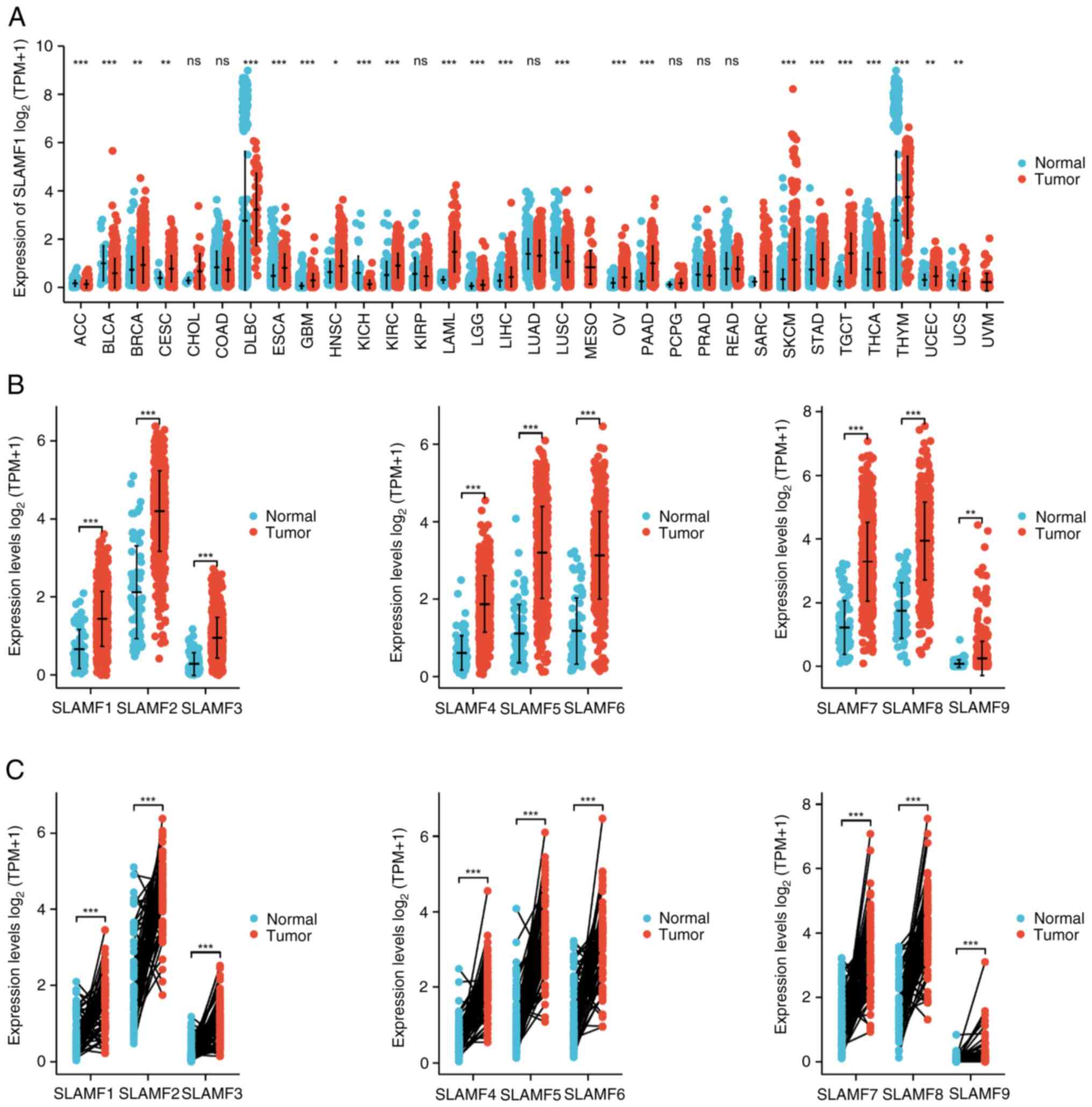
Increased transcription levels of
SLAMFs in ccRCC
To compare the expression of the SLAMFs in tumor
with normal tissues, the mRNA levels of the SLAMFs were assessed
using TCGA and GTEx data. The results revealed significantly
elevated expression of SLAMFs in esophageal carcinoma, glioblastoma
multiforme, KIRC, low-grade glioma, pancreatic adenocarcinoma, skin
cutaneous melanoma and stomach adenocarcinoma compared with
corresponding normal tissues (Figs.
1A and SI). Additionally,
SLAMFs (excluding SLAMF4) demonstrated significantly high
expression in ovarian cancer and testicular cancer compared with
normal tissue (Figs. 1A and
S1). Moreover, the expression of
SLAMFs in ccRCC was analyzed using both paired and unpaired
methods. The results demonstrated that the mRNA expression level of
SLAMFs in tumor tissues were significantly higher than those in
normal tissues (Fig. 1B and C).
SLAMFs are highly associated with
clinical features
To assess the association between SLAMFs with
clinical characteristics, two online analysis tools were used. The
results from the GEPIA2 database indicated that SLAMF3 and 5
demonstrated no statistically significant association with ccRCC
staging; however, SLAMF1, 2, 4, 6, 7, 8 and 9 were significantly
associated with the staging of ccRCC (Fig. 2A-I). Additionally, results from the
UALCAN database indicated that SLAMFs were significantly associated
with ethnicity, sex, age, tumor grade, ccRCC subtypes and nodal
metastasis status. Compared with lower grade and lower age group,
higher expression of SLAMFs was observed with significantly higher
tumor grade and age. The ccB subtype in ccRCC demonstrated a
significantly stronger association with SLAMF expression compared
with the ccA subtype (SLAMF1, 3, 7, 8 and 9) (Table SI). These findings suggest that
SLAMFs may contribute to the progression of ccRCC.
Prognosis of SLAMFs and associations
among members
The diagnostic value of SLAMFs was assessed using
ROC analysis, revealing high diagnostic values for SLAMF1-8 with
corresponding AUC values of 0.820, 0.892, 0.884, 0.928, 0.924,
0.907, 0.913 and 0.920, respectively. By contrast, SLAMF9
demonstrated a relatively low diagnostic value, with an AUC value
of 0.614 (Fig. 2J-L). Furthermore,
to assess the effect of SLAMFs on the survival rate of patients
with ccRCC, the effect of high and low expression of SLAMFs on OS
