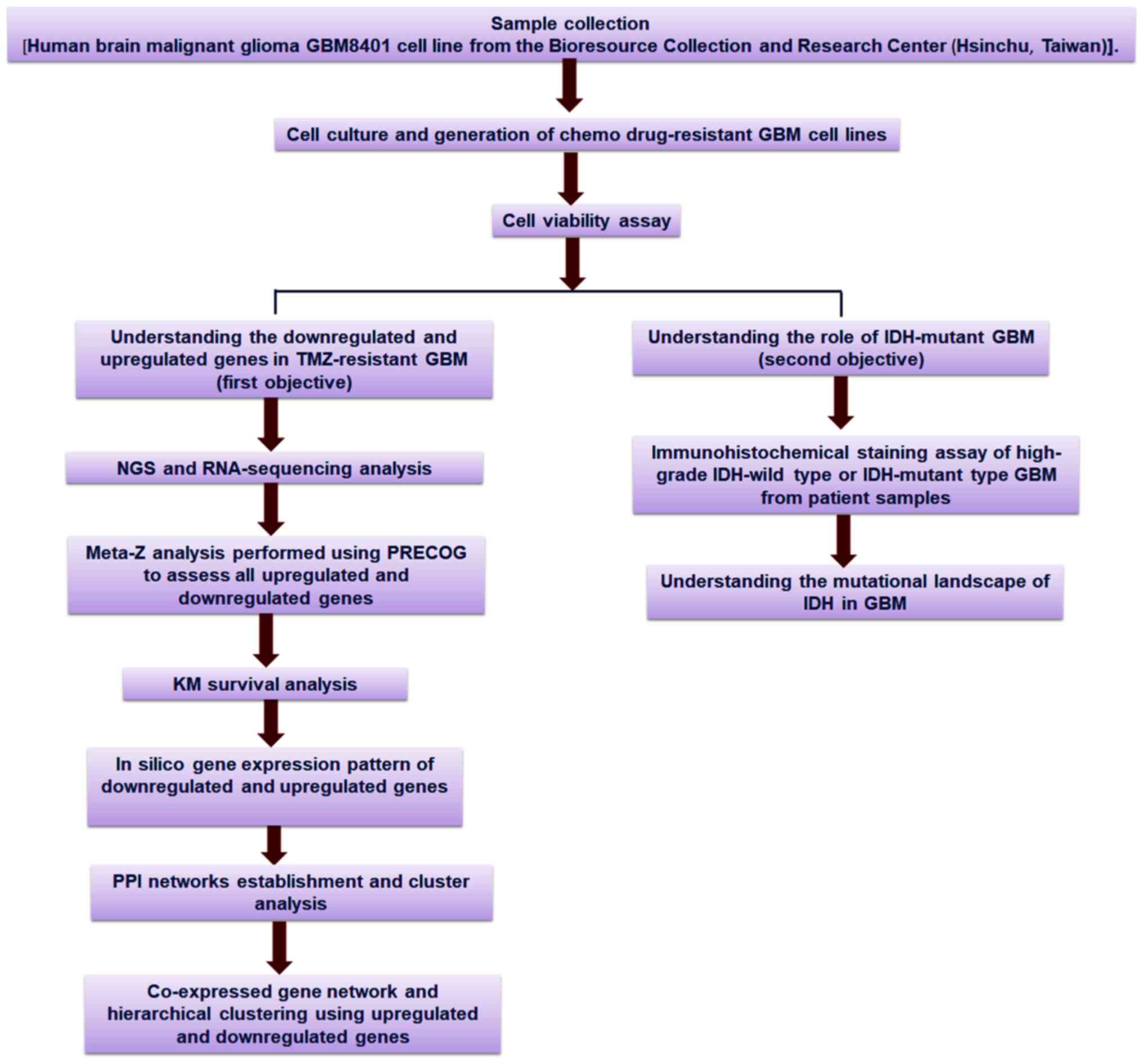
