Introduction
Lung cancer is characterized by its extreme
invasiveness and metastatic nature, contributing to the highest
incidences of cancer and mortality rates worldwide (1). Histologically, lung cancer is
categorized into two main types: Small cell lung cancer and
non-small cell lung cancer, the latter of which represents ~85% of
all cases (2), with lung
adenocarcinoma (LUAD) emerging as the predominant subtype (3). The widespread adoption of low-dose
computed tomography has notably increased the detection rate of
lung cancer (4). However, despite
recent advances in the diagnosis and treatment of lung cancer, a
significant number of patients with LUAD ultimately succumb to the
disease. Reports indicate a high mortality rate for LUAD, with a
5-year overall survival (OS) rate of <15% (5,6).
Therefore, there is an urgent need to explore novel biomarkers and
develop effective therapeutic approaches to improve the diagnosis
and prognosis of patients with LUAD.
MIS18 Kinetochore Protein A (MIS18A), also known as
Mis18α or C21orf45, is a crucial component of the Mis18 protein
family, which also includes Mis18β and M18BP1. MIS18A is pivotal
for the recruitment of centromere protein (CENP) A within the Mis18
complex and is essential for centromeric chromatin organization
(7). MIS18A localizes to the
centromere in a cell cycle-dependent manner, with its centromeric
signals notably intensifying during a specific phase that spans
from late anaphase-telophase to early G1, encompassing the
post-segregation and pre-replication periods (8). Additionally, MIS18A and MIS18B form a
heterotetramer complex through their C-terminal coiled-coil
domains, which is pivotal for centromere recognition (9). CENPA, which encodes a centromeric
protein with a histone fold domain closely related to histone H3,
is crucial for the localization of this protein to the centromere
(10). Elevated levels of CENPA
have been identified as a potential diagnostic biomarker for LUAD
and have been linked to poor OS rates (11). However, the precise roles and
implications of MIS18A, which is intricately linked to CENPA
expression, as well as the significance of the MIS18A complex in
cancer, remain unclear. Nuclear MIS18A may function as a histone
modifier, potentially influencing chromatin hypermethylation
through interactions with DNA methyltransferase (DNMT) 3A and
DNMT3B (12). Furthermore, the
upregulation of MIS18A has been linked to high microsatellite
instability in colorectal cancer, highlighting the therapeutic
potential of targeting autophagy protein 5-MIS18A or MIS18A, as
their interaction promotes the hypermethylation of the hMLH1
promoter CpG island (13).
Therefore, further exploration of the involvement of MIS18A in
various cancer types is essential to comprehensively understand its
clinical significance and potential mechanisms in diverse cancer
types. The present study reports a primary effort to investigate
MIS18A in the context of LUAD with the aim of unraveling its
expression profile, prognostic relevance and underlying
mechanisms.
Materials and methods
Data acquisition
Gene expression data, including LUAD mRNA count and
Fragments Per Kilobase per Million mapped reads data, were
collected from UCSC Xena (14)
(https://xenabrowser.net/), for a cohort
comprising 510 LUAD samples and 58 normal samples. This dataset
also included clinical annotations and survival outcomes. To
standardize the comparability and facilitate analysis, all data
underwent a logarithmic transformation using the formula
log2(x+1). Long non-coding RNA (lncRNA), microRNA
(miRNA) and nucleotide variation datasets pertinent to LUAD were
obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Additionally,
the GSE30219 (15) (293 LUAD
samples and 14 normal samples), GSE10072 (16) (58 LUAD samples and 49 normal
samples) and GSE27262 (17) (25
LUAD tissues and 25 adjacent normal tissues) datasets were
retrieved from the Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/geo/).
Discrepancies in MIS18A expression and
its diagnostic potential in LUAD
This investigation focused on comparing the MIS18A
expression levels between normal and tumor tissues based on data
from TCGA and GEO databases. Patients with LUAD were categorized
into low and high expression groups based on the median MIS18A
expression level. The expression patterns of MIS18A across various
cancer types were investigated using the TIMER database (18) (https://cistrome.shinyapps.io/timer/). The diagnostic
utility of MIS18A in LUAD was assessed using Receiver Operating
Characteristic (ROC) curve analysis. Validation of differential
expression was conducted through reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) using our own collected samples
(described later). The elevated MIS18A protein expression in LUAD
was also confirmed through analysis of the Human Protein Atlas
(HPA) (19) (http://www.proteinatlas.org/) and UALCAN (20) datasets (http://ualcan.path.uab.edu/).
Prognostic value analysis of MIS18A in
LUAD
The prognostic significance of MIS18A in the
TCGA-LUAD and GSE30219 cohorts was assessed using the R packages,
‘survival’ (version 3.6.4) (21)
and ‘survminer’ (version 0.4.9) (22). Validation was conducted by analyzing
the OS time using the Kaplan-Meier Plotter database (https://kmplot.com/analysis/) and the Gene Expression
Profiling Interactive Analysis (GEPIA) dataset (23) (http://gepia.cancer-pku.cn/). Univariate and
multivariate Cox proportional hazard regression analyses were
performed to evaluate the independent prognostic significance of
MIS18A in LUAD.
Identification of differentially
expressed genes (DEGs) and functional enrichment analysis of
MIS18A
Differential analysis of the TCGA dataset was
performed using the ‘DESeq2’ (version 1.44.0) (24) package in R (version 4.2.2), where
DEGs were characterized by genes with an absolute log2(fold change)
>1 and adjusted P<0.05. The identification of these DEGs was
visually depicted through Volcano plots. Subsequently, the Weighted
Gene Co-expression Network Analysis (WGCNA) method, implemented via
the R package ‘WGCNA’, was used to identify gene modules correlated
with the expression of the MIS18A gene (25). The resultant gene module underwent
enrichment analyses, including Gene Ontology (GO), Kyoto
Encyclopedia of Genes and Genomes (KEGG) and Gene Set Enrichment
Analysis (GSEA). To comprehensively elucidate the biological
functions associated with MIS18A, enrichment analyses were
performed using the R packages, ‘clusterProfiler’ (version 4.12.0)
and ‘org.Hs.eg.db’ (version 3.19.1).
Diagnostic value and survival analysis
of the hub Genes
During the GSEA, 6 hub genes were identified through
the intersection of the protein-protein interaction (PPI) network
associated with MIS18A, obtained from the GeneMANIA (26) database (http://www.genemania.org), with the turquoise module.
Subsequently, a comprehensive investigation into the diagnostic and
prognostic significance of these hub genes was conducted, as
aforementioned.
Immune infiltration analysis of MIS18A
in LUAD
The ESTIMATE algorithm (27) was used to calculate estimate scores,
immune scores, stromal scores and tumor purity for LUAD samples.
Subsequently, the differences in these scores between samples with
high and low MIS18A expression were analyzed. The single-sample
GSEA (ssGSEA) algorithm, implemented through the ‘GSVA’ package
(version 1.52.2) (28), was adopted
to evaluate 28 subtypes of immune cells in LUAD. Additionally, the
relationship between MIS18A and chemokines, as well as chemokine
receptors, was explored using the Tumor-Immune System Interaction
Database (TISIDB; http://cis.hku.hk/TISIDB/index.php).
Examination of the correlation between
tumor mutation burden (TMB) and drug sensitivity with MIS18A in
LUAD
The ‘maftools’ package (version 2.20.0) was utilized
for the comprehensive analysis and visualization of Mutation
Annotation Format data (29). The
Spearman's correlation coefficient test was used to examine the
correlation between MIS18A and TMB. To predict the potential
efficacy of chemotherapy and targeted treatments for LUAD cases
with upregulated MIS18A, semi-inhibitory concentration
(IC50) data were obtained from the Genomics of Drug
Sensitivity in Cancer database (https://www.cancerrxgene.org/) (30). The analysis was conducted using the
‘OncoPredict’ (version 1.2) (31),
‘ggpubr’ (version 0.6.0) and ‘ggplot2’ (version 3.5.1) packages
within the R software.
Pathological sample collection
The samples and data from 30 pathological LUAD cases
were collected from the First Affiliated Hospital of Guangxi
Medical University (Nanning, China). A total of 30 LUAD and 30
adjacent non-cancerous tissue samples were collected from July 2022
to September 2022. The included patients comprised 15 males and 15
females, with an average age of 63 years and an age range of 32 to
78 years. The inclusion criteria required patients to have
undergone surgical treatment at the First Affiliated Hospital of
Guangxi Medical University and to have received a confirmed
pathological diagnosis of LUAD. Patients with other types of lung
cancer or immune system disorders, or those who had received
chemotherapy or radiation therapy before surgery were excluded from
the present study. All participants provided written informed
consent, and the study protocol was approved by the Ethics
Committee of the First Affiliated Hospital of Guangxi Medical
University, guaranteeing adherence to strict ethical standards to
protect patient privacy and rights. These meticulous data
collection methods and ethical considerations have established a
solid foundation for the subsequent data analysis in this
study.
Cell culture and RNA interference
The A549 LUAD cell line was obtained from The Cell
Bank of Type Culture Collection of The Chinese Academy of Sciences.
These cells were cultured in Dulbecco's Modified Eagle Medium
(DMEM) supplemented with 10% fetal bovine serum (FBS) and 1%
penicillin and streptomycin (all sourced from Gibco; Thermo Fisher
Scientific, Inc.), and were maintained at 37°C in a 5%
CO2 atmosphere. Lipofectamine® 8000 (Beyotime
Institute of Biotechnology) was used to transfect small interfering
RNA (siRNA) into the cells in a serum-free culture medium according
to the manufacturer's instructions. For the CCK-8 assay performed
in a 96-well plate, 5 µl serum-free medium, 4 pmol siRNA and 0.16
µl Lipofectamine 8000 transfection reagent were used. For the wound
healing and Transwell invasion assays performed in 6-well plates,
125 µl serum-free medium, 100 pmol siRNA and 4 µl Lipofectamine
8000 transfection reagent were used. After adding siRNA, the
mixture was gently mixed, followed by the addition of Lipofectamine
8000 transfection reagent and gentle mixing again. The mixtures
were incubated at room temperature for 20 min before adding to the
cells. The cells were then incubated at 37°C in a 5% CO2
atmosphere for 2 days before subsequent experiments. Specific
details regarding the siRNA sequences can be found in Table I.
 | Table I.siRNA sequences. |
Table I.
siRNA sequences.
| Primer | Sequence
(5′-3′) |
|---|
| si-NC |
UUCUCCGAACGUGUCACGU |
| si-MIS18A-1 |
GCCGAAUCCAAAUUGUCCUUU |
| si-MIS18A-2 |
GCCCAAGAAUCUUGAUUACAA |
| si-MIS18A-3 |
CUUCGCUGUGUUUCCUGUAAU |
RNA extraction and RT-qPCR
The aforementioned patient tissue samples were
immediately flash-frozen in liquid nitrogen upon collection and
subsequently stored at −80°C. Total RNA was extracted from these
samples using RNAiso Plus (Takara Bio, Inc.) at 4°C. cDNA was
synthesized from 1.0 µg total RNA using the Prime Script RT Master
Mix (Takara Bio, Inc.), as per the manufacturer's instructions.
qPCR was conducted using the Fast Start Universal SYBR Green Master
(Roche Diagnostics) and 2X Q3 SYBR qPCR Master Mix (Tolo Biotech
Co., Ltd.) to quantify gene expression. The amplification protocol
involved an initial denaturation at 95°C for 30 sec, followed by 40
cycles of 95°C for 10 sec and 60°C for 30 sec. Relative gene
expression was determined using the 2−ΔΔCq method
(17), with GAPDH as the
normalization control. The primer sequences (synthesized by Nanning
Genesis Biotechnology Co., Ltd.) were as follows: GAPDH forward,
5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, 5′-ATGGTGGTGAAGACGCCAGT-3′;
and MIS18A forward, 5′-TGCTTCGCTGTGTTTCCTGT-3′ and reverse,
5′-TGACACAATTTGCTTTTCAGAGGAC-3′. The same protocol was followed for
the MIS18A siRNA-transfected cells.