were compared (Fig. 3). The results
indicated that a high expression of SLAMF1, 4, 7, 8 and 9 was
significantly associated with a worse prognosis in ccRCC compared
with the low expression groups; however, the expression of SLAMF2,
3, 5 and 6 had no significant association with the prognosis of
ccRCC. These findings suggest that certain members of SLAMFs,
particularly SLAMF1, 4, 7, 8 and 9, may contribute to a poor
prognosis in ccRCC.
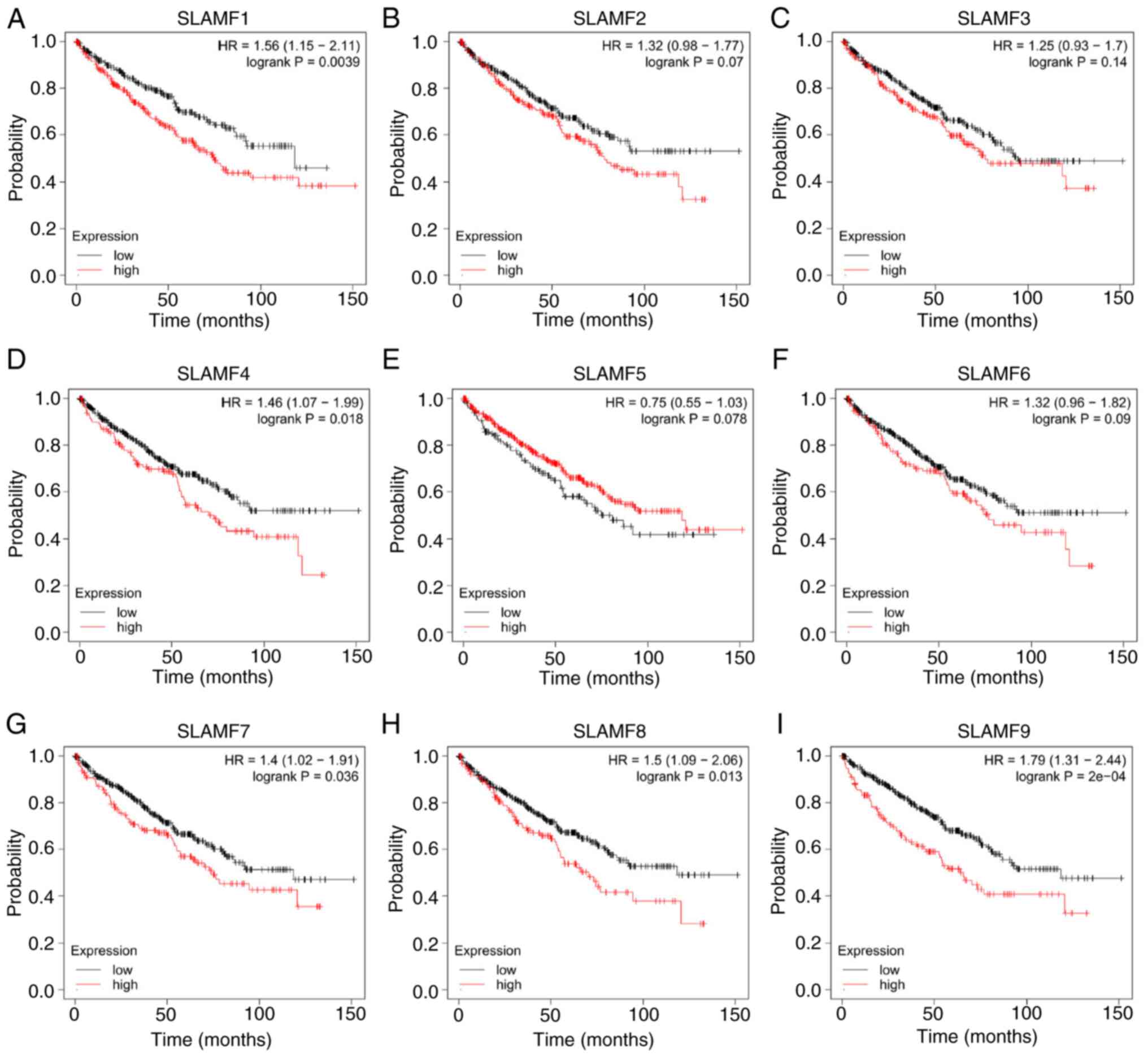 | Figure 3.Effects of SLAMF expression on
overall survival in patients with ccRCC. Association between the
prognosis of patients with ccRCC and the high and low expression of
(A) SLAMF1, (B) SLAMF2, (C) SLAMF3, (D) SLAMF4, (E) SLAMF5, (F)
SLAMF6, (G) SLAMF7, (H) SLAMF8 and (I) SLAMF9. SLAMF, signaling
lymphocyte activation molecule family; ccRCC, clear cell renal cell
carcinoma; HR, hazard ratio. |
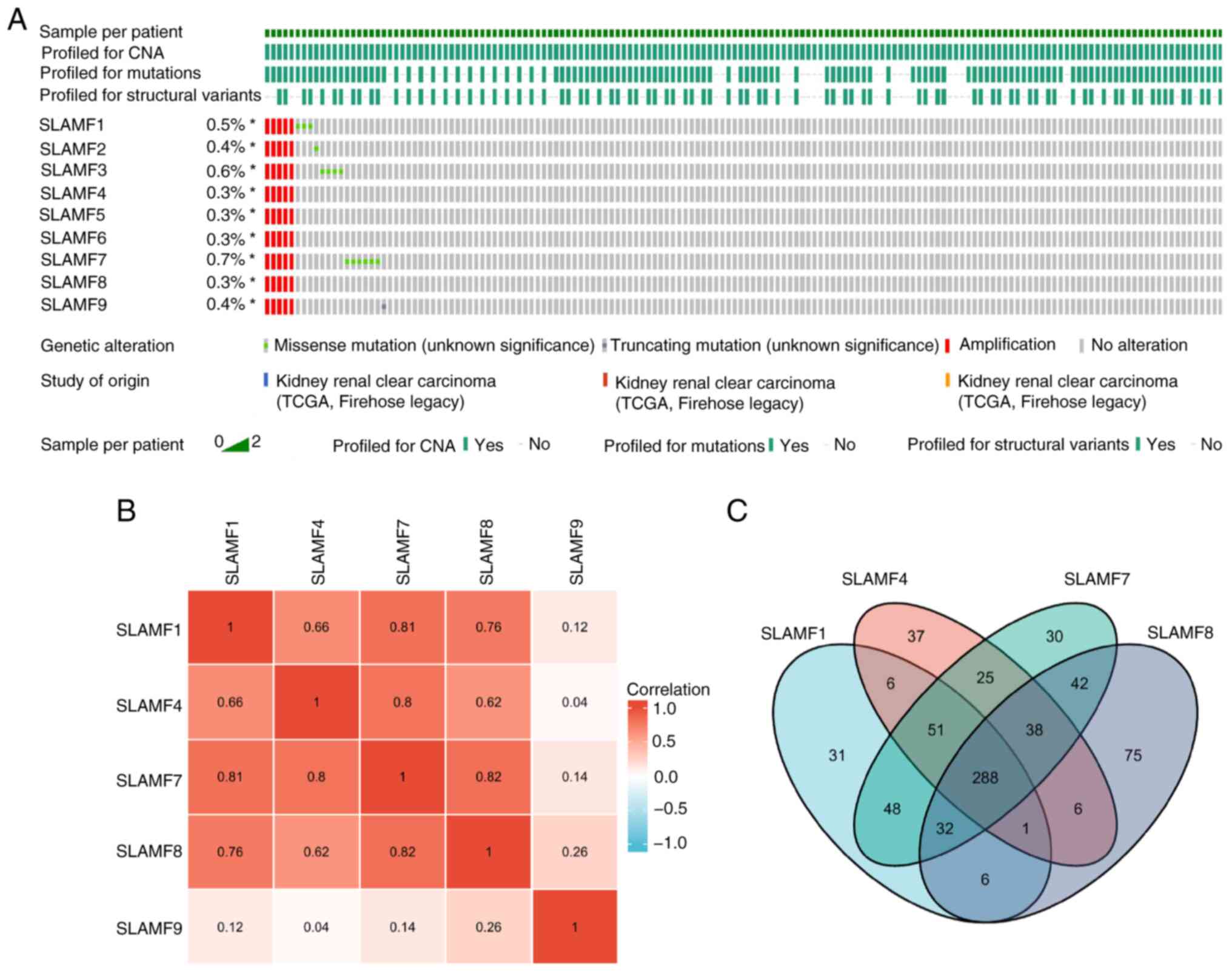
Mutation frequency and related genes
in SLAMFs
A comprehensive analysis of the mutation, copy
number and structure of the SLAMFs was performed, revealing their
primary involvement in missense mutations and amplifications
(Fig. 4A). According to the
analysis results, the main alteration of SLAMFs gene in ccRCC was
amplification, that is, the increase in gene copy number, which was
associated with an increase in SLAMFs gene expression. This was
consistent with the results shown in Fig. 1. Since SLAMFs have a significant
effect on the prognosis of patients with ccRCC, the co-expressed
genes of SLAMF1, 4, 7, 8 and 9 were screened. A notably strong
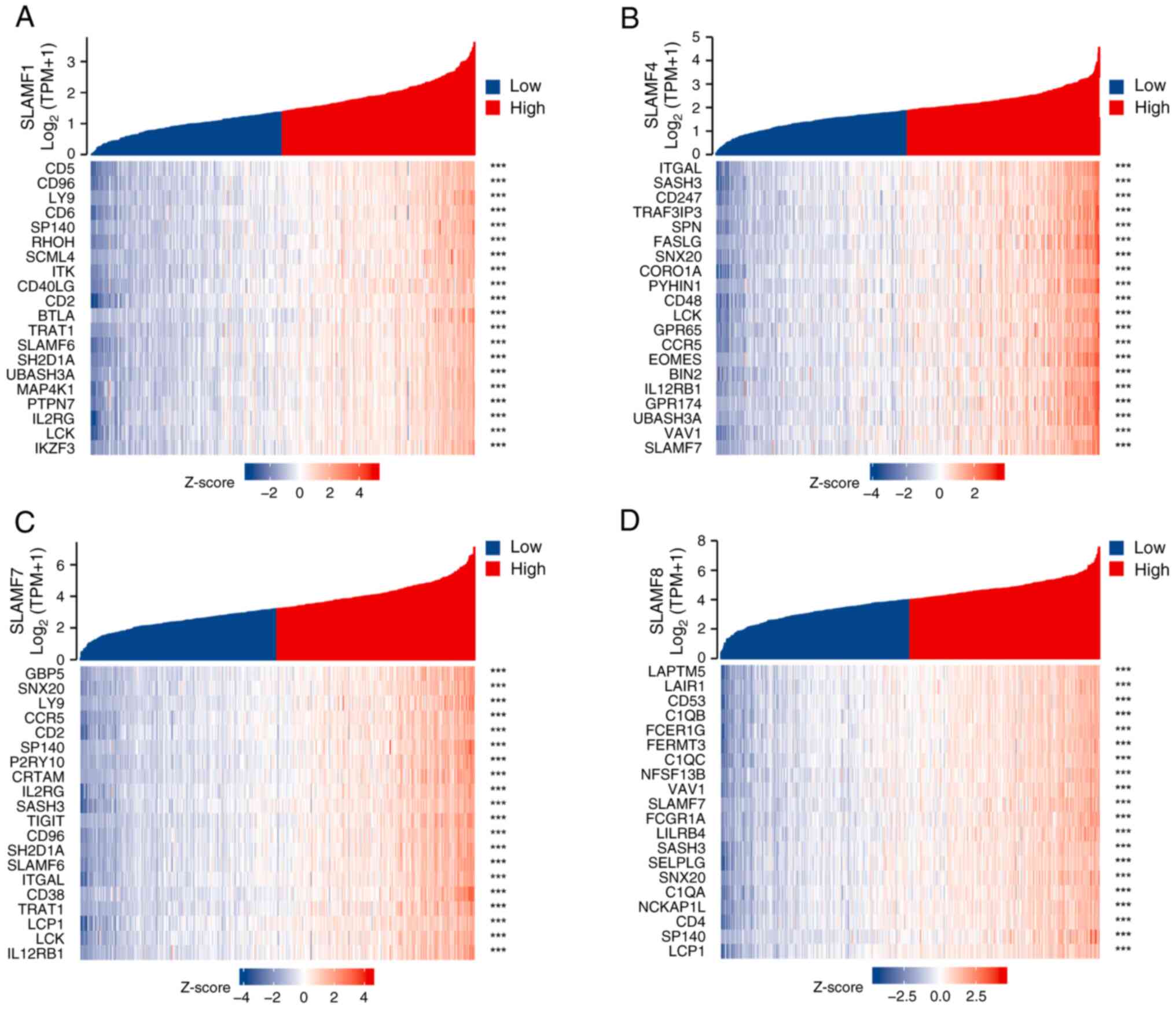
correlation was revealed for SLAMF1, 4, 7 and 8 (Fig. 4B). Subsequently, co-expressed genes
were screened for each SLAMF in ccRCC, resulting in 463
co-expressed genes identified for SLAMF1, 452 for SLAMF4, 544 for
SLAMF7, and 488 co-expressed genes for SLAMF8. In total, 288
co-expressed genes were identified for SLAMF1, 4, 7 and 8 (Fig. 4C). Additionally, the association
between SLAMF1, 4, 7 and 8 expression levels with different
tumor-node-metastasis staging and grading was analyzed (Fig. 5A-D). In addition, the expression of
SLAMF1, 4, 7 and 8 in ccRCC cell lines (786-O and 769-P) was higher
compared with that in a normal renal epithelial cell line (HK-2)
(Fig. 5E and F). Furthermore, the
top 20 genes that were among the 288 co-expressed genes associated
with SLAMF1, 4, 7 and 8 were assessed (Fig. 6) (P<0.05).