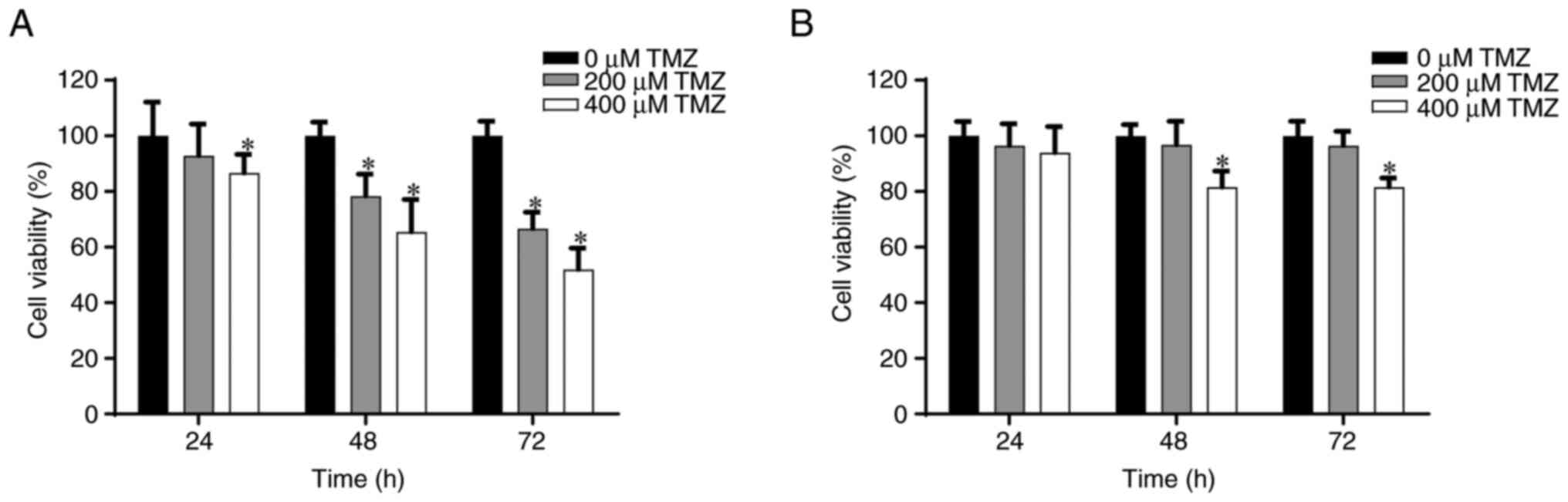