Cell Counting Kit-8 (CCK-8) assay
The transfected cells were seeded into 96-well
plates and incubated for 48 h. Subsequently, 10 µl CCK-8 reagent
(Dojindo Laboratories, Inc.) was added to each well, and the cells
were incubated at 37°C for 2 h. The absorbance at 450 nm was
measured using a microplate reader (Thermo Fisher Scientific,
Inc.).
Wound healing assay
Cells were seeded into 6-well plates and transfected
upon reaching 80–90% confluency. A precise vertical wound was
created using a sterile 200 µl pipette tip. The plate was
subsequently washed with phosphate buffered saline to remove any
detached cells, then further cultured in serum-free medium. Images
were collected at 0, 24 and 48 h following wounding using a light
microscope (Nikon Corporation), and the wound closure rate was
subsequently analyzed by measuring the wound area at each time
point using ImageJ (version 1.53q; National Institutes of Health)
software and calculating the percentage of wound closure at 24 and
48 h relative to the initial wound area at 0 h.
Transwell invasion assay
Invasion assays were performed using Transwell
plates with a 8-µm pore size (LabSelect; Beijing Lanjieke
Technology Co., Ltd.). The Transwell chambers were precoated with
Matrigel; all operations were carried out on ice, and all pipette
tips were pre-cooled. Matrigel was diluted with serum-free culture
medium at a ratio of 9:1 (culture medium: Matrigel) and mixed
thoroughly. Matrigel mixture (100 µl) was carefully added to the
polycarbonate membrane in each Transwell chamber, forming a layer
of artificial extracellular matrix while avoiding bubbles. The
chambers were then incubated at 37°C for 2 h to allow the Matrigel
to solidify. Cells, collected 24 h after transfection, were
resuspended and 200 µl containing 20,000 cells was placed into the
upper chamber of the Transwell plate. The lower chamber contained
500 µl DMEM supplemented with 10% FBS. The cells were then
incubated at 37°C for 24 h. Following incubation, the cells were
stained with crystal violet (Beyotime Institute of Biotechnology)
for 10 min at room temperature and quantified in various randomly
selected regions using a light microscope (Nikon Corporation).
Construction of the competing
endogenous RNA (ceRNA) network for MIS18A
An analysis using the ENCORI database (https://starbase.sysu.edu.cn/) was performed to
predict non-coding RNAs that may regulate MIS18A expression through
the ceRNA mechanism. The criteria for miRNA selection included a
Spearman's correlation coefficient of <-0.2 and P<0.05,
enabling the identification of miRNAs targeting MIS18A.
Subsequently, emphasis was placed on miRNAs that were downregulated
in LUAD and exhibited a positive correlation with patient
prognosis. Regarding lncRNAs, two criteria were employed:
Spearman's correlation coefficient of <-0.2 and P<0.05 for a
negative correlation with miRNA, and a Spearman's correlation
coefficient of >0.2 and P<0.05 for a positive correlation
with mRNA. lncRNAs meeting these criteria were considered as those
that target MIS18A, while those upregulated in LUAD with a negative
correlation with patient prognosis were identified as core lncRNAs
associated with MIS18A.
Statistical analysis
Statistical analyses were performed using R (version
4.2.2) and GraphPad Prism (version 7.0; Dotmatics). Differential
expression analysis between two groups was performed using the
Wilcoxon rank-sum test. Survival analysis was conducted using the
Kaplan-Meier method to plot survival curves, with the log-rank test
to assess differences between groups. For cases where survival
curves exhibited late crossover, the two-stage method provided by
the ‘TSHRC’ package (version 0.1.6) was employed for further
comparison. Univariate and multivariate Cox regression analyses
were used to identify independent prognostic factors. Correlation
analysis was conducted using the Spearman's correlation test.
Multiple group comparisons were conducted using one-way ANOVA
followed by the Tukey post hoc test. Each assay was replicated in
at least three independent experiments. P<0.05 was considered to
indicate a statistically significant difference.
Results
Diagnostic potential of MIS18A in
LUAD
Pan-cancer expression analysis using the TIMER
database revealed a significant increase in MIS18A mRNA expression
in 19 different cancer tissue types (Fig. 1A). Analysis of the TCGA dataset and
validation across three GEO datasets confirmed the upregulation of
MIS18A in LUAD (Fig. 1B-E).
Furthermore, the differential expression of MIS18A in LUAD was
validated using RT-qPCR with tissue samples collected from patients
at the First Affiliated Hospital of Guangxi Medical University
(Fig. 1J). The area under the curve
(AUC) values were 0.962 for the TCGA dataset and 0.917, 0.890 and
0.971 for the GSE30219, GSE10072 and GSE27262 GEO datasets,
respectively (Fig. 1F-I),
suggesting that MIS18A could serve as a reliable diagnostic marker
for LUAD. Examination using the HPA (Fig. 1K) and UALCAN (Fig. 1L) databases revealed a significant
upregulation of MIS18A protein expression in LUAD samples.
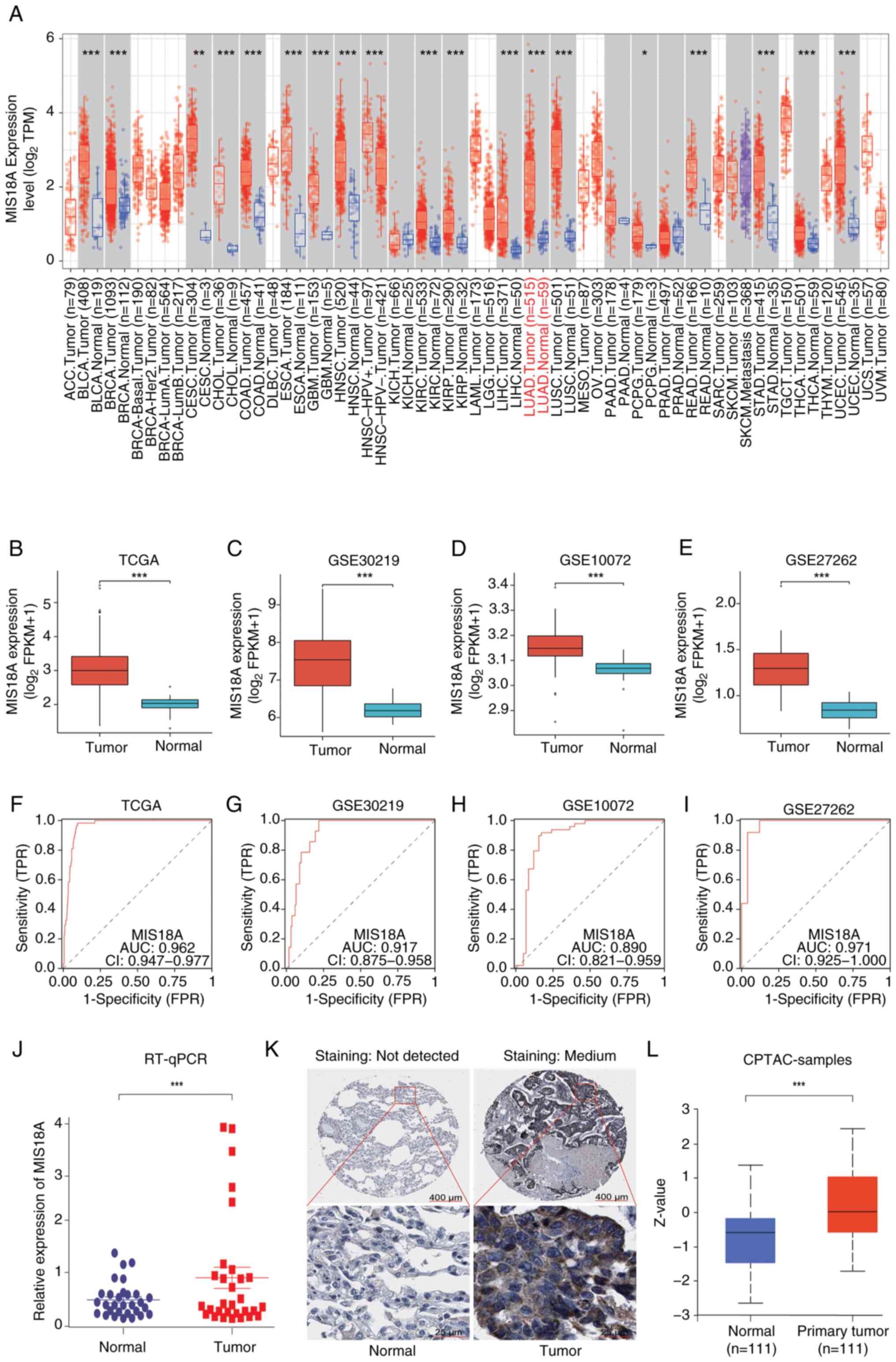 | Figure 1.MIS18A upregulation in LUAD and its
diagnostic significance. (A) Data regarding the upregulation of
MIS18A mRNA levels in various malignancies, including LUAD, was
retrieved from the TIMER database. Comparative analysis of MIS18A
between tumor and normal groups in (B) TCGA and three GEO datasets,
(C) GSE30219, (D) GSE10072 and (E) GSE27262. Diagnostic Receiver
Operator Characteristic curve analysis of MIS18A in (F) TCGA and
the three GEO datasets, (G) GSE30219, (H) GSE10072 and (I)
GSE27262. (J) Comparison of MIS18A expression levels between tumor
and adjacent lung tissues using RT-qPCR. (K) Representative
immunohistochemistry images of MIS18A in LUAD and normal lung
tissues from the Human Protein Atlas database. (L) Upregulation of
MIS18A protein levels in LUAD tissues according to UALCAN.
*P<0.05, **P<0.01, ***P<0.001. LUAD, lung adenocarcinoma;
MIS18A, MIS18 kinetochore protein A; TCGA, The Cancer Genome Atlas;
GEO, Gene Expression Omnibus; TPM, transcripts per million; FPKM,
Fragments Per Kilobase per Million mapped reads; TPR, true-positive
rate; FPR, false-positive rate; CPTAC, Clinical Proteomic Tumor
Analysis Consortium; AUC, area under the curve; CI, confidence
interval; RT-qPCR, reverse transcription-quantitative PCR. |
Prognostic significance of MIS18A in
LUAD
Next, the correlation between MIS18A expression and
various clinical characteristics, including prognosis and tumor (T)
and node stage were investigated. Analysis of the TCGA cohort
indicated that high MIS18A expression was correlated with a poorer
prognosis (Fig. 2A). This
association was validated following analyses of the GSE30219
dataset (Fig. 2B) and GEPIA
(Fig. 2C) and Kaplan-Meier Plotter
(Fig. 2D) databases. Within the
TCGA-LUAD cohort, both the Cox univariate (Fig. 2E) and multivariate (Fig. 2F) analyses demonstrated that MIS18A
expression could independently predict a poor prognosis [hazard
ratio (HR)>1; P<0.05]. Moreover, elevated MIS18A expression
was detected in patients with LUAD who exhibited a higher clinical
T stage (Fig. 2G), the presence of
lymph node metastases (Fig. 2H) and
advanced pathological stages (Fig.
2I).