SLAMFs are associated with immune
activation
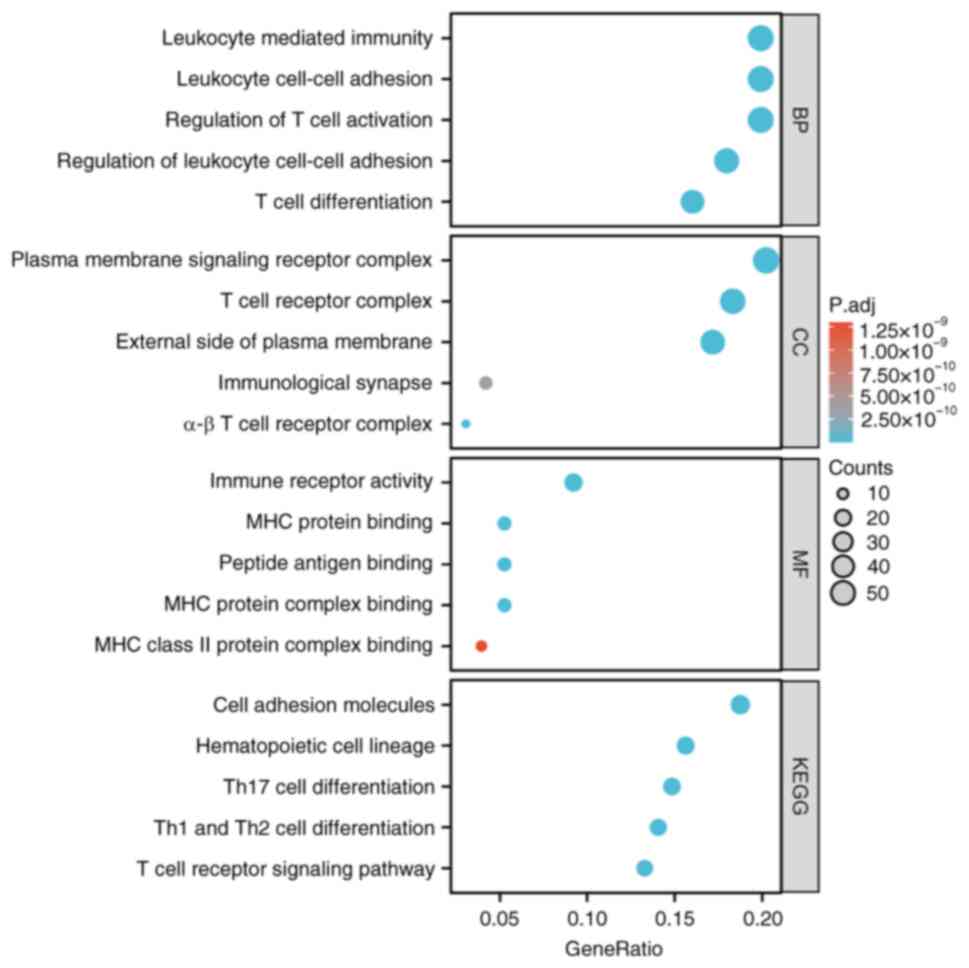
GO/KEGG analysis was performed on 288 co-expressed
genes associated with SLAMF1, 4, 7 and 8. The GO and BP analysis
demonstrated significant associations between T cell activation,
regulation of T cell activation, regulation of lymphocyte
activation, leukocyte cell-cell adhesion and positive regulation of
cell activation and SLAMF co-expression genes. CC analysis
highlighted a significant association between T cell receptors,
whilst MF analysis reveal a significant association with
cytokines/cytokines receptor activity and SLAMF co-expression genes
(P.adj<0.05; Fig. 7 and Table SII). KEGG pathway enrichment
analysis demonstrated that SLAMFs co-expression genes were
significantly associated with cell adhesion molecules (CAMs),
hematopoietic cell and helper T 1, 2 and 17 immune cell
differentiation, and T cell receptor signaling pathway (Fig. 7). In addition, KEGG enrichment
analysis was performed using the top 20 co-expressed genes
associated with SLAMF1, 4, 7 and 8 in (P.adj<0.05; Table SII). GSEA indicated involvement in
pathways such as the Jak-stat signaling pathway, toll like receptor
signaling pathway, B cell receptor signaling pathway and nod like
receptor signaling pathway (Table
SIII). These findings suggested that SLAMFs and their
co-expressed genes may participate in the regulating of the immune
system function and contribute to the development of ccRCC.
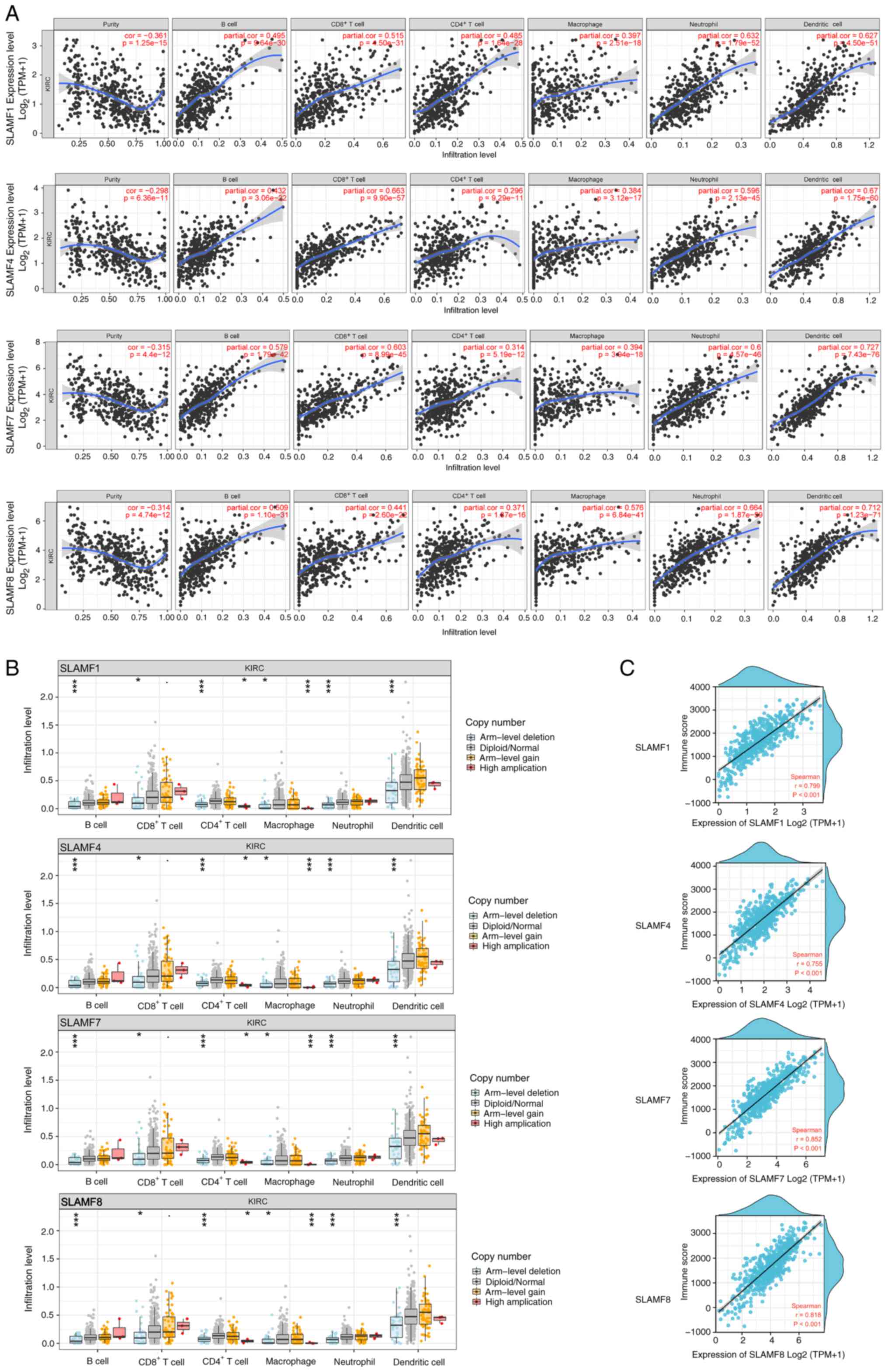
SLAMFs promote the infiltration of
immune cells
The TIMER results indicated that SLAMF1, 4, 7 and 8
were positively correlated with infiltration levels of B cells,
CD4+ T cells, CD8+ T cells, macrophages,
neutrophils and dendritic cells in ccRCC (Fig. 8A). To further assess these findings,
the correlations between SLAMF1, 4, 7 and 8 and immune cell gene
markers were analyzed, including B cells, CD8+ T cells,
T cells, monocytes, tumor-associated macrophages (TAMs), M1
macrophages, M2 macrophages and T cell exhaustion (Table SIV, Table SV, Table SVI, Table SVII). The results demonstrated that
SLAMF1, 4, 7 and 8 were significantly positively correlated with
gene markers of immune cells. This suggests that SLAMFs can promote
immune cell infiltration in the tumor microenvironment (TME).
Analysis of SCNA revealed that arm-level deletion of SLAMF1, 4, 7
and 8 was significantly associated with decreased infiltration
levels of B cells, CD4+ T cells, CD8+ T
cells, macrophages, neutrophils and dendritic cells (Fig. 8B; P<0.001). Moreover, the level
of immune infiltration in the ccRCC strongly and significantly
correlated with the expression of SLAMF1, 4, 7 and 8 (Fig. 8C). In summary, the results indicate
that SLAMF1, 4, 7 and 8 can promote immune cells infiltration in
ccRCC.