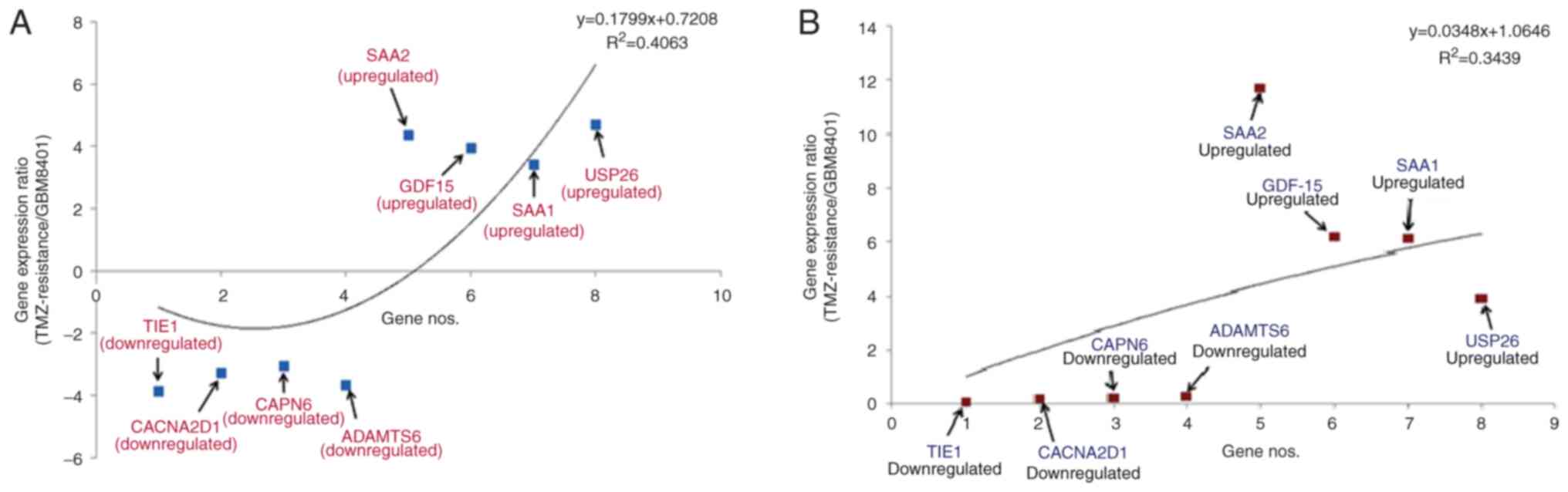
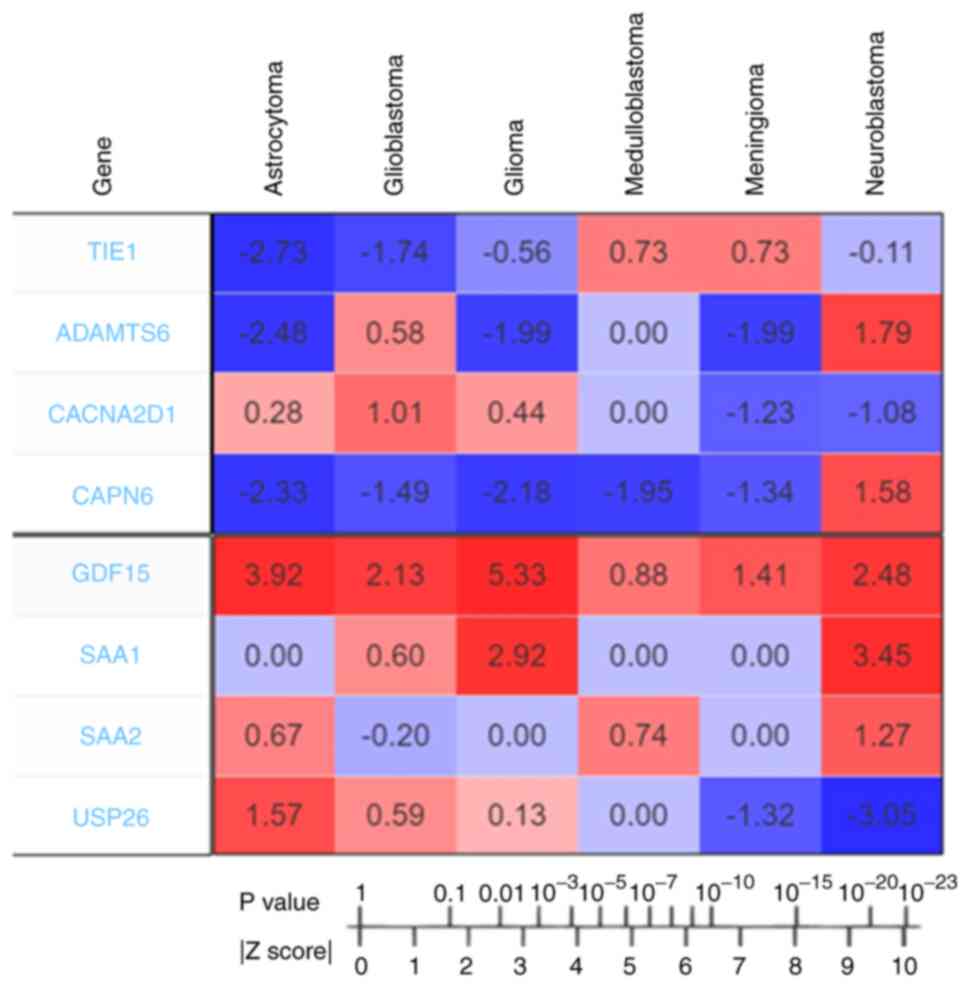