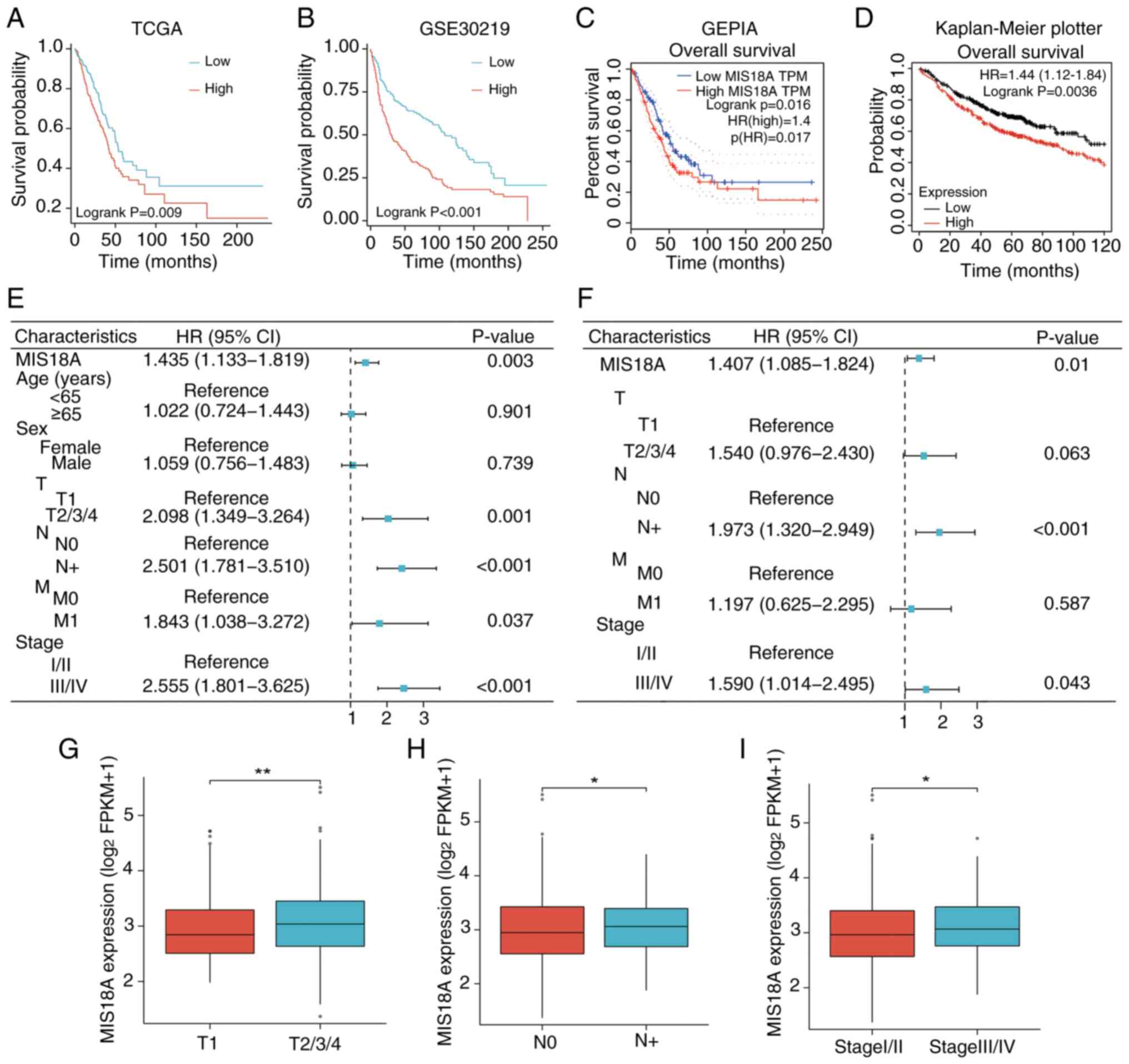 | Figure 2.Correlation of upregulated MIS18A
expression with poor prognosis. Kaplan-Meier survival analysis
comparing distinct MIS18A expression levels in the (A) TCGA and (B)
GSE30219 datasets. (C) Kaplan-Meier survival curve of MIS18A from
the GEPIA database. (D) Kaplan-Meier survival curve of MIS18A from
the Kaplan-Meier Plotter database. (E) Univariate and (F)
multivariate Cox regression analysis using the TCGA dataset.
Association of the MIS18A mRNA levels in samples from patients
across various (G) clinical T, (H) N and (I) pathological stages.
*P<0.05, **P<0.01. TCGA, The Cancer Genome Atlas; GEPIA, Gene
Expression Profiling Interactive Analysis; T, tumor; N, node; M,
metastasis; TPM, transcripts per million; FPKM, Fragments Per
Kilobase per Million mapped reads; HR, hazard ratio; CI, confidence
interval; MIS18A, MIS18 kinetochore protein A. |
Identification of DEGs and enrichment
analysis to explore MIS18A-related signaling pathways in LUAD
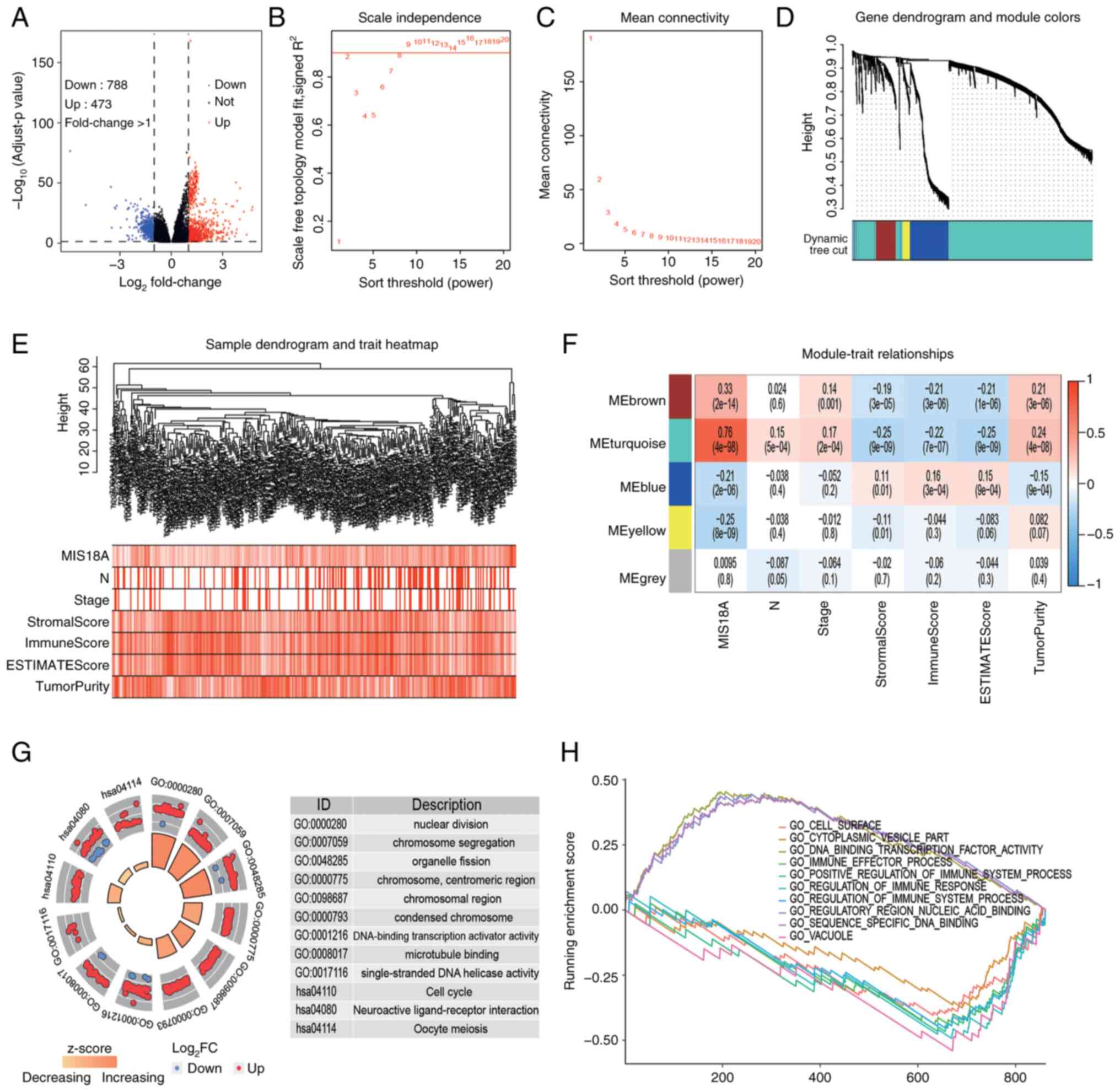
To investigate the potential roles of MIS18A in
LUAD, 788 upregulated and 473 downregulated DEGs were identified by
analyzing the gene expression profiles from TCGA database (Fig. 3A). Cluster analysis of these genes
can provide insights into the functions of MIS18A. Utilizing the
average linkage hierarchical clustering method, DEGs were
effectively classified into five modules (Fig. 3B-E). The heatmap revealed that the
genes within the turquoise module (n=899) exhibited a strong
positive correlation with MIS18A expression (ρ=0.76;
P=4×10−98) and a weak correlation with tumor purity
(ρ=0.24; P=4×10−8). Additionally, MIS18A expression was
negatively correlated with the immune score (ρ=−0.22;
P=7×10−7) (Fig. 3F).
Subsequent functional enrichment analysis of the turquoise module
genes identified significant associations with the cell cycle and
metabolic pathways (Fig. 3G). These
included terms such as ‘nuclear division’, ‘chromosome
segregation’, ‘chromosomal region’, ‘DNA-binding transcription
activator activity’, ‘single-stranded DNA helicase activity’, ‘Cell
cycle’ and ‘neuroactive ligand-receptor interaction’. Additionally,
GSEA highlighted the involvement of genes from the turquoise module
in a variety of biological terms, including ‘GO_CELL_SURFACE’,
‘GO_CYTOPLASMIC_VESICLE_PART’,
‘GO_DNA_BINDING_TRANSCRIPTION_FACTOR_ACTIVITY’,
‘GO_IMMUNE_EFFECTOR_PROCESS’,
‘GO_POSITIVE_REGULATION_OF_IMMUNE_SYSTEM_PROCESS’,
‘GO_REGULATION_OF_IMMUNE_RESPONSE’,
‘GO_REGULATION_OF_IMMUNE_SYSTEM_PROCESS’,
‘GO_REGULATORY_REGION_NUCLEIC_ACID_BINDING’,
‘GO_SEQUENCE_SPECIFIC_DNA_BINDING’ and ‘GO_VACUOLE’ (Fig. 3H). These terms were primarily
associated with cell cycle and immune-related pathways. Enrichment
analysis therefore suggested that MIS81A may promote cell division
and influence the tumor immune microenvironment.
Identification and analysis of the
diagnostic and prognostic value of the hub genes
To investigate the relationship between MIS18A and
other genes, the MIS18A PPI network was examined using GeneMANIA,
which predicted its interactions with 20 proteins. These proteins
collectively form a multifaceted PPI network characterized by
physical interactions, co-expression patterns, predicted
interactions, co-localization, genetic interactions and pathway
associations. These proteins were implicated in various biological
processes, including ‘CENP-A containing chromatin organization’,
‘DNA replication-independent nucleosome assembly’, ‘chromatin
remodeling at centromere’, ‘histone exchange’, ‘DNA
replication-independent nucleosome organization’, ‘centromere
complex assembly’ and ‘nucleosome assembly’ (Fig. 4A). Overlapping the PPI network with
genes from the turquoise module identified 6 DEGs, namely MIS18A,
Holliday junction recognition protein (HJURP), Opa interacting
protein 5 (OIP5), CENPA, CENPK and CENPU, as hub genes (Fig. 4B). Subsequent analyses of these hub
genes were performed. Cox analysis identified all hub genes as risk
factors (HR >1; Fig. 4C),
contributing to an unfavorable prognosis in LUAD. This was further
corroborated by the Kaplan-Meier survival analysis using the GEPIA
database (Fig. 4N-R). Additionally,
the hub genes were found to be upregulated in tumor tissues
(Fig. 4D-H). ROC curves were
generated using the TCGA cohort to evaluate the diagnostic
efficacies of HJURP, OIP5, CENPA, CENPK and CENPU for LUAD, with
corresponding AUCs of 0.984, 0.949, 0.967, 0.943 and 0.958,
respectively (Fig. 4I-M). These
results demonstrated the notable diagnostic potential of the hub
genes identified in LUAD.