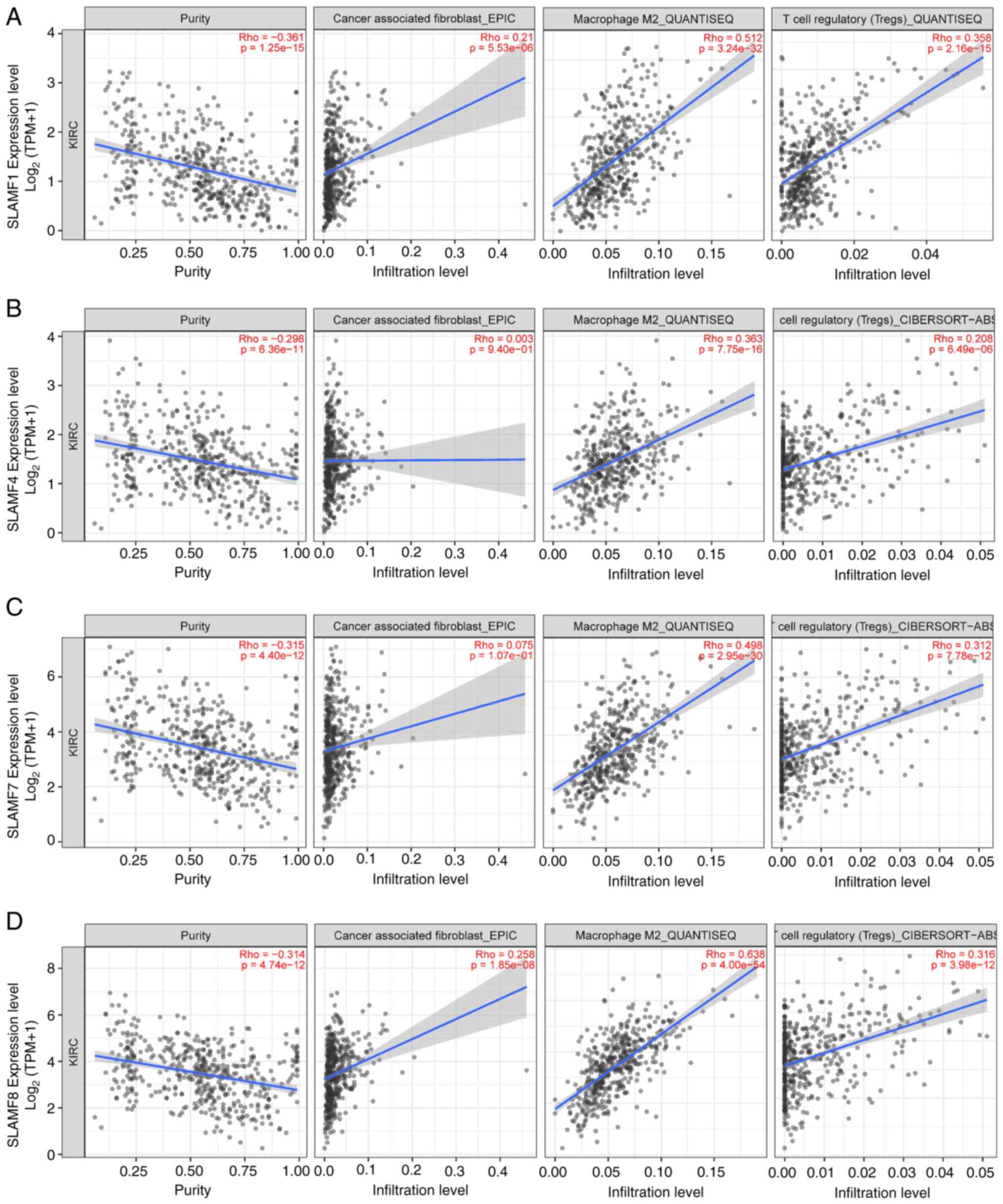
Association of the SLAMFs with
immunoinhibitors
A total of three algorithms were used (EPIC,
QUANTISEQ and CIBERSORT-ABS) based on the TIMER database to analyze
the correlations between the infiltration levels of M2 macrophages,
Treg cells and CAFs in ccRCC and the expression of SLAMFs (Fig. 9). The infiltration levels of M2
macrophages and Treg cells were significantly positively correlated
with the expressions of SLAMF1, 4, 7 and 8. Meanwhile, the
infiltration level of CAFs was only significantly correlated with
the expression of SLAMF1 and 8. Nevertheless, the overall trend
indicates a notable upregulation of tumor-related immune cells with
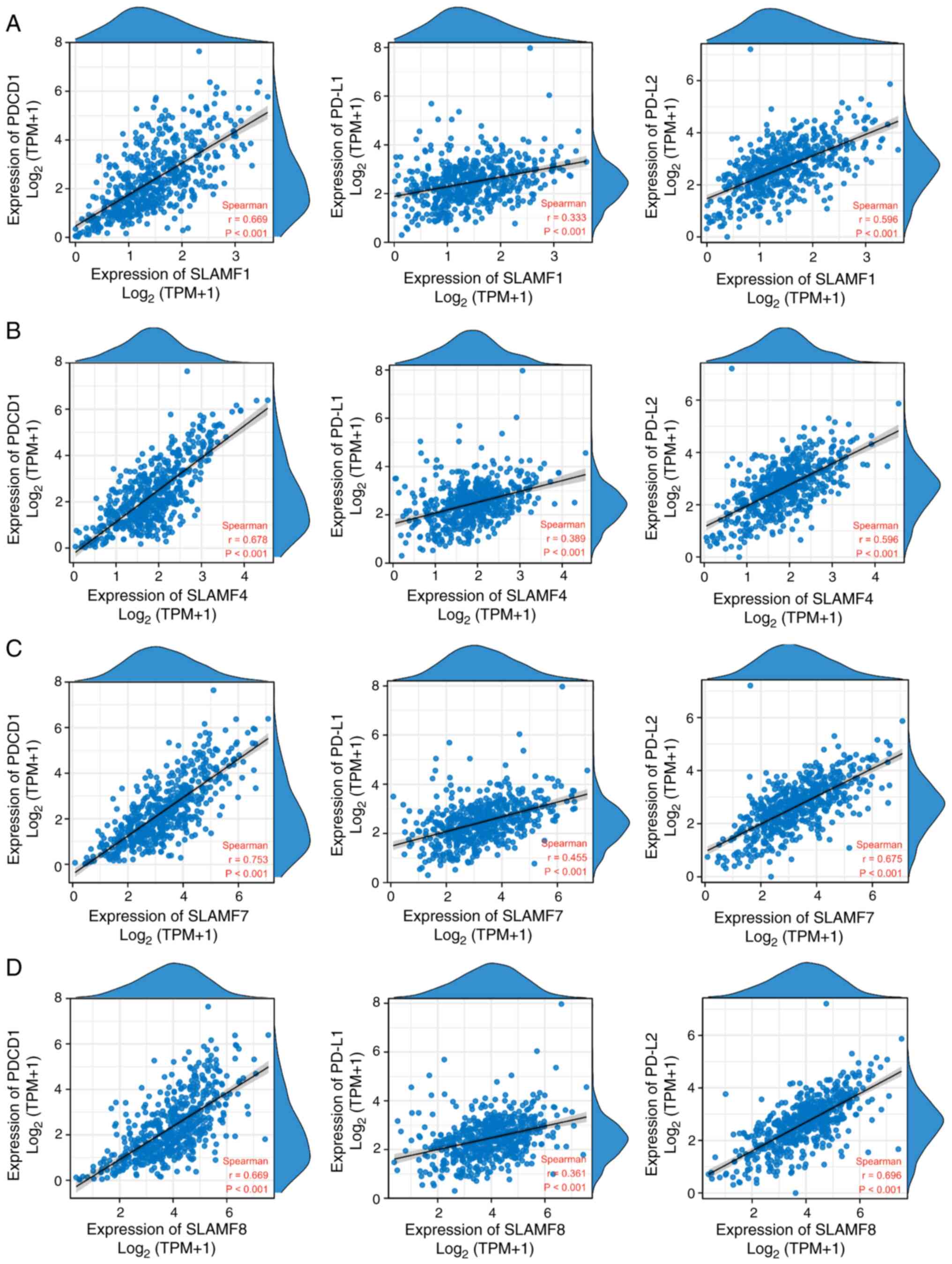
increased expression of SLAMF members. Additionally, PD-1 is a
crucial immunosuppressive molecule (47), and the results of the present study
demonstrated that the expression of SLAMF1, 4, 7 and 8 was
significantly positively correlated with the expression of PD-1 and
its ligands PD-L1 and PD-L2 in ccRCC (Fig. 10). These findings suggest that
SLAMF may inhibit the tumor-killing effect of immune cells via PD-1
and tumor-associated immune cells.
Association between
chemokines/chemokine receptors with SLAMFs
Based on the results of the functional analysis of
SLAMFs, the association between cytokines and SLAMFs were further
assessed. The findings revealed that the SLAMF1, 4, 7 and 8 were
generally positively associated with chemokines/chemokine receptors
in ccRCC (Fig. 11A and B).
Moreover, C-C motif chemokine ligand (CCL)4, CCL5, C-X-C motif
chemokine ligand (CXCL)9, CXCL10, CXCL11, X-C motif chemokine
ligand (XCL)1, XCL2, C-C motif chemokine receptor (CCR)1, CCR2,
CCR4, CCR5, CCR8, C-X-C motif chemokine receptor (CXCR)3, CXCR6 and
X-C motif chemokine receptor 1 were significantly correlated with
SLAMF1, 4, 7 and 8 exhibited (r>0.5; P<0.001). As the main
molecules in antigen presentation, MHCs were also demonstrated to
be significantly positively correlated with SLAMF1, 4, 7 and 8
(P<0.001; Fig. 11C) (48). These results suggest that SLAMFs may
promote immune cell infiltration in ccRCC through interactions with
chemokines and MHCs.