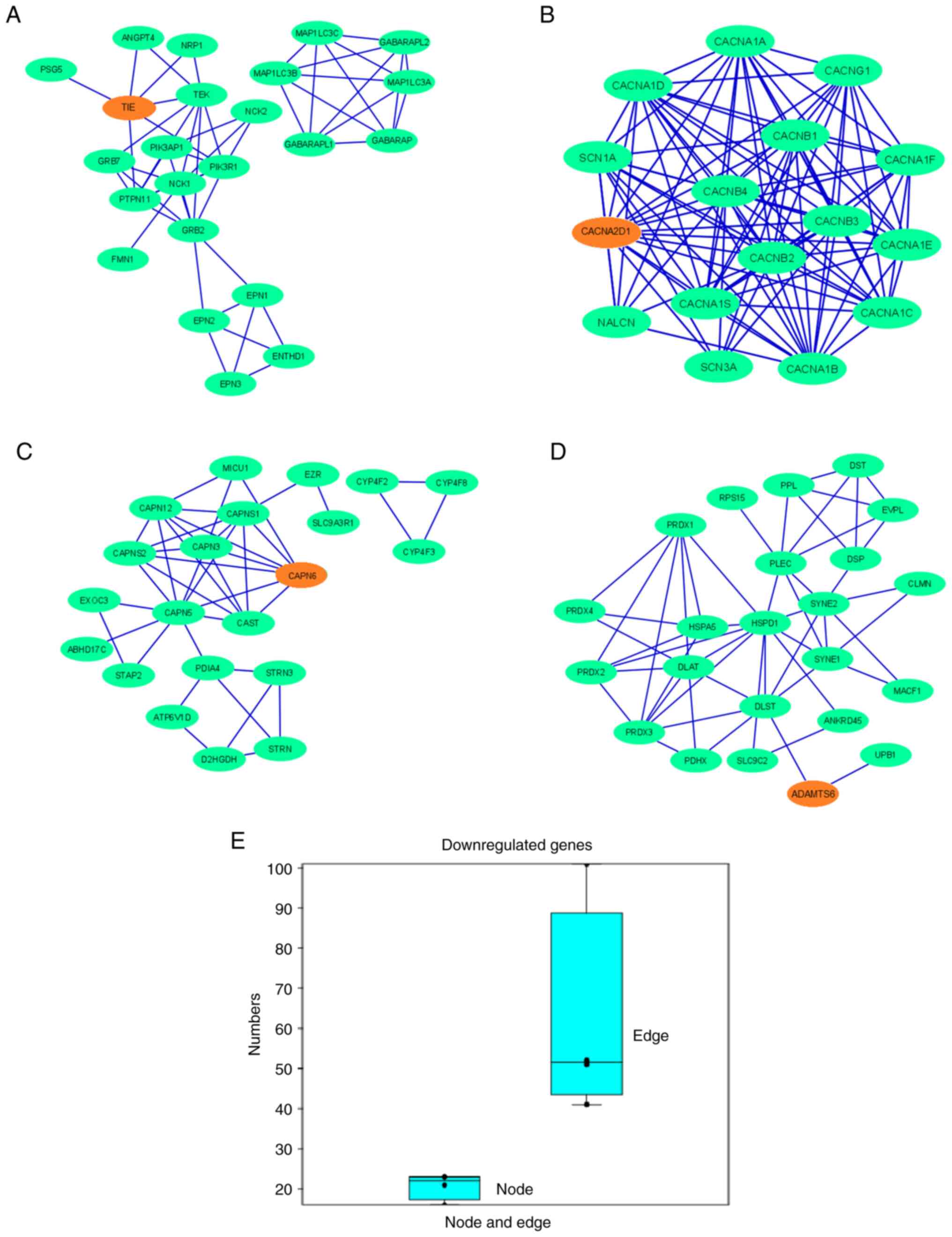