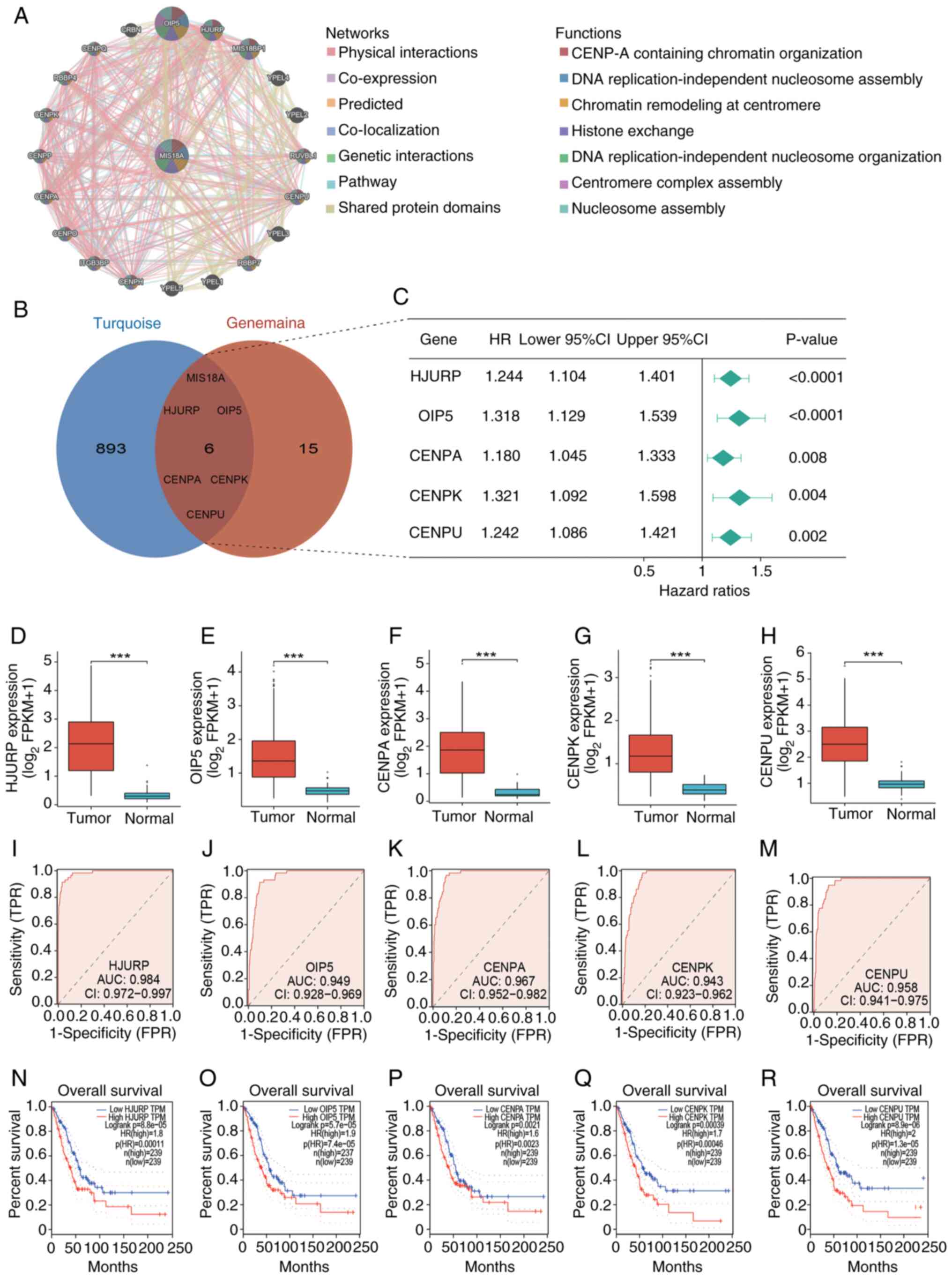 | Figure 4.Identification of hub genes and their
diagnostic and prognostic value in LUAD. (A) Protein-protein
interaction network of MIS18A with interactive genes from the
GeneMANIA database. Each point represents a gene. In the network
section, green lines represent genetic interactions and in the
functions section, the green parts within the genes signify DNA
replication-independent nucleosome organization. (B) Identification
of the 6 hub genes: MIS18A, HJURP, OIP5, CENPA, CENPK and CENPU.
(C) Forest plot of Cox analysis of the hub gene expression. (D-H)
Differential expression of the hub genes between tumor and normal
groups. (I-M) Diagnostic Receiver Operator Characteristic curves of
the hub genes for differentiating LUAD from normal tissue. (N-R)
High expression of the hub genes was associated with decreased
overall survival in patients with LUAD from the Gene Expression
Profiling Interactive Analysis database. ***P<0.001. LUAD, lung
adenocarcinoma; MIS18A, MIS18 kinetochore protein A; HJURP,
holliday junction recognition protein; OIP5, Opa interacting
protein 5; CENPA, centromere protein A; HR, hazard ratio; CI,
confidence interval; AUC, area under the curve; FPR, false-positive
rate; TPR, true-positive rate. |
Relationship between MIS18A and the
tumor microenvironment (TME)
The TME plays a crucial role in clonal evolution,
growth, metastasis, prognosis, drug resistance and tumor treatment
outcomes. Consequently, the immune characteristics of MIS18A in
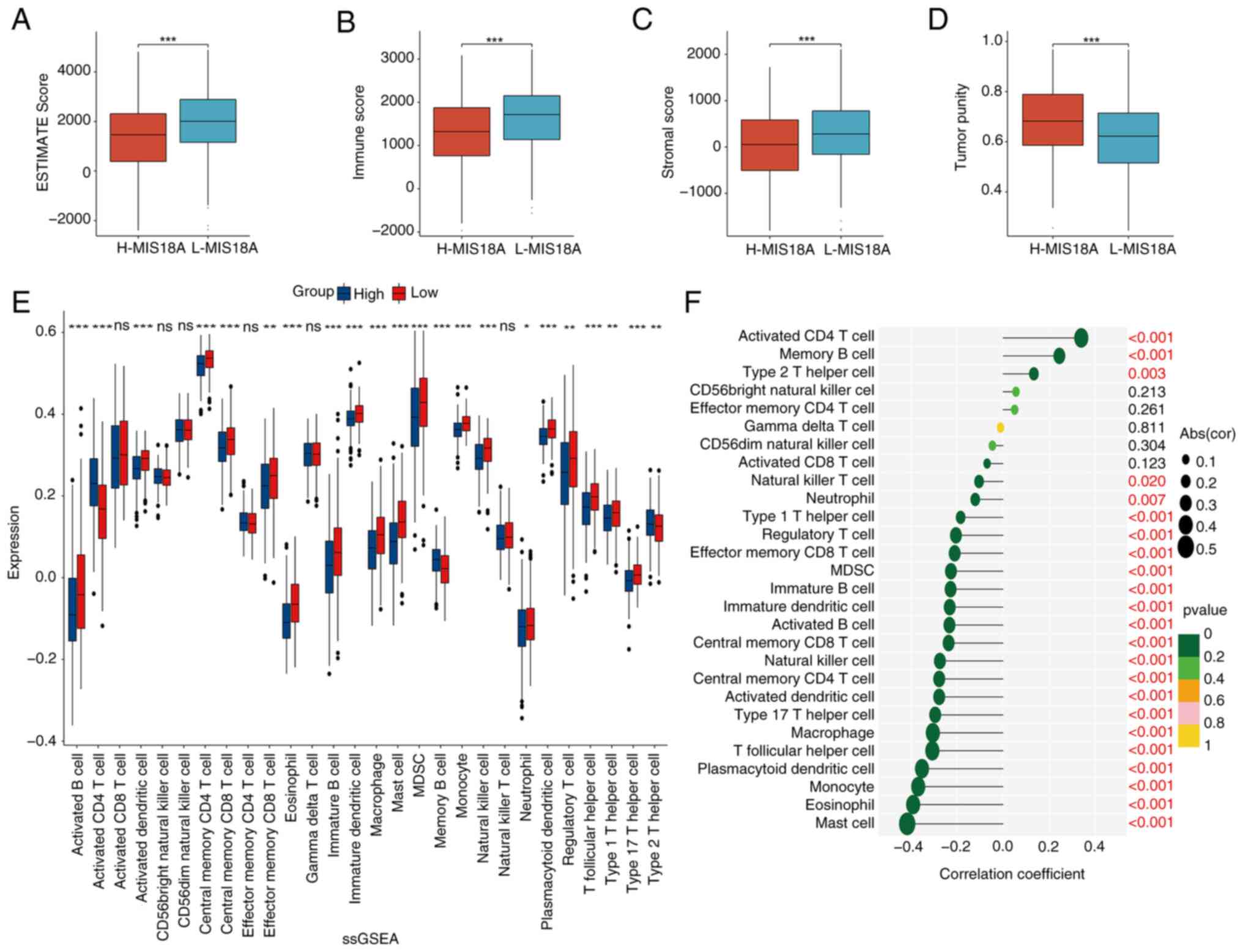
LUAD were analyzed. The ESTIMATE algorithm was used to calculate
the stromal, immune and ESTIMAE scores, which were subsequently
used to estimate tumor purity. The results indicated that elevated
MIS18A expression was associated with a significant decrease in the
ESTIMATE (Fig. 5A), immune
(Fig. 5B) and stromal (Fig. 5C) scores, which was accompanied by a
higher tumor purity (Fig. 5D). The
ssGSEA revealed a significant increase in the enrichment of
activated CD4+ T cells, memory B cells and Type 2 T
helper cells in the high MIS18A expression group. By contrast, the
low MIS18A expression group exhibited higher enrichment levels in
19 immune cell subtypes, including activated B cells, activated
dendritic cells (DCs), central memory CD4+ T cells and
natural killer cells (Fig. 5E).
Furthermore, MIS18A expression demonstrated positive correlations
with activated CD4+ T cells (ρ=0.34; P<0.001), memory
B cells (ρ=0.24; P<0.001) and Type 2 T helper cells (ρ=0.13;
P=0.003), but negative correlations with mast cells (ρ=−0.42;
P<0.001), eosinophils (ρ=−0.39; P<0.001), monocytes (ρ=−0.37;
P<0.001) and plasmacytoid DCs (ρ=−0.35; P<0.001), among
others (Fig. 5F). Additionally,
analysis of the TISIDB revealed a negative correlation between
MIS18A and various immune-related chemokines, further
substantiating the role of MIS18A as an immune regulator in LUAD
(Fig. S1).