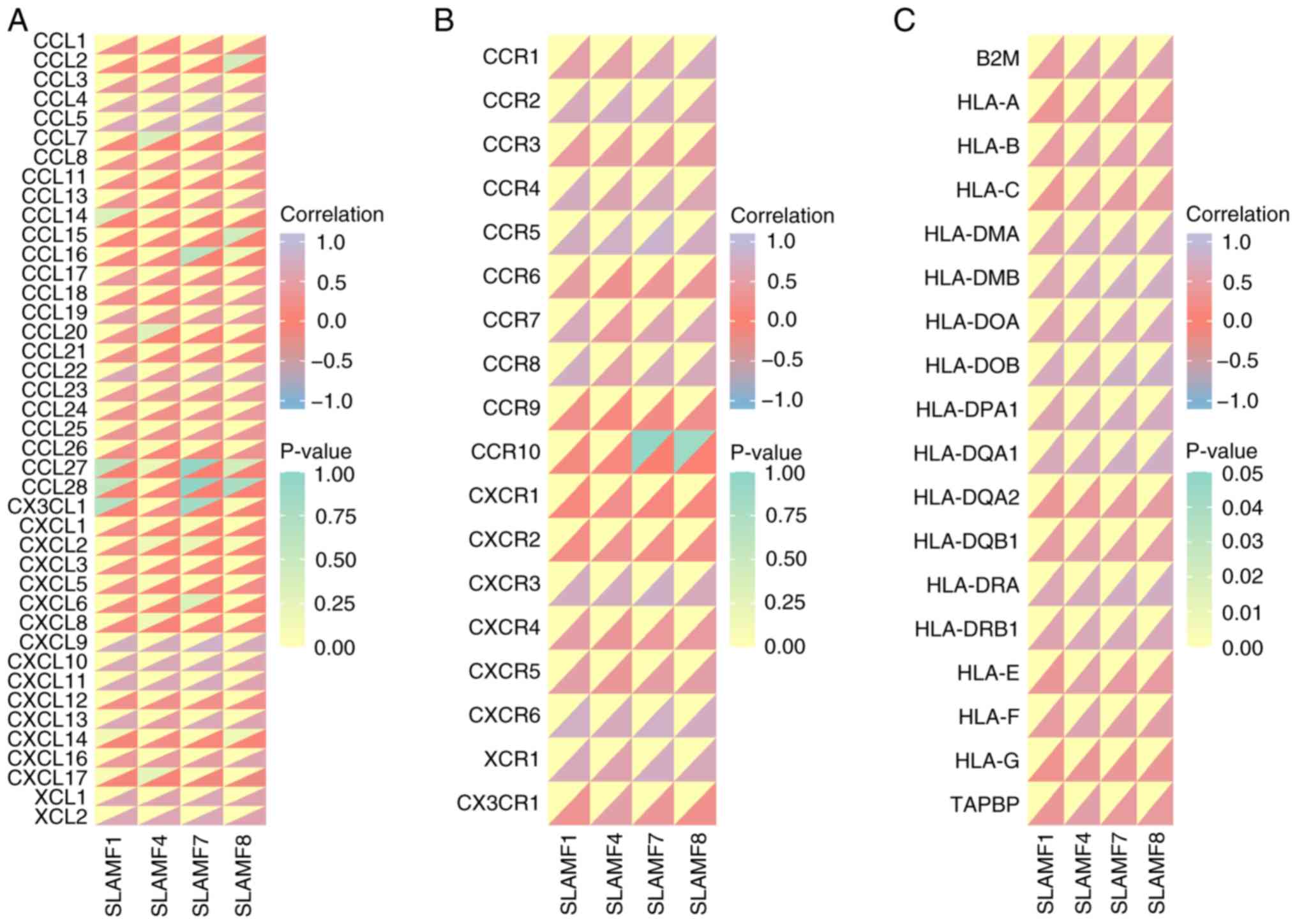 | Figure 11.Correlation of SLAMF members with
immune-related molecules. Correlation between SLAMF1, SLAMF4,
SLAMF7, SLAMF8 and (A) chemokines, (B) chemokine receptors and (C)
major histocompatibility complex molecules. SLMAF, signaling
lymphocyte activation molecule family; CCL, chemokine (C-C motif)
ligand; CXCL, chemokine (C-X-C motif) ligand; CX3CL, chemokine
(C-X3-C motif) ligand; XCL, chemokine (X-C motif) ligand; HLA,
human leukocyte antigen; B2M, β-2 microglobulin; TABPB, transporter
associated with antigen processing binding protein. |
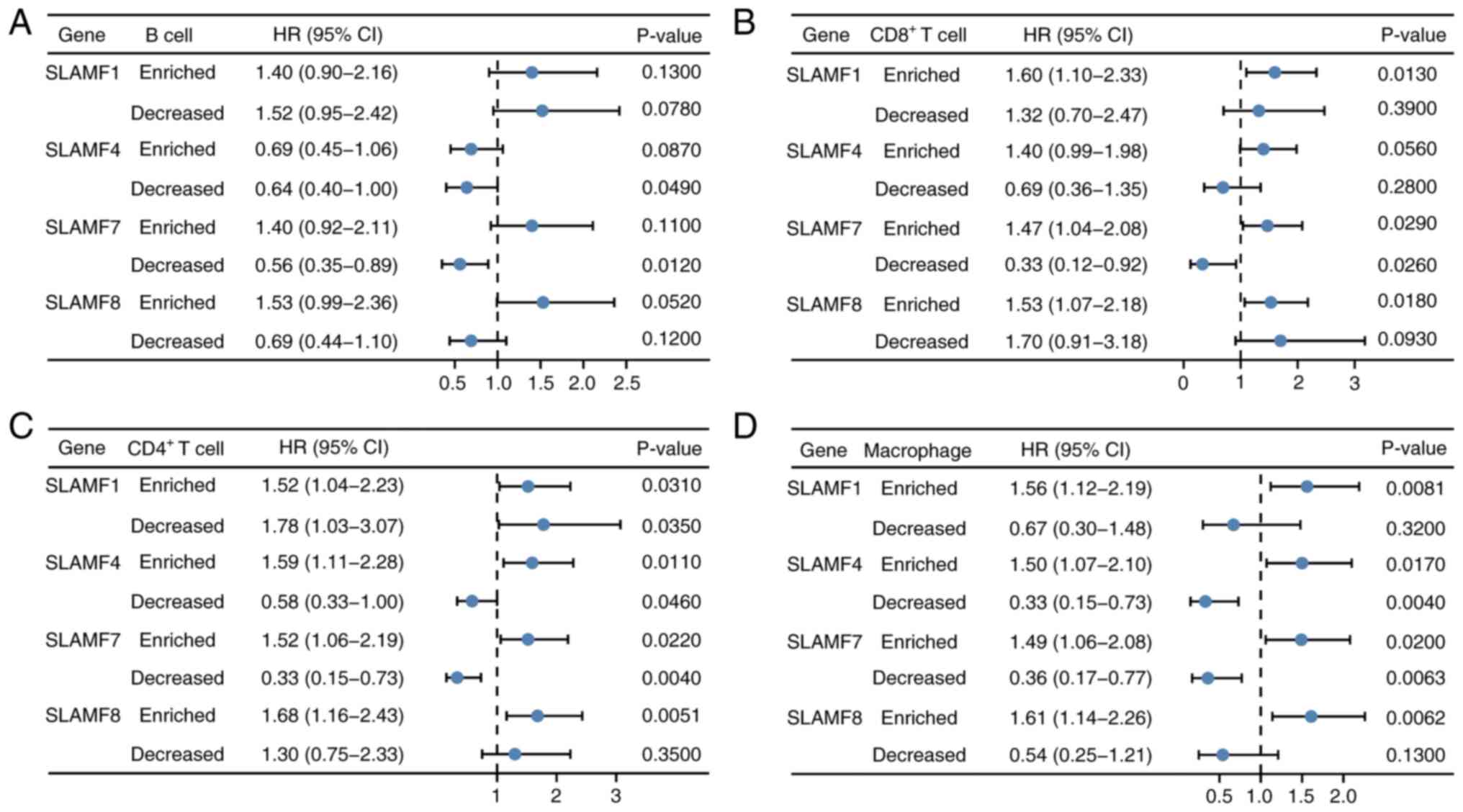
Increased level of immune cell
infiltration affects the prognosis of SLAMFs in ccRCC
A prognosis analysis of SLAMFs were performed in
combination with B cells, CD4+ T cells, CD8+
T cells and macrophages in ccRCC (Fig.
12). Forest plots containing hazard ratio (HR) and P-values
were used to present the results. The results showed that, in the
group with decreased B cell infiltration levels, ccRCC patients
with high expression of SLAMF4 and 7 had a better prognosis than
those with low expression of SLAMFs. Patients with ccRCC with high
expression of SLAMF1, 4, 7 and 8 had a worse prognosis than those
with low expression of SLAMFs in the group with enriched
CD4+ T cell infiltration levels, whilst those with high
expression of SLAMF1, 4 and 7 had better prognosis in the group
with decreased CD4+ T cell infiltration levels. In the
group with enriched CD8+ T cell infiltration levels,
patients with ccRCC had a worse prognosis with high expression of
SLAMF1, 7 and 8 than those with low expression of SLAMFs, whilst
those with high expression of SLAMF7 had a better prognosis in the
group with decreased CD8+ T cells infiltration levels.