Mutation analysis and assessment of
drug sensitivity benefits
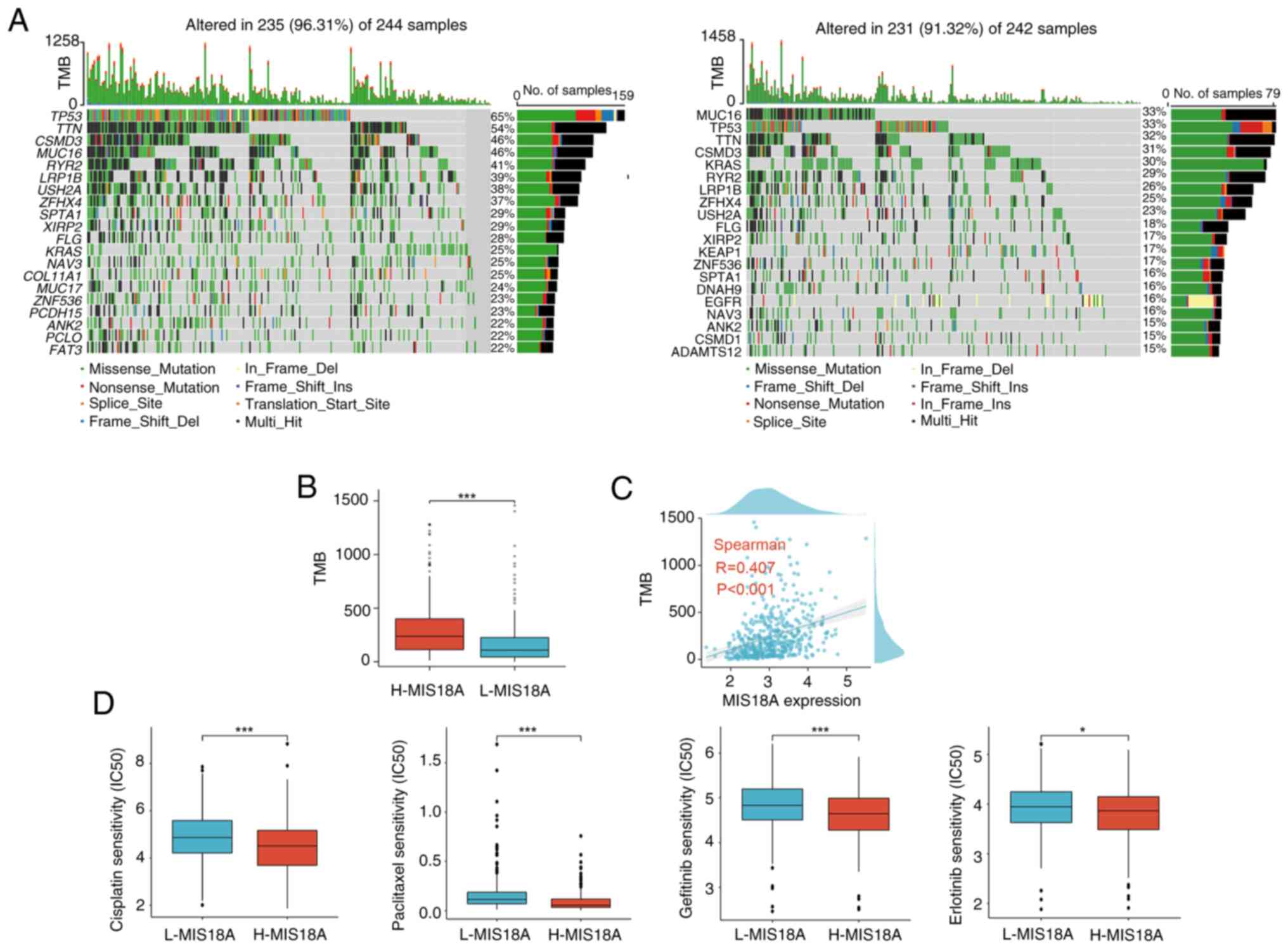
To assess the correlation between MIS18A expression
and the immune therapeutic response, gene mutation maps for
different MIS18A expression groups were presented using a waterfall
plot. Elevated MIS18A expression was associated with an increased
frequency of TP53 and TTN mutations (Fig. 6A). Moreover, the high MIS18A
expression cohort demonstrated a significantly higher TMB and a
robust correlation between MIS18A expression and the TMB
(P<0.01; Fig. 6B and C). With
the view of improving treatment outcomes for patients with LUAD,
variations in sensitivity to commonly used chemotherapeutic agents
and targeted drugs between the high and low MIS18A expression
groups were meticulously examined. The results demonstrated that
the high MIS18A expression group displayed a lower IC50
value than the low MIS18A expression group in response to
cisplatin, paclitaxel, gefitinib and erlotinib (Fig. 6D).
Experimental verification
Based on the aforementioned evidence suggesting the
potential involvement of MIS18A in LUAD progression, gene knockdown
experiments were conducted in A549 cells. Initially, knocking down
MIS18A at three distinct sites significantly reduced its
expression, consequently leading to the selection of si-MIS18A-2
for further experiments (Fig. 7A).
Subsequent analysis using the CCK-8 assay demonstrated a
significant reduction in A549 cell viability at 48 h after MIS18A
downregulation (Fig. 7B).
Furthermore, the results of the wound healing assay revealed a
significant inhibition of A549 cell migration following MIS18A
knockdown (Fig. 7C). Finally, the
Transwell invasion assay demonstrated a significant reduction in
A549 cell invasion following MIS18A knockdown (Fig. 7D). Collectively, these results
confirmed a reduction in cellular viability, migration and invasion
upon transfection and subsequent gene knockdown with si-MIS18A.
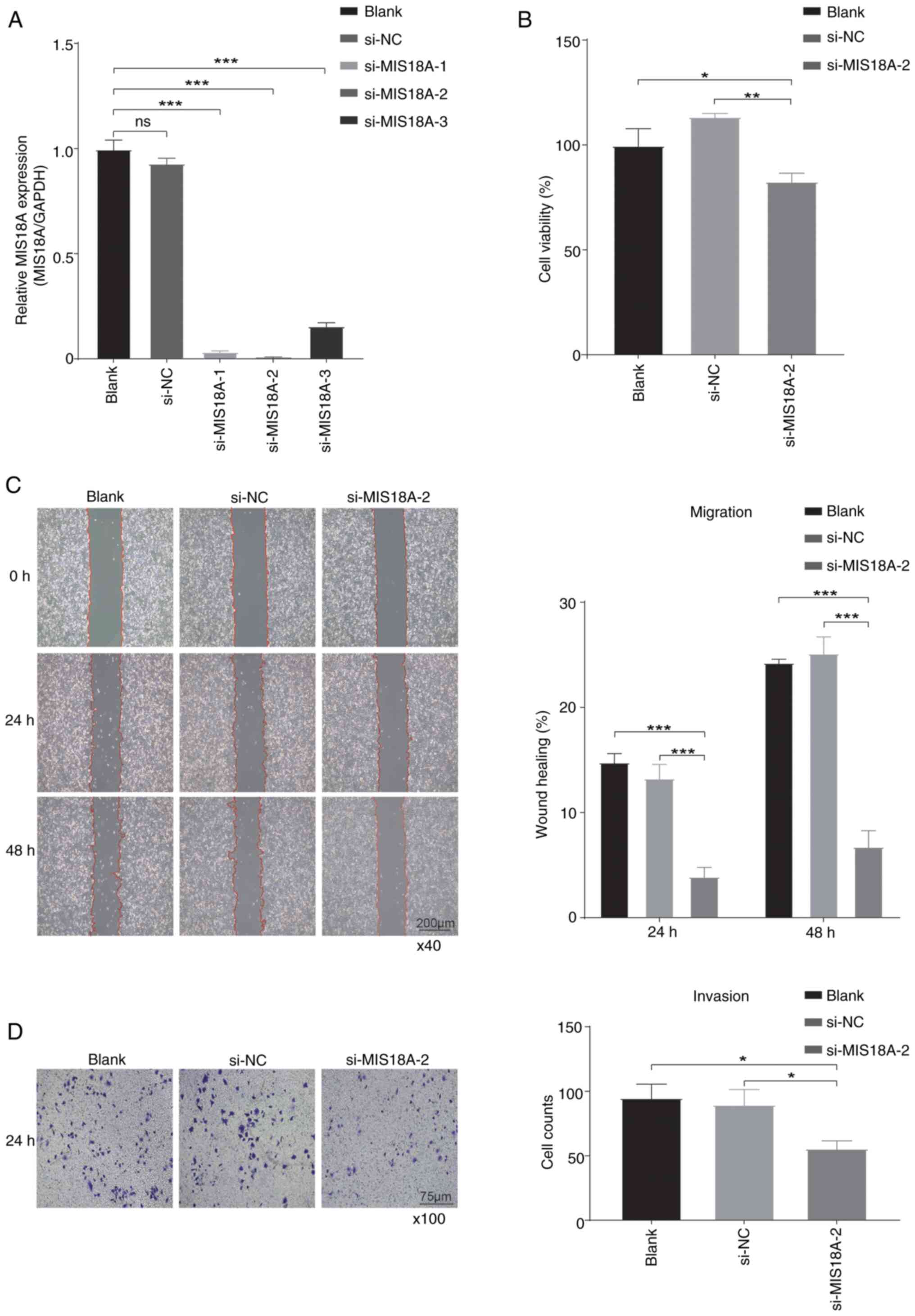 | Figure 7.Suppression of cell proliferation,
migration and invasion by MIS18A knockdown in LUAD. (A) MIS18A mRNA
expression in A549 cells following transfection with si-MIS18A. (B)
Cell Counting Kit-8 assay showing a reduction in LUAD cell
viability following transfection with si-MIS18A-2. (C) Wound
healing assays revealed a decrease in the wound healing rate of
LUAD cells following transfection with si-MIS18A-2. Magnification,
×40. (D) Transwell assays showing the suppression of LUAD cell
invasion following transfection with si-MIS18A-2. Magnification,
×100. *P<0.05, **P<0.01, ***P<0.001. LUAD, lung
adenocarcinoma; MIS18A, MIS18 kinetochore protein A; si, small
interfering; NC, negative control; ns, not significant. |
Establishment of a ceRNA network
associated with MIS18A
The aforementioned findings suggested that MIS18A
enhances the viability, migration and invasion of LUAD cells,
prompting an in-depth investigation into its multifaceted nature.
Increasing evidence suggests that alterations and dysfunctions in
lncRNAs contribute to abnormal gene expression and promote the
development, progression and metastasis of various cancer types
(32,33). lncRNAs sequester miRNAs, thereby
influencing their mRNA expression (34). Therefore, a ceRNA regulatory network
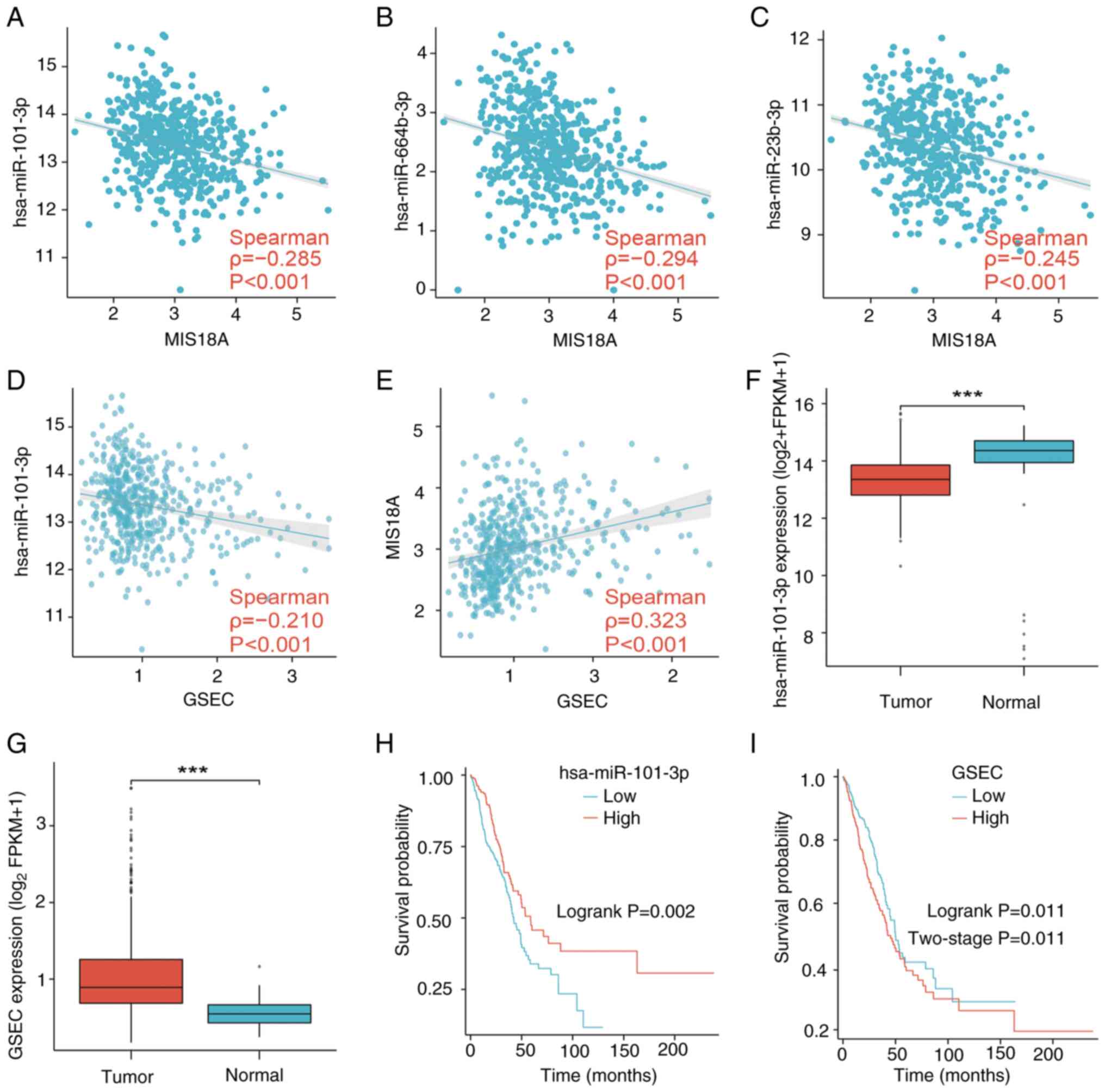
of MIS18A in LUAD was constructed to explore the relationships
between MIS18A, lncRNAs and miRNAs. The three predicted miRNAs in
the network demonstrated correlation coefficients with MIS18A
expression of approximately-0.3, indicating a weak correlation
(Fig. 8A-C). Despite this,
hsa-miR-101-3p was selected as the principal miRNA for further
study, primarily due to its observed upregulation in lung tissues
(Fig. 8F) and the association of
high hsa-miR-101-3p expression with a favorable prognosis in
patients (Fig. 8H), underscoring
its significant biological relevance in LUAD. GSEC, the lncRNA
predicted by hsa-miR-101-3p, showed an inverse correlation with
hsa-miR-101-3p (Fig. 8D) and a
positive correlation with MIS18A (Fig.
8E). The upregulation of GSEC in LUAD demonstrated a
significant association with an unfavorable prognosis (Fig. 8G and I), resulting in the
identification of GSEC as the principal lncRNA in the ceRNA network
associated with MIS18A in LUAD.
Discussion
Lung cancer is a common and lethal malignancy
worldwide (35). The emergence of
precision medicine has steered tumor treatment strategies towards
minimally invasive, efficient and personalized approaches,
progressively advancing cancer therapeutics (36). In terms of advancement in cancer
research, Wang et al (37)
demonstrated the development of integrative serum metabolic
fingerprint-based multimodal platforms designed for lung nodule
characterization and the early detection of LUAD. Another study
underscored the innovative application of mass
spectrometry/spectroscopy and machine learning in in vitro
diagnostics, demonstrating their potential to enhance diagnostic
accuracy and efficiency (38).
Additionally, Liang et al (39) delved into the field of
nanozyme-based clinical biomarker assays, which hold promising
potential in disease diagnosis and personalized medicine. These
research findings not only highlight the cutting-edge technologies
shaping the landscape of cancer diagnostics but also provide
valuable insights for improving disease detection.
At present, there is limited understanding of the
role of MIS18A in tumors. As a critical component of the Mis18
complex, MIS18A is crucial for the intricate process of chromosome
segregation and the precise positioning of the centromere protein,
CENPA (9). An investigations has
revealed that the deletion of MIS18A causes significant chromosomal
misalignment, CENPA depletion and cell death (40). Notably, MIS18A and CENPA were
significantly downregulated in a murine model of colorectal cancer,
providing additional evidence that the aberrant functionality of
MIS18A leads to erroneous chromosome segregation (41). These findings revealed the
indispensable role of MIS18A in cellular division and provided
crucial insights into its potential implications in
tumorigenesis.
In the present study, a comprehensive analysis of
MIS18A expression and its clinical implications in patients with
LUAD were conducted using diverse data sources and bioinformatic
methods. The results revealed the upregulated expression of MIS18A
in LUAD, highlighting its potential as a diagnostic marker.
Additionally, upregulation of MIS18A in patients with LUAD showed a
strong correlation with poor prognosis, suggesting its potential
involvement in driving tumor progression. Therefore, MIS18A may
function as an independent prognostic indicator of LUAD. In
addition, a significant correlation was observed between MIS18A
levels and immune characteristics. To further investigate the
biological functions of MIS18A in LUAD, 899 genes in the turquoise
module were analyzed. GO enrichment analysis confirmed the previous
observations, establishing a significant association between MIS18A
and processes related to cell division and chromosomes.
Additionally, KEGG enrichment analysis further revealed a robust
correlation between MIS18A and pathways such as neuroactive
ligand-receptor interactions and the cell cycle. Notably, the
chromosome 15q25.1 locus has been identified as a significant
susceptibility region for lung cancer through a genome-wide
association study (42). The study
indicated that common genetic variations in this region may affect
the structure or expression of genes involved in the neuroactive
ligand-receptor interaction pathway, potentially influencing lung
cancer susceptibility. Consequently, the neuroactive
ligand-receptor interaction pathway has been identified as a risk
factor for lung cancer development. Dysregulation of the cell cycle
is a fundamental mechanism in tumor development and presents
numerous potential targets for therapeutic intervention (43,44).
In the present study, validation through GSEA reinforced these
findings and suggested the potential involvement of MIS18A in
processes related to the immune system. In summary, the functional
enrichment analysis results highlighted the significant involvement
of MIS18A in the initiation and progression of LUAD.
In the present study, WGCNA and PPI network analysis
identified 6 hub genes in LUAD: MIS18A, HJURP, OIP5, CENPA, CENPK
and CENPU. Cox regression analysis demonstrated that all these hub
genes were associated with an elevated risk of LUAD. Furthermore,
upregulation of each hub gene was indicative of an unfavorable
prognosis in patients with LUAD, emphasizing their diagnostic
significance. Prior studies have highlighted the significance of
HJURP in LUAD, indicating that elevated HJURP expression is
associated with poor prognosis (45,46).
In addition, HJURP is known to facilitate tumor cell proliferation,
migration and invasion via the Wnt/β-catenin signaling pathway
(47). OIP5, a member of the cancer
testis antigen family (48), plays
a pivotal role in the structure and function of kinetochores and
centromeric regions (49).
Associations with the mutant genes TP53, SMARCA4 and SCN1A suggest
potential pathogenic roles for OIP5 in the development of LUAD
(50). Additionally, components of
the CENPA-nucleosome associated complex, such as CENPA and CENPU,
are essential for the development and evolution of LUAD (51,52).
The upregulation of CENPK in LUAD tissues and cells has also been
associated with increased cell viability, migration, invasion and
epithelial-mesenchymal transition (53,54).
These findings collectively reinforce the pivotal roles of the
identified hub genes, including MIS18A, in driving LUAD
progression, and provide additional evidence of its impact on
disease progression.
The TME plays a crucial role in tumor development by
influencing key processes such as growth, invasion, metastasis and
immune evasion (55,56). The results of the present study
revealed an association between elevated MIS18A expression and
lower immune and stromal scores, along with increased tumor purity.
The ssGSEA revealed a negative correlation between MIS18A
expression and various tumor-infiltrating immune cells, including B
cells, CD4+ T cells, CD8+ T cells, DCs,
macrophages, natural killer cells, monocytes, eosinophils and mast
cells. Notably, among the cytotoxic T lymphocytes, CD8+
T cells have emerged as the primary driving force in the immune
response against cancer, owing to their unique ability to directly
identify and eliminate malignant cells (57). Specifically, CD8+ T cells
recognize major histocompatibility complex class I molecules that
present tumor antigens on the surface of malignant cells. Upon
recognition, CD8+ T cells release cytotoxic substances
such as perforin, granzymes and cytokines, resulting in the
elimination of the targeted tumor cells. Additionally,
CD4+ T cells are crucial in tumor immunity, modulating
immune responses via cytokine secretion and interactions with other
immune cells, fostering an environment conducive to effective
immune surveillance and tumor elimination (58). DCs possess unique abilities for
antigen uptake and processing, which enable the identification and
internalization of various antigens, including those present on
tumor cell surfaces (59). During
cancer cell invasion, chemokines and their receptors orchestrate
the migration of malignant cells (60). The results of the present study
identified a negative correlation between MIS18A expression and
several chemokines, including chemokine (C-C motif) ligand (CCL)
14, chemokine (C-X-C motif) ligand (CXCL) 16, CCL23, CCL17, CCL19
and CCL13, as well as significant associations with diverse
chemokine receptors, including CX3CR, CCR6, CXCR2, CCR4 and CCR7.
The activation of monocytes, macrophages and THP-1 cells is
attributable to the binding of CCL14 to CCR1, CCR3 and CCR5
(61). In addition, the
Wnt/β-catenin pathway has been found to be carcinogenic in various
cancer types, including hepatocellular carcinoma (HCC) (62) and LUAD (63,64).
Zhu et al (61) demonstrated
the activation of the Wnt/β-catenin pathway in HCC by CCL14. By
knocking down CCL14 in HCC cells, an increase in
phosphorylated-GSK3β (S9) and β-catenin (S33/S37) levels was
observed, leading to the upregulation of target genes of the
Wnt/β-catenin pathway. This suggested a potential pathway through
which CCL14 inhibits the proliferation or apoptosis of LUAD cells.
Additionally, CCR7 and CCL19 have been identified as favorable
prognostic factors for patients with LUAD (65). The findings of the present study
therefore underscored the significant role of MIS18A in shaping the
immune microenvironment within tumors, suggesting that elevated
MIS18A expression suppresses cancer immunity, thereby promoting
cancer progression.
Considering the functional characteristics of MIS18A
in LUAD and its impact on tumor-infiltrating immune cells, the
association between MIS18A expression and the sensitivity to
chemotherapy and targeted therapy was analyzed in the present
study. In recent years, TMB has emerged as a focal point in the
study of biomarkers associated with immune checkpoint inhibitors
(ICIs) (66). Despite the ongoing
debate regarding its reliability as a predictive marker (67–71),
elevated TMB has been demonstrated to stimulate the generation of
novel immune antigens, enhancing tumor immunogenicity and the
efficacy of ICIs (72–74). In the present study, it was noted
that patients with LUAD exhibiting high MIS18A expression
demonstrated a heightened TMB compared with patients with low
MIS18A expression, suggesting a potential link between MIS18A and
the immune response to treatment. However, further experimental
validation is required to confirm this finding. In addition, the
results of the OncoPredict analysis suggested that patients with
LUAD and elevated MIS18A expression may benefit from treatment
regimens involving cisplatin, paclitaxel, erlotinib and gefitinib.
At present, cisplatin-based chemotherapy is the primary treatment
for lung cancer owing to the rapid growth and metabolism of tumor
cells and functions, by disrupting DNA replication and
transcription and causing apoptosis in tumor cells (75). Paclitaxel, an established anticancer
agent, disrupts the dynamic equilibrium of tubulin proteins,
promotes tubulin protein aggregation and microtubule assembly and
inhibits depolymerization, thereby stabilizing microtubules and
impeding cancer cell mitosis, leading to apoptosis and effectively
preventing cancer cell proliferation (76). Epidermal growth factor receptor
(EGFR) is a membrane-bound receptor that is widely expressed in
human epidermal and stromal cells and is known for its tyrosine
kinase activity. The tyrosine kinase activity of EGFR is precisely
regulated in normal cells. However, gene mutations can lead to the
sustained activation of EGFR and contribute to tumorigenesis
(77). Gefitinib and erlotinib,
first-generation small molecule EGFR tyrosine kinase inhibitor
targeted drugs extensively used in clinical settings for patients
with advanced LUAD, have demonstrated promising efficacy (78,79).
Gefitinib and erlotinib act by effectively binding to EGFR,
inhibiting tyrosine kinase activity, blocking downstream signal
transduction, suppressing angiogenesis and inducing apoptosis in
tumor cells (80,81).
To validate the findings regarding the role MIS18A
in LUAD, a series of cell experiments were also conducted in the
present study. MIS18A knockdown significantly reduced the
viability, migration and invasion of LUAD cells, highlighting its
crucial role in promoting cell viability and potentially enhancing
the metastatic capacity of tumor cells. These results emphasized
the potential of MIS18A as a novel predictive biomarker of
LUAD.