In the group with enriched macrophage infiltration levels, patients
with ccRCC with high expression of SLAMF1, 4, 7 and 8 had a worse
prognosis, whereas those with high expression of SLAMF4 and 7 had a
better prognosis in the group with decreased macrophages
infiltration levels. These results suggest that SLAMFs may impact
the prognosis of patients through the level of immune cell
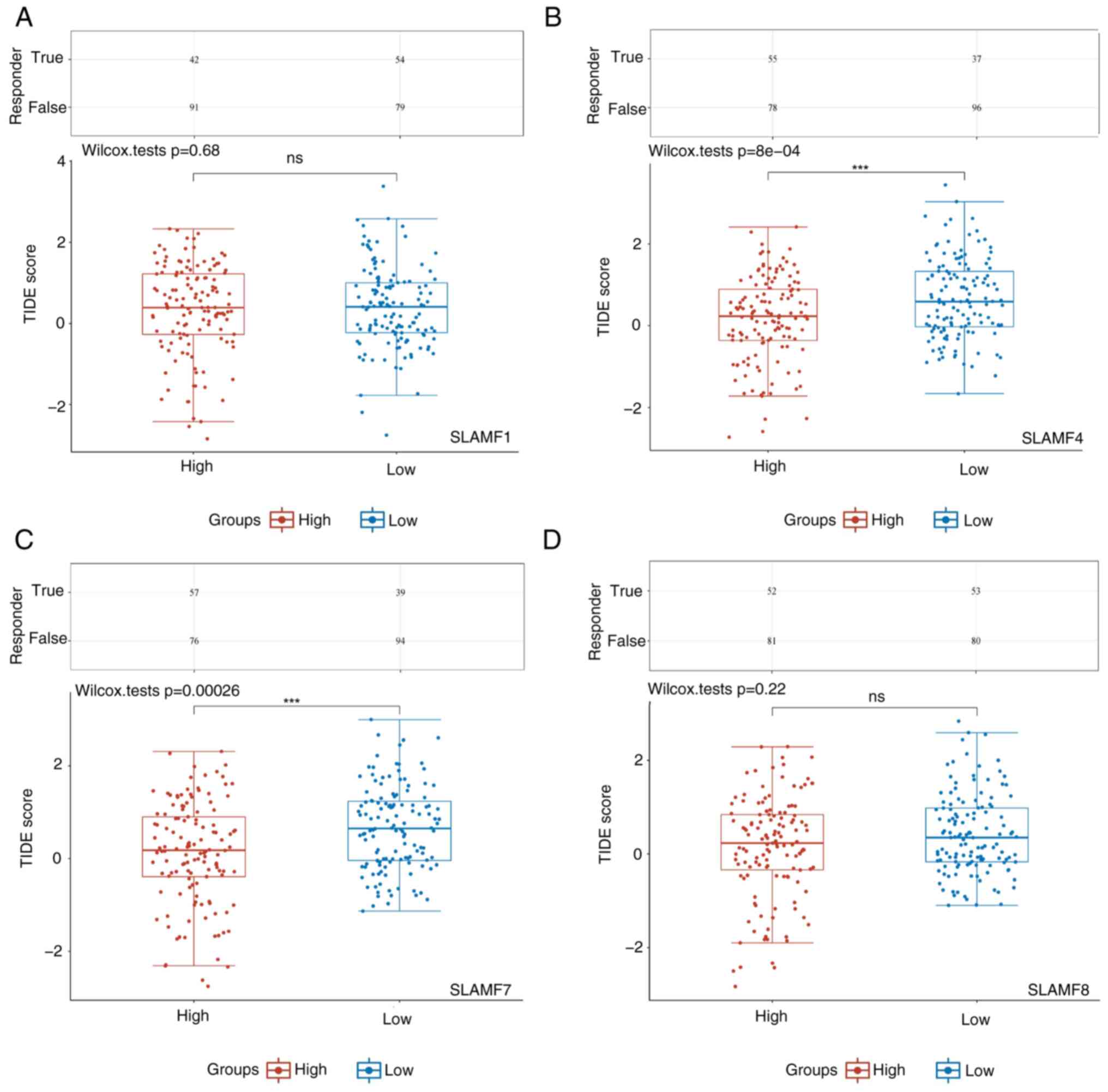
infiltration in ccRCC. Furthermore, the high-expression SLAMF4 and
7 groups had significantly lower TIDE scores, compared with the
low-expression groups (Fig. 13).
The TIDE score was inversely proportional to ICB efficacy,
indicating that a high expression of SLAMF4 and 7 was associated
with an improved response to ICB therapy in patients with
ccRCC.
Discussion
With the advent of the era of immune-targeted tumor
therapy, an increasing number of SLAMF members, which are
associated with the immune system, have been extensively studied in
several tumors, including head and neck squamous cell carcinoma,
gastric cancer, colorectal cancer, ovarian cancer and chronic
lymphocytic leukemia (25,49–53).
However, research on the role of SLAMF members in ccRCC has been
limited. In the present study, a flow chart of the present research
is shown in Fig. 14. In addition,
it was demonstrated that SLAMF members were differentially
expressed in several cancers relative to normal tissues, and
notably, the expression of nine SLAMF members was upregulated in
ccRCC, providing a compelling rationale for further investigation.
Moreover, the high expression of SLAMF was associated with cancer
stage, tumor grade and ccRCC subtype. The results also indicated
that a high expression of SLAMF members was associated with poor
prognosis in patients with ccRCC.
Previous studies reported that SLAMF members
strongly bind SLAMF-associated proteins (SAP). They have also
identified mutations in SAP in patients with X-linked
lymphoproliferative immunodeficiency disease, and reported that
reduced SAP exerts inhibitory effects on SLAMF members (54,55).
These findings suggest that SLAMF has a regulatory role in the
immune system. In the field of cancer, SLAMF members have been
reported to have an inhibitory effect on immune responses in the
TME, contributing to the immune escape of tumors (50). However, the molecular mechanism of
SLAMF members in ccRCC has been rarely studied.
Based on data from TCGA and GTEx, the present study
comprehensively explored the biological role of SLAMF in ccRCC for
the first time, to the best of our knowledge, including: SLAMF
expression and gene changes, clinical characteristics and
diagnostic significance, correlation of immune cell infiltration,
and prediction of immune checkpoint inhibitor efficacy. The present
study lays a theoretical foundation for the future research of
SLAMF in ccRCC, especially in immunotherapy. The present study
demonstrated that the molecular mechanisms of SLAMF members are
associated with immunity, as expected. Furthermore, the pathway
enrichment of SLAMF members was associated with CAMs and leukocyte
cell-cell adhesion. Cancer migration is a process wherein cancer
cells are shed from the primary tumor site and metastasize to
distant sites, in which CAMs serve an essential role in this
process (56). The metastasis of
ccRCC is associated with a poor prognosis for patients with cancer
(57), and blocking the metastasis
of ccRCC may improve the prognosis. The high expression of SLAMF
members may be a factor contributing to ccRCC metastasis and there
is evidence that SLAMF members are involved in several
physiological and pathological processes, including the regulation
of immune responses (58).
In the present study, the high expression of SLAMF
members was highly correlated with several immune cells, a finding
which is consistent with previous results. There is evidence to
suggest that SLAMF1 serves a co-stimulatory role in the activation
and differentiation of T and B cells (19), the formation of germinal centers,
and antibody production (59,60).
Moreover, SLAMF1 is markedly upregulated in CD8+ T cells compared
with CD4+ T cells (61). However,
in tumor cells, a high expression of SLAMF1 can promote the growth
and survival of tumor cells. There are reports that SLAMF1 can
mediate the survival and proliferation of tumor cells through the
PI3K/Akt signaling pathway, and participate in regulating cell
metabolism, proliferation and survival (62,63).
SLAMF4 serves a complex role in cancer immunity, acting as an
inhibitory immune checkpoint and ‘don't eat me’ receptor on
macrophages, hindering tumor cell phagocytosis (64). In CD8+ T cells, particularly in
cancers like head and neck squamous cell carcinoma, SLAMF4
expression is notably increased, especially in exhausted T cells
expressing other co-inhibitory receptors like PD-1 (50,65).
Immunogenic peptides from SLAMF7 antigens activate specific
cytotoxic T lymphocyte clones against multiple myeloma (66). In mouse models lacking SLAMF7, tumor
growth is slower, and CD8+ T cells express lower levels of PD-1
(23). SLAMF8 is expressed in
anaplastic large-cell lymphoma cell lines and serves a role in
oncogenic signaling pathways, with knockdown of SLAMF8 associated
with a reduction in cell proliferation and an increase in apoptosis
(67). High SLAMF8 expression is
associated with a worse prognosis in glioma, including reduced OS
and chemotherapy resistance (68),
but it is correlated with improved efficacy of anti-PD-1
immunotherapy in gastrointestinal cancers (69).
In the present study, based on the gene mutations of
SLAMF members, it was demonstrated that the arm-level deletion of
SLAMF members was associated with the level of immune cell
infiltration in tumors. Furthermore, the effect of the expression
of SLAMF members on the prognosis of patients with ccRCC was
regulated by immune cells. Specifically, enriched immune cells in
patients with ccRCC and high SLAMF expression were associated with
a worse prognosis relative to decreased immune cells. Although
immune cells resisted tumor cells under normal conditions, the
prognosis of ccRCC was poor. Therefore, we hypothesize that tumor
cells may inhibit immune cells through several mechanisms to
achieve immune escape. The results of the present study
demonstrated that the expression of SLAMF was generally positively
correlated with the infiltration of CAFs, M2 macrophages and Tregs
cells, that is, high expression of SLAMF can promote the
infiltration of these cells in the TME. Existing studies had
reported that immunosuppressive suppressor cell subsets are
transferred to the TME through the secretion of cytokines
(including chemokines and interleukins) by CAFs (48,70).
Interferon-γ secretion by CD8+ T cells would be inhibited by Tregs,
which are immunosuppressive regulatory molecules (71). M2 macrophages, which function as
tumor-associated immune macrophages, have also been reported to
promote cancer progression in many cancers (72,73).
Tregs, CAFs and M2 cells differ from normal immune cells, exerting
a negative effect on the immune response. The present study
demonstrated that certain SLAMF members inhibited tumor-related
immune cell function by promoting the infiltration of tumor immune
negative regulatory cells, such as CAFs, M2 macrophages and Tregs
cells.
Moreover, PD-1 immunotherapy has made considerable
progress in the treatment of several cancers (74). PD-1 mainly binds to two ligands,
PD-L1 and PD-L2, participating in the activation of the PD-1
signaling pathway. The activation of the PD-1 signaling pathway can
inhibit the activity of T cells, and prevent dendritic cells (DCs)
from activating T cells. Therefore, anti-PD-1 treatment is highly
beneficial for tumor treatment (75). In the present study, SLAMF members
were positively correlated with PD-1 and its ligands PD-L1/2.
Chemokines serve a crucial role in guiding the
migration of immune cells. Recent studies have demonstrated that
chemokine receptors induce CD8+ T cells and DCs to migrate to the
TME by binding to ligands (76).
However, Tregs and TAMs are also induced to migrate to the TME,
leading to immune tolerance (76).