The ceRNA regulatory networks are widely
acknowledged as crucial post-transcriptional regulators of gene
expression. Mounting evidence indicates that ceRNA regulatory
networks contribute to the regulation of various biological
processes, particularly tumorigenesis (82–84).
In the present study, the potential miRNAs that target MIS18A were
initially predicted. A marked decrease in hsa-miR-101-3p levels was
noted, which was inversely related to MIS18A expression, indicating
a favorable OS in LUAD. Notably, hsa-miR-101-3p serves as a
biomarker for various cancer types, including bladder cancer
(85), prostate cancer (86), HCC (87) and colorectal cancer (88). Additionally, the potential upstream
lncRNAs that may regulate hsa-miR-101-3p expression was predicted
in the present study. The results demonstrated a significant
upregulation of GSEC, which exhibited a positive correlation with
MIS18A expression and was associated with a poor OS in LUAD.
Existing data indicate that GSEC plays a notable role in
carcinogenesis by affecting multiple signaling pathways across
various cancer types, including LUAD (89), triple negative breast cancer
(90) and osteosarcoma (91). However, to the best of our
knowledge, the significance of the GSEC/hsa-miR-101-3p/MIS18A ceRNA
regulatory network in LUAD has not yet been explored. The present
study investigated the prognostic implications of the
GSEC/hsa-miR-101-3p/MIS18A network, thereby providing a fresh
perspective for LUAD treatment.
The present study has several limitations. First,
although the expression levels and biological functions of MIS18A
were successfully validated in LUAD cells, the efficiency of the
si-MIS18A knockdown was not assessed by measuring MIS18A protein
expression. Second, the potential molecular mechanisms by which
MIS18A functions in tumor progression were not investigated.
Further studies based on animal models are warranted to
comprehensively elucidate the functional roles of MIS18A in
vivo. Third, although the bioinformatics analyses revealed
significant associations between MIS18A, immune infiltration and
chemokines, these findings lacked experimental validation in
vitro. In future, investigations will focus on the molecular
mechanisms and immunoregulatory functions of MIS18A in LUAD.
In summary, the results of the present study
suggested that MIS18A may serve as a potential diagnostic and
prognostic marker in patients with LUAD. Furthermore, MIS18A has
the potential to influence the biological activity of immune cells,
affect the cell cycle and mitigate clinical drug resistance,
suggesting its potential role in tumor immunotherapy. Notably, the
experimental knockdown of MIS18A resulted in a significant
reduction in cell viability, migration and invasion, emphasizing
the potential of MIS18A as a biomarker and possible therapeutic
target for LUAD.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YZ contributed to the conceptualization of the
present study, methodology, software (coding and implementing data
analysis tools), investigation (conducting and designing
experiments), validation, data curation and writing of the original
manuscript draft. ZL contributed to the conceptualization of the
present study, methodology, investigation (data collection and
experimental execution), validation and data curation. ZW
contributed to the methodology, software (coding and maintaining
analysis software), investigation (performing laboratory
experiments) and validation. TZ contributed to the software
(software testing and debugging), validation and investigation
(sample analysis and data collection). LD contributed to the
methodology and investigation (conducting sample analyses). GL
contributed to the investigation (data gathering and preliminary
analysis) and reviewing and editing the manuscript. HP contributed
to the investigation (participating in experimental work) and
reviewing and editing the manuscript. HL contributed to the design
and optimization of experimental methodologies, and the reviewing
and editing of the manuscript. YW contributed to the organization
and management of data (systematically collecting, cleaning, and
preparing the data to ensure its integrity and readiness for
analysis), the reviewing and editing of the manuscript, supervision
of the overall research project, securing funding, and project
administration. YZ and YW confirm the authenticity of all the raw
data and revised the final version of the manuscript. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The research protocol and the process of collecting
human samples received approval from The Medical Ethics Committee
of The First Affiliated Hospital of Guangxi Medical University
(Nanning, China; approval no. 2023-E508-01). All participants
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang W, Zhang R, Zeng Y, Li Y, Chen Y,
Zhou J, Zhang Y, Wang A, Zhu J, Liu Z, et al: ALCAP2 inhibits lung
adenocarcinoma cell proliferation, migration and invasion via the
ubiquitination of β-catenin by upregulating the E3 ligase NEDD4L.
Cell Death Dis. 12:7552021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim JW, Marquez CP, Kostyrko K, Koehne AL,
Marini K, Simpson DR, Lee AG, Leung SG, Sayles LC, Shrager J, et
al: Antitumor activity of an engineered decoy receptor targeting
CLCF1-CNTFR signaling in lung adenocarcinoma. Nat Med.
25:1783–1795. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oudkerk M, Liu S, Heuvelmans MA, Walter JE
and Field JK: Lung cancer LDCT screening and mortality
reduction-evidence, pitfalls and future perspectives. Nat Rev Clin
Oncol. 18:135–151. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fang C, Liang Y, Huang Y, Jiang D, Li J,
Ma H, Guo L, Jiang W and Feng Y: P3H4 promotes malignant
progression of lung adenocarcinoma via interaction with EGFR.
Cancers (Basel). 14:32432022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, et al: Mapping the hallmarks of lung adenocarcinoma
with massively parallel sequencing. Cell. 150:1107–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pan D, Klare K, Petrovic A, Take A,
Walstein K, Singh P, Rondelet A, Bird AW and Musacchio A:
CDK-regulated dimerization of M18BP1 on a Mis18 hexamer is
necessary for CENP-A loading. Elife. 6:e233522017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fujita Y, Hayashi T, Kiyomitsu T, Toyoda
Y, Kokubu A, Obuse C and Yanagida M: Priming of centromere for
CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1.
Dev Cell. 12:17–30. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nardi IK, Zasadzińska E, Stellfox ME,
Knippler CM and Foltz DR: Licensing of centromeric chromatin
assembly through the Mis18α-Mis18β heterotetramer. Mol Cell.
61:774–787. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sullivan KF, Hechenberger M and Masri K:
Human CENP-A contains a histone H3 related histone fold domain that
is required for targeting to the centromere. J Cell Biol.
127:581–592. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu WT, Wang Y, Zhang J, Ye F, Huang XH,
Li B and He QY: A novel strategy of integrated microarray analysis
identifies CENPA, CDK1 and CDC20 as a cluster of diagnostic
biomarkers in lung adenocarcinoma. Cancer Lett. 425:43–53. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim IS, Lee M, Park KC, Jeon Y, Park JH,
Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH and Kim KI: Roles of Mis18α
in epigenetic regulation of centromeric chromatin and CENP-A
loading. Mol Cell. 46:260–273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun SY, Hu XT, Yu XF, Zhang YY, Liu XH,
Liu YH, Wu SH, Li YY, Cui SX and Qu XJ: Nuclear translocation of
ATG5 induces DNA mismatch repair deficiency (MMR-D)/microsatellite
instability (MSI) via interacting with Mis18α in colorectal cancer.
Br J Pharmacol. 178:2351–2369. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:186ra1662013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Landi MT, Dracheva T, Rotunno M, Figueroa
JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et
al: Gene expression signature of cigarette smoking and its role in
lung adenocarcinoma development and survival. PLoS One.
3:e16512008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei TYW, Hsia JY, Chiu SC, Su LJ, Juan CC,
Lee YC, Chen JM, Chou HY, Huang JY, Huang HM and Yu CT: Methylosome
protein 50 promotes androgen- and estrogen-independent
tumorigenesis. Cell Signal. 26:2940–2950. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor- infiltrating immune cells. Cancer Res. 77:e108–e110.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Therneau T: A package for survival
analysis in R. R package version 3.6–4. 2024.
|
|
22
|
Kassambara A, Kosinski M and Biecek P:
Survminer: Drawing survival curves using ‘ggplot2’. R package
version 0.4.9. 2021.
|
|
23
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mostafavi S, Ray D, Warde-Farley D,
Grouios C and Morris Q: GeneMANIA: A real-time multiple association
network integration algorithm for predicting gene function. Genome
Biol. 9 (Suppl 1):S42008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mayakonda A, Lin DC, Assenov Y, Plass C
and Koeffler HP: Maftools: Efficient and comprehensive analysis of
somatic variants in cancer. Genome Res. 28:1747–1756. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang W, Soares J, Greninger P, Edelman EJ,
Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et
al: Genomics of drug sensitivity in cancer (GDSC): A resource for
therapeutic biomarker discovery in cancer cells. Nucleic Acids Res.
41((Database Issue)): D955–D961. 2013.PubMed/NCBI
|
|
31
|
Stridfeldt F, Cavallaro S, Hååg P,
Lewensohn R, Linnros J, Viktorsson K and Dev A: Analyses of single
extracellular vesicles from non-small lung cancer cells to reveal
effects of epidermal growth factor receptor inhibitor treatments.
Talanta. 259:1245532023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang K, Zhang L, Mi Y, Tang Y, Ren F, Liu
B, Zhang Y and Zheng P: A ceRNA network and a potential regulatory
axis in gastric cancer with different degrees of immune cell
infiltration. Cancer Sci. 111:4041–4050. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song YX, Sun JX, Zhao JH, Yang YC, Shi JX,
Wu ZH, Chen XW, Gao P, Miao ZF and Wang ZN: Non-coding RNAs
participate in the regulatory network of CLDN4 via ceRNA mediated
miRNA evasion. Nat Commun. 8:2892017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Malhotra J, Malvezzi M, Negri E, La
Vecchia C and Boffetta P: Risk factors for lung cancer worldwide.
Eur Respir J. 48:889–902. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cucchiara F, Petrini I, Romei C, Crucitta
S, Lucchesi M, Valleggi S, Scavone C, Capuano A, De Liperi A,
Chella A, et al: Combining liquid biopsy and radiomics for
personalized treatment of lung cancer patients. State of the art
and new perspectives. Pharmacol Res. 169:1056432021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang L, Zhang M, Pan X, Zhao M, Huang L,
Hu X, Wang X, Qiao L, Guo Q, Xu W, et al: Integrative serum
metabolic fingerprints based multi-modal platforms for lung
adenocarcinoma early detection and pulmonary nodule classification.
Adv Sci (Weinh). 9:e22037862022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen X, Shu W, Zhao L and Wan J: Advanced
mass spectrometric and spectroscopic methods coupled with machine
learning for in vitro diagnosis. VIEW. 4:202200382023. View Article : Google Scholar
|
|
39
|
Liang D, Wang Y and Qian K: Nanozymes:
Applications in clinical biomarker detection. Interdiscip Med.
1:e202300202023. View Article : Google Scholar
|
|
40
|
Baumann K: Keeping centromeric identity.
Nat Rev Mol Cell Biol. 13:3402012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pussila M, Törönen P, Einarsdottir E,
Katayama S, Krjutškov K, Holm L, Kere J, Peltomäki P, Mäkinen MJ,
Linden J and Nyström M: Mlh1 deficiency in normal mouse colon
mucosa associates with chromosomally unstable colon cancer.
Carcinogenesis. 39:788–797. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ji X, Bossé Y, Landi MT, Gui J, Xiao X,
Qian D, Joubert P, Lamontagne M, Li Y, Gorlov I, et al:
Identification of susceptibility pathways for the role of
chromosome 15q25.1 in modifying lung cancer risk. Nat Commun.
9:32212018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Carroll B and Korolchuk VI: Nutrient
sensing, growth and senescence. FEBS J. 285:1948–1958. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kamal MA, Al-Zahrani MH, Khan SH, Al-Subhi
HA, Kuerban A, Aslam M, Al-Abbasi FA and Anwar F: Tubulin proteins
in cancer resistance: A review. Curr Drug Metab. 21:178–185. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen L, Zeng C, Yan L, Liao W, Zhen C and
Yao J: Prognostic value of holliday junction-recognizing protein
and its correlation with immune infiltrates in lung adenocarcinoma.
Oncol Lett. 24:2322022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yin Q, Chen W, Zhang C and Wei Z: A
convolutional neural network model for survival prediction based on
prognosis-related cascaded Wx feature selection. Lab Invest.