Additionally, chemokines promote tumor progression through several
mechanisms, including angiogenesis and metastasis (77). In the present study, the expression
of chemokines/chemokine receptors and SLAMF members were positively
correlated. Past research has reported that the CCL17/CCL22-CCR4
axis migrated Tregs to the TME; CXCL9/10/11/12-CXCR3/CXCR4 axis
migrated CD8+ T cells to the TME; and the CCL2/CCR2 axis migrated
TAM to the TME (76). CXCL13+CD8+ T
cells could not only exhaust immune cells but also damage the
function of CD8+ T cells (78).
Furthermore, CCL4 and CCL5 have been reported participate in the
proliferation, metastasis and invasion of ccRCC cells (79,80).
These findings indicate that although immune cells migrate to the
TME, they may inhibit immune function to promote immune escape via
chemokines.
In summary, through the analysis of the biological
function of SLAMF members in ccRCC, it was demonstrated that
SLAMF1, 4, 7 and 8 may serve an important role in ccRCC: They may
promote the progression of ccRCC through immune-related pathways
and may become new immunotherapeutic targets in the future. The
current study has certain limitations that need to be addressed.
First, the results are mainly based on bioinformatic analysis of
public datasets, and further experimental validation, especially
in vivo, as well as validation of clinical samples, are
needed to confirm the findings. Second, the expression of SLAMF
members, especially SLAMF1, 4, 7 and 8, maybe used as clinical
prognostic biomarkers of ccRCC, but the appropriate threshold of
their expression levels need to be determined, and the accuracy,
feasibility and clinical practicability of expression detection
needs to be fully demonstrated in a larger patient population.
Third, more clinical trials and population data are needed to
further validate the expression of SLAMF members and the efficacy
of ICI therapy. Nevertheless, the present study provides valuable
insight into the potential role of SLAMF members as prognostic
biomarkers for ccRCC and promotes further research into their
clinical relevance and therapeutic potential.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present work was supported by the Program for Young Key
Teachers in Colleges and Universities in Henan Province (grant nos.
2020GGJS150 and 2021GGJS104) and the Key Medical Science and
Technology Research Program Project of Henan Province (grant no.
20232028).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
HW and NS conceived and coordinated the study. HW
and NS confirm the authenticity of all the raw data. KC and NS
wrote the manuscript. KC, LZ and YF analyzed the data, and made the
figures and tables. ZW, WD, JL, WS and PS processed the data. WS
and HW reviewed and revised the article. All authors contributed to
interpretation of data, manuscript revision and critical
discussion. All authors contributed to the article and have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing of interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AUC
|
area under curve
|
|
BP
|
biological process
|
|
CAF
|
cancer-associated fibroblast
|
|
CAM
|
cell adhesion molecule
|
|
CC
|
cellular component
|
|
RCC
|
renal cell carcinoma
|
|
ccRCC
|
clear cell RCC
|
|
GEPIA
|
Gene Expression Profiling Interactive
Analysis
|
|
GO
|
Gene Ontology
|
|
GSEA
|
gene set enrichment analysis
|
|
GTEx
|
Genotype-Tissue Expression
|
|
ICB
|
immune checkpoint blockade
|
|
ICIs
|
immune checkpoint inhibitors
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
KIRC
|
kidney renal clear cell carcinoma
|
|
MF
|
molecular function
|
|
MHC
|
major histocompatibility complex
|
|
OS
|
overall survival
|
|
ROC
|
receiver operating characteristic
|
|
SCNA
|
somatic copy number alteration
|
|
SLAMF
|
signaling lymphocyte activation
molecule family member
|
|
TAM
|
tumor-associated macrophage
|
|
TCGA
|
The Cancer Genome Atlas
|
|
TIDE
|
Tumor Immune Dysfunction and
Exclusion
|
|
TME
|
tumor microenvironment
|
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moch H, Cubilla AL, Humphrey PA, Reuter VE
and Ulbright TM: The 2016 WHO classification of tumours of the
urinary system and male genital organs-part A: Renal, penile, and
testicular tumours. Eur Urol. 70:93–105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marchetti A, Rosellini M, Mollica V, Rizzo
A, Tassinari E, Nuvola G, Cimadamore A, Santoni M, Fiorentino M,
Montironi R and Massari F: The molecular characteristics of
non-clear cell renal cell carcinoma: What's the story morning
glory? Int J Mol Sci. 22:62372021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiang J, Han P, Qian J, Zhang S, Wang S,
Cao Q and Shao P: Knockdown of ALPK2 blocks development and
progression of renal cell carcinoma. Exp Cell Res. 392:1120292020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He YH, Chen C and Shi Z: The biological
roles and clinical implications of microRNAs in clear cell renal
cell carcinoma. J Cell Physiol. 233:4458–4465. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao S, Yan L, Zhang H, Fan X, Jiao X and
Shao F: Identification of a metastasis-associated gene signature of
clear cell renal cell carcinoma. Front Genet. 11:6034552021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cochetti G, Cari L, Nocentini G, Maulà V,
Suvieri C, Cagnani R, Rossi De Vermandois JA and Mearini E:
Detection of urinary miRNAs for diagnosis of clear cell renal cell
carcinoma. Sci Rep. 10:212902020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Obeng RC, Arnold RS, Ogan K, Master VA,
Pattaras JG, Petros JA and Osunkoya AO: Molecular characteristics
and markers of advanced clear cell renal cell carcinoma: Pitfalls
due to intratumoral heterogeneity and identification of genetic
alterations associated with metastasis. Int J Urol. 27:790–797.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cros J, Sbidian E, Posseme K, Letierce A,
Guettier C, Benoît G and Ferlicot S: Nestin expression on tumour
vessels and tumour-infiltrating macrophages define a poor prognosis
subgroup of pt1 clear cell renal cell carcinoma. Virchows Arch.
469:331–337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Janowitz T, Welsh SJ, Zaki K, Mulders P
and Eisen T: Adjuvant therapy in renal cell carcinoma-past,
present, and future. Semin Oncol. 40:482–491. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Motzer RJ, Jonasch E, Agarwal N, Alva A,
Baine M, Beckermann K, Carlo MI, Choueiri TK, Costello BA, Derweesh
IH, et al: Kidney cancer, version 3.2022, NCCN clinical practice
guidelines in oncology. J Natl Compr Canc Netw. 20:71–90. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rosellini M, Marchetti A, Mollica V, Rizzo
A, Santoni M and Massari F: Prognostic and predictive biomarkers
for immunotherapy in advanced renal cell carcinoma. Nat Rev Urol.
20:133–157. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang B, Chen D and Hua H: TBC1D3 family is
a prognostic biomarker and correlates with immune infiltration in
kidney renal clear cell carcinoma. Mol Ther Oncolytics. 22:528–538.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Santoni M, Rizzo A, Mollica V, Matrana MR,
Rosellini M, Faloppi L, Marchetti A, Battelli N and Massari F: The
impact of gender on the efficacy of immune checkpoint inhibitors in
cancer patients: The MOUSEION-01 study. Crit Rev Oncol Hematol.
170:1035962022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rizzo A, Mollica V, Dall'Olio FG, Ricci
AD, Maggio I, Marchetti A, Rosellini M, Santoni M, Ardizzoni A and
Massari F: Quality of life assessment in renal cell carcinoma phase
II and III clinical trials published between 2010 and 2020: A
systematic review. Future Oncol. 17:2671–2681. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mollica V, Rizzo A, Marchetti A, Tateo V,
Tassinari E, Rosellini M, Massafra R, Santoni M and Massari F: The
impact of ECOG performance status on efficacy of immunotherapy and
immune-based combinations in cancer patients: The MOUSEION-06
study. Clin Exp Med. 23:5039–5049. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dragovich MA and Mor A: The SLAM family
receptors: Potential therapeutic targets for inflammatory and
autoimmune diseases. Autoimmun Rev. 17:674–682. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gunes M, Rosen ST, Shachar I and Gunes EG:
Signaling lymphocytic activation molecule family receptors as
potential immune therapeutic targets in solid tumors. Front
Immunol. 15:12974732024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Farhangnia P, Ghomi SM, Mollazadehghomi S,
Nickho H, Akbarpour M and Delbandi AA: SLAM-family receptors come
of age as a potential molecular target in cancer immunotherapy.
Front Immunol. 14:11741382023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tojjari A, Giles FJ, Vilbert M, Saeed A
and Cavalcante L: SLAM modification as an immune-modulatory
therapeutic approach in cancer. Cancers (Basel). 15:48082023.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Su R, Jin C, Zhou L, Cao Y, Kuang M, Li L
and Xiang J: Construction of a ceRNA network of hub genes affecting
immune infiltration in ovarian cancer identified by WGCNA. BMC
Cancer. 21:9702021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lewinsky H, Gunes EG, David K, Radomir L,
Kramer MP, Pellegrino B, Perpinial M, Chen J, He TF, Mansour AG, et
al: CD84 is a regulator of the immunosuppressive microenvironment
in multiple myeloma. JCI Insight. 6:e1416832021.PubMed/NCBI
|
|
23
|
O'Connell P, Hyslop S, Blake MK, Godbehere
S, Amalfitano A and Aldhamen YA: SLAMF7 Signaling reprograms t
cells toward exhaustion in the tumor microenvironment. J Immunol.
206:193–205. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Agresta L, Hoebe KHN and Janssen EM: The
emerging role of CD244 signaling in immune cells of the tumor
microenvironment. Front Immunol. 9:28092018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yigit B, Wang N, Ten Hacken E, Chen SS,
Bhan AK, Suarez-Fueyo A, Katsuyama E, Tsokos GC, Chiorazzi N, Wu
CJ, et al: SLAMF6 as a regulator of exhausted CD8+ T cells in
cancer. Cancer Immunol Res. 7:1485–1496. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
GTEx Consortium: The genotype-tissue
expression (GTEx) project. Nat Genet. 45:580–585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vivian J, Rao AA, Nothaft FA, Ketchum C,
Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD,
Musselman-Brown A, et al: Toil enables reproducible, open source,
big biomedical data analyses. Nat Biotechnol. 35:314–316. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47((W1)): W556–W560.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Obuchowski NA and Bullen JA: Receiver
operating characteristic (ROC) curves: Review of methods with
applications in diagnostic medicine. Phys Med Biol. 63:07TR012018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lánczky A and Győrffy B: Web-based
survival analysis tool tailored for medical research (KMplot):
Development and implementation. J Med Internet Res. 23:e276332021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: an open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48((W1)): W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hughes CE and Nibbs RJB: A guide to
chemokines and their receptors. FEBS J. 285:2944–2971. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kakinuma T and Hwang ST: Chemokines,
chemokine receptors, and cancer metastasis. J Leukoc Biol.
79:639–651. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kotsias F, Cebrian I and Alloatti A:
Antigen processing and presentation. Int Rev Cell Mol Biol.
348:69–121. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang X, Spandidos A, Wang H and Seed B:
PrimerBank: A PCR primer database for quantitative gene expression
analysis, 2012 update. Nucleic Acids Res. 40((Database Issue)):
D1144–D1149. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang H, Song C, Ding Y, Pan X, Ge Z, Tan
BH, Gowda C, Sachdev M, Muthusami S, Ouyang H, et al:
Transcriptional regulation of JARID1B/KDM5B histone demethylase by
ikaros, histone deacetylase 1 (HDAC1), and casein kinase 2 (CK2) in
B-cell acute lymphoblastic leukemia. J Biol Chem. 291:4004–4018.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chan BKC: Data analysis using R
programming. Adv Exp Med Biol. 1082:47–122. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Han Y, Liu D and Li L: PD-1/PD-L1 pathway:
current researches in cancer. Am J Cancer Res. 10:727–742.
2020.PubMed/NCBI
|
|
48
|
Wen M, Li Y, Qin X, Qin B and Wang Q:
Insight into cancer immunity: MHCs, immune cells and commensal
microbiota. Cells. 12:18822023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ma R, Qu X, Che X, Yang B, Li C, Hou K,
Guo T, Xiao J and Liu Y: Comparative analysis and in vitro
experiments of signatures and prognostic value of immune checkpoint
genes in colorectal cancer. Onco Targets Ther. 14:3517–3534. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Agresta L, Lehn M, Lampe K, Cantrell R,
Hennies C, Szabo S, Wise-Draper T, Conforti L, Hoebe K and Janssen
EM: CD244 represents a new therapeutic target in head and neck
squamous cell carcinoma. J Immunother Cancer. 8:e0002452020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lewinsky H, Barak AF, Huber V, Kramer MP,
Radomir L, Sever L, Orr I, Mirkin V, Dezorella N, Shapiro M, et al:
CD84 regulates PD-1/PD-L1 expression and function in chronic
lymphocytic leukemia. J Clin Invest. 128:5465–5478. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wu D, Zhang P, Ma J, Xu J, Yang L, Xu W,
Que H, Chen M and Xu H: Serum biomarker panels for the diagnosis of
gastric cancer. Cancer Med. 8:1576–1583. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Quan Q, Xiong X, Wu S and Yu M:
Identification of immune-related key genes in ovarian cancer based
on WGCNA. Front Genet. 12:7602252021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Veillette A and Latour S: The SLAM family
of immune-cell receptors. Curr Opin Immunol. 15:277–285. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wu N and Veillette A: SLAM family
receptors in normal immunity and immune pathologies. Curr Opin
Immunol. 38:45–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Smart JA, Oleksak JE and Hartsough EJ:
Cell adhesion molecules in plasticity and metastasis. Mol Cancer
Res. 19:25–37. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ishihara M, Hu J, Zhang X, Choi Y, Wong A,
Cano-Ruiz C, Zhao R, Tan P, Tso JL and Wu L: Comparing metastatic
clear cell renal cell carcinoma model established in mouse kidney
and on chicken chorioallantoic membrane. J Vis Exp. 10.3791/60314.
2020. View Article : Google Scholar
|
|
58
|
Fouquet G, Marcq I, Debuysscher V, Bayry
J, Rabbind Singh A, Bengrine A, Nguyen-Khac E, Naassila M and
Bouhlal H: Signaling lymphocytic activation molecules Slam and
cancers: Friends or foes? Oncotarget. 9:16248–16262. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
De Salort J, Sintes J, Llinàs L,
Matesanz-Isabel J and Engel P: Expression of SLAM (CD150)
cell-surface receptors on human B-cell subsets: From pro-B to
plasma cells. Immunol Lett. 134:129–136. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Karampetsou MP, Comte D, Suárez-Fueyo A,
Katsuyama E, Yoshida N, Kono M, Kyttaris VC and Tsokos GC:
Signaling lymphocytic activation molecule family member 1
engagement inhibits T cell-B cell interaction and diminishes
interleukin-6 production and plasmablast differentiation in
systemic lupus erythematosus. Arthritis Rheumatol. 71:99–108. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang N, Morra M, Wu C, Gullo C, Howie D,
Coyle T, Engel P and Terhorst C: CD150 is a member of a family of
genes that encode glycoproteins on the surface of hematopoietic
cells. Immunogenetics. 53:382–394. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gordiienko I, Shlapatska L, Kholodniuk V,
Sklyarenko L, Gluzman DF, Clark EA and Sidorenko SP: The interplay
of CD150 and CD180 receptor pathways contribute to the pathobiology
of chronic lymphocytic leukemia B cells by selective inhibition of
Akt and MAPK signaling. PLoS One. 12:e01859402017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yurchenko M, Shlapatska LM, Romanets OL,
Ganshevskiy D, Kashuba E, Zamoshnikova A, Ushenin YV, Snopok BA and
Sidorenko SP: CD150-mediated Akt signalling pathway in normal and
malignant B cells. Exp Oncol. 33:9–18. 2011.PubMed/NCBI
|
|
64
|
Li D, Xiong W, Wang Y, Feng J, He Y, Du J,
Wang J, Yang M, Zeng H, Yang YG, et al: SLAMF3 and SLAMF4 are
immune checkpoints that constrain macrophage phagocytosis of
hematopoietic tumors. Sci Immunol. 7:eabj55012022.PubMed/NCBI
|
|
65
|
Mittal R, Wagener M, Breed ER, Liang Z,
Yoseph BP, Burd EM, Farris AB III, Coopersmith CM and Ford ML:
Phenotypic T cell exhaustion in a murine model of bacterial
infection in the setting of pre-existing malignancy. PLoS One.
9:e935232014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Bae J, Song W, Smith R, Daley J, Tai YT,
Anderson KC and Munshi NC: A novel immunogenic CS1-specific peptide
inducing antigen-specific cytotoxic T lymphocytes targeting
multiple myeloma. Br J Haematol. 157:687–701. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Sugimoto A, Kataoka TR, Ito H, Kitamura K,
Saito N, Hirata M, Ueshima C, Takei Y, Moriyoshi K, Otsuka Y, et
al: SLAM family member 8 is expressed in and enhances the growth of
anaplastic large cell lymphoma. Sci Rep. 10:25052020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zou CY, Guan GF, Zhu C, Liu TQ, Guo Q,
Cheng W and Wu AH: Costimulatory checkpoint SLAMF8 is an
independent prognosis factor in glioma. CNS Neurosci Ther.
25:333–342. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang Q, Cheng L, Qin Y, Kong L, Shi X, Hu
J, Li L, Ding Z, Wang T, Shen J, et al: SLAMF8 expression predicts
the efficacy of anti-PD1 immunotherapy in gastrointestinal cancers.
Clin Transl Immunology. 10:e13472021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
De Jaeghere EA, Denys HG and De Wever O:
Fibroblasts fuel immune escape in the tumor microenvironment.
Trends Cancer. 5:704–723. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Langhans B, Nischalke HD, Krämer B, Dold
L, Lutz P, Mohr R, Vogt A, Toma M, Eis-Hübinger AM, Nattermann J,
et al: Role of regulatory T cells and checkpoint inhibition in
hepatocellular carcinoma. Cancer Immunol Immunother. 68:2055–2066.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Sumitomo R, Hirai T, Fujita M, Murakami H,
Otake Y and Huang CL: M2 tumor-associated macrophages promote tumor
progression in non-small-cell lung cancer. Exp Ther Med.
18:4490–4498. 2019.PubMed/NCBI
|
|
73
|
Tu D, Dou J, Wang M, Zhuang H and Zhang X:
M2 macrophages contribute to cell proliferation and migration of
breast cancer. Cell Biol Int. 45:831–838. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wu X, Gu Z, Chen Y, Chen B, Chen W, Weng L
and Liu X: Application of PD-1 blockade in cancer immunotherapy.
Comput Struct Biotechnol J. 17:661–674. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ai L, Xu A and Xu J: Roles of PD-1/PD-L1
pathway: Signaling, cancer, and beyond. Adv Exp Med Biol.
1248:33–59. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kohli K, Pillarisetty VG and Kim TS: Key
chemokines direct migration of immune cells in solid tumors. Cancer
Gene Ther. 29:10–21. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Marcuzzi E, Angioni R, Molon B and Calì B:
Chemokines and chemokine receptors: Orchestrating tumor
metastasization. Int J Mol Sci. 20:962018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Dai S, Zeng H, Liu Z, Jin K, Jiang W, Wang
Z, Lin Z, Xiong Y, Wang J, Chang Y, et al: Intratumoral
CXCL13+CD8+T cell infiltration determines poor clinical outcomes
and immunoevasive contexture in patients with clear cell renal cell
carcinoma. J Immunother Cancer. 9:e0018232021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang L, Zhang M, Wang L, Li J, Yang T,
Shao Q, Liang X, Ma M, Zhang N, Jing M, et al: Identification of
CCL4 as an immune-related prognostic biomarker associated with
tumor proliferation and the tumor microenvironment in clear cell
renal cell carcinoma. Front Oncol. 11:6946642021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Lin J, Yu M, Xu X, Wang Y, Xing H, An J,
Yang J, Tang C, Sun D and Zhu Y: Identification of biomarkers
related to CD8+ T cell infiltration with gene co-expression network
in clear cell renal cell carcinoma. Aging (Albany NY).
12:3694–3712. 2020. View Article : Google Scholar : PubMed/NCBI
|