102:1064–1074. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wei Y, Ouyang GL, Yao WX, Zhu YJ, Li X,
Huang LX, Yang XW and Jiang WJ: Knockdown of HJURP inhibits
non-small cell lung cancer cell proliferation, migration, and
invasion by repressing Wnt/β-catenin signaling. Eur Rev Med
Pharmacol Sci. 23:3847–3856. 2019.PubMed/NCBI
|
|
48
|
Afsharpad M, Nowroozi MR, Mobasheri MB,
Ayati M, Nekoohesh L, Saffari M, Zendehdel K and Modarressi MH:
Cancer-testis antigens as new candidate diagnostic biomarkers for
transitional cell carcinoma of bladder. Pathol Oncol Res.
25:191–199. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Naetar N, Hutter S, Dorner D, Dechat T,
Korbei B, Gotzmann J, Beug H and Foisner R: LAP2alpha-binding
protein LINT-25 is a novel chromatin-associated protein involved in
cell cycle exit. J Cell Sci. 120:737–747. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Abdel-Maksoud MA, Hassan F, Mubarik U,
Mubarak A, Farrag MA, Alghamdi S, Atuahene SA, Almekhlafi S and
Aufy M: An in-silico approach leads to explore six genes as a
molecular signatures of lung adenocarcinoma. Am J Cancer Res.
13:727–757. 2023.PubMed/NCBI
|
|
51
|
Zhou H, Bian T, Qian L, Zhao C, Zhang W,
Zheng M, Zhou H, Liu L, Sun H, Li X, et al: Prognostic model of
lung adenocarcinoma constructed by the CENPA complex genes is
closely related to immune infiltration. Pathol Res Pract.
228:1536802021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wu Q, Chen YF, Fu J, You QH, Wang SM,
Huang X, Feng XJ and Zhang SH: Short hairpin RNA-mediated
down-regulation of CENP-A attenuates the aggressive phenotype of
lung adenocarcinoma cells. Cell Oncol (Dordr). 37:399–407. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang Y, Wang Y, Ren C, Wang H, Zhang Y and
Xiu Y: Upregulation of centromere protein K is crucial for lung
adenocarcinoma cell viability and invasion. Adv Clin Exp Med.
30:691–699. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lai H, Wen X, Peng Y and Zhang L:
Identification of stem cell-related gene markers by comprehensive
transcriptome analysis to predict the prognosis and immunotherapy
of lung adenocarcinoma. Curr Stem Cell Res Ther. 19:743–754. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Monteran L, Zait Y and Erez N: It's all
about the base: Stromal cells are central orchestrators of
metastasis. Trends Cancer. 10:208–229. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Gao D, Fang L, Liu C, Yang M, Yu X, Wang
L, Zhang W, Sun C and Zhuang J: Microenvironmental regulation in
tumor progression: Interactions between cancer-associated
fibroblasts and immune cells. Biomed Pharmacother. 167:1156222023.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hebeisen M, Oberle SG, Presotto D, Speiser
DE, Zehn D and Rufer N: Molecular insights for optimizing T cell
receptor specificity against cancer. Front Immunol. 4:1542013.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Speiser DE, Chijioke O, Schaeuble K and
Münz C: CD4+ T cells in cancer. Nat Cancer. 4:317–329.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cardenas MA, Prokhnevska N and Kissick HT:
Organized immune cell interactions within tumors sustain a
productive T-cell response. Int Immunol. 33:27–37. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Laurent V, Guérard A, Mazerolles C, Le
Gonidec S, Toulet A, Nieto L, Zaidi F, Majed B, Garandeau D,
Socrier Y, et al: Periprostatic adipocytes act as a driving force
for prostate cancer progression in obesity. Nat Commun.
7:102302016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhu M, Xu W, Wei C, Huang J, Xu J, Zhang
Y, Zhao Y, Chen J, Dong S, Liu B and Liang C: CCL14 serves as a
novel prognostic factor and tumor suppressor of HCC by modulating
cell cycle and promoting apoptosis. Cell Death Dis. 10:7962019.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen J, Rajasekaran M, Xia H, Zhang X,
Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BK, Ooi LL,
et al: The microtubule-associated protein PRC1 promotes early
recurrence of hepatocellular carcinoma in association with the
Wnt/β-catenin signalling pathway. Gut. 65:1522–1534. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jiang N, Zou C, Zhu Y, Luo Y, Chen L, Lei
Y, Tang K, Sun Y, Zhang W, Li S, et al: HIF-1α-regulated miR-1275
maintains stem cell-like phenotypes and promotes the progression of
LUAD by simultaneously activating Wnt/β-catenin and Notch
signaling. Theranostics. 10:2553–2570. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cao Y, Geng J, Wang X, Meng Q, Xu S, Lang
Y, Zhou Y, Qi L, Wang Z, Wei Z, et al: RNA-binding motif protein 10
represses tumor progression through the Wnt/β-catenin pathway in
lung adenocarcinoma. Int J Biol Sci. 18:124–139. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Itakura M, Terashima Y, Shingyoji M, Yokoi
S, Ohira M, Kageyama H, Matui Y, Yoshida Y, Ashinuma H, Moriya Y,
et al: High CC chemokine receptor 7 expression improves
postoperative prognosis of lung adenocarcinoma patients. Br J
Cancer. 109:1100–1108. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Choucair K, Morand S, Stanbery L, Edelman
G, Dworkin L and Nemunaitis J: TMB: A promising immune-response
biomarker, and potential spearhead in advancing targeted therapy
trials. Cancer Gene Ther. 27:841–853. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hellmann MD, Nathanson T, Rizvi H, Creelan
BC, Sanchez-Vega F, Ahuja A, Ni A, Novik JB, Mangarin LMB,
Abu-Akeel M, et al: Genomic features of response to combination
immunotherapy in patients with advanced non-small-cell lung cancer.
Cancer Cell. 33:843–852.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Samstein RM, Lee CH, Shoushtari AN,
Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ,
Omuro A, et al: Tumor mutational load predicts survival after
immunotherapy across multiple cancer types. Nat Genet. 51:202–206.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Goodman AM, Kato S, Bazhenova L, Patel SP,
Frampton GM, Miller V, Stephens PJ, Daniels GA and Kurzrock R:
Tumor mutational burden as an independent predictor of response to
immunotherapy in diverse cancers. Mol Cancer Ther. 16:2598–2608.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Rizvi H, Sanchez-Vega F, La K, Chatila W,
Jonsson P, Halpenny D, Plodkowski A, Long N, Sauter JL, Rekhtman N,
et al: Molecular determinants of response to anti-programmed cell
death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in
patients with non-small-cell lung cancer profiled with targeted
next-generation sequencing. J Clin Oncol. 36:633–641. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Hellmann MD, Ciuleanu TE, Pluzanski A, Lee
JS, Otterson GA, Audigier-Valette C, Minenza E, Linardou H, Burgers
S, Salman P, et al: Nivolumab plus ipilimumab in lung cancer with a
high tumor mutational burden. N Engl J Med. 378:2093–2104. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Havel JJ, Chowell D and Chan TA: The
evolving landscape of biomarkers for checkpoint inhibitor
immunotherapy. Nat Rev Cancer. 19:133–150. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Jardim DL, Goodman A, de Melo Gagliato D
and Kurzrock R: The challenges of tumor mutational burden as an
immunotherapy biomarker. Cancer Cell. 39:154–173. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chan TA, Yarchoan M, Jaffee E, Swanton C,
Quezada SA, Stenzinger A and Peters S: Development of tumor
mutation burden as an immunotherapy biomarker: Utility for the
oncology clinic. Ann Oncol. 30:44–56. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yang Y, Adebali O, Wu G, Selby CP, Chiou
YY, Rashid N, Hu J, Hogenesch JB and Sancar A: Cisplatin-DNA adduct
repair of transcribed genes is controlled by two circadian programs
in mouse tissues. Proc Natl Acad Sci USA. 115:E4777–E4785.
2018.PubMed/NCBI
|
|
76
|
Zhu L and Chen L: Progress in research on
paclitaxel and tumor immunotherapy. Cell Mol Biol Lett. 24:402019.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Ahmadi A, Mohammadnejadi E and
Razzaghi-Asl N: Gefitinib derivatives and drug-resistance: A
perspective from molecular dynamics simulations. Comput Biol Med.
163:1072042023. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zou J, Lan H, Li W, Xie S, Tong Z, Song X
and Wang C: Comprehensive analysis of circular RNA expression
profiles in gefitinib-resistant lung adenocarcinoma patients.
Technol Cancer Res Treat. 21:153303382211391672022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang Q and Xu K: Advances in the research
of autophagy in EGFR-TKI treatment and resistance in lung cancer.
Zhongguo Fei Ai Za Zhi. 19:607–614. 2016.(In Chinese). PubMed/NCBI
|
|
80
|
Fukuoka M, Wu YL, Thongprasert S,
Sunpaweravong P, Leong SS, Sriuranpong V, Chao TY, Nakagawa K, Chu
DT, Saijo N, et al: Biomarker analyses and final overall survival
results from a phase III, randomized, open-label, first-line study
of gefitinib versus carboplatin/paclitaxel in clinically selected
patients with advanced non-small-cell lung cancer in Asia (IPASS).
J Clin Oncol. 29:2866–2874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Chen C, Wan M, Peng X, Zhang Q and Liu Y:
GPR37-centered ceRNA network contributes to metastatic potential in
lung adenocarcinoma: Evidence from high-throughput sequencing.
Transl Oncol. 39:1018192024. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wu X, Sui Z, Zhang H, Wang Y and Yu Z:
Integrated analysis of lncRNA-Mediated ceRNA network in lung
adenocarcinoma. Front Oncol. 10:5547592020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Feng W, Gong H, Wang Y, Zhu G, Xue T, Wang
Y and Cui G: circIFT80 functions as a ceRNA of miR-1236-3p to
promote colorectal cancer progression. Mol Ther Nucleic Acids.
18:375–387. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Rao X, Cao H, Yu Q, Ou X, Deng R and Huang
J: NEAT1/MALAT1/XIST/PKD-Hsa-Mir-101-3p-DLGAP5 axis as a novel
diagnostic and prognostic biomarker associated with immune cell
infiltration in bladder cancer. Front Genet. 13:8925352022.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Duca RB, Massillo C, Dalton GN, Farré PL,
Graña KD, Gardner K and De Siervi A: MiR-19b-3p and miR-101-3p as
potential biomarkers for prostate cancer diagnosis and prognosis.
Am J Cancer Res. 11:2802–2820. 2021.PubMed/NCBI
|
|
87
|
Chen Z, Lin X, Wan Z, Xiao M, Ding C, Wan
P, Li Q and Zheng S: High expression of EZH2 mediated by ncRNAs
correlates with poor prognosis and tumor immune infiltration of
hepatocellular carcinoma. Genes (Basel). 13:8762022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Tao L, Xu C, Shen W, Tan J, Li L, Fan M,
Sun D, Lai Y and Cheng H: HIPK3 inhibition by exosomal
hsa-miR-101-3p is related to metabolic reprogramming in colorectal
cancer. Front Oncol. 11:7583362022. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Song J, Sun Y, Cao H, Liu Z, Xi L, Dong C,
Yang R and Shi Y: A novel pyroptosis-related lncRNA signature for
prognostic prediction in patients with lung adenocarcinoma.
Bioengineered. 12:5932–5949. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhang J, Du C, Zhang L, Wang Y, Zhang Y
and Li J: lncRNA GSEC promotes the progression of triple negative
breast cancer (TNBC) by targeting the miR-202-5p/AXL axis. Onco
Targets Ther. 14:2747–2759. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Liu R, Ju C, Zhang F, Tang X, Yan J, Sun
J, Lv B, Guo Y, Liang Y, Lv XB and Zhang Z: LncRNA GSEC promotes
the proliferation, migration and invasion by sponging
miR-588/EIF5A2 axis in osteosarcoma. Biochem Biophys Res Commun.
532:300–307. 2020. View Article : Google Scholar : PubMed/NCBI
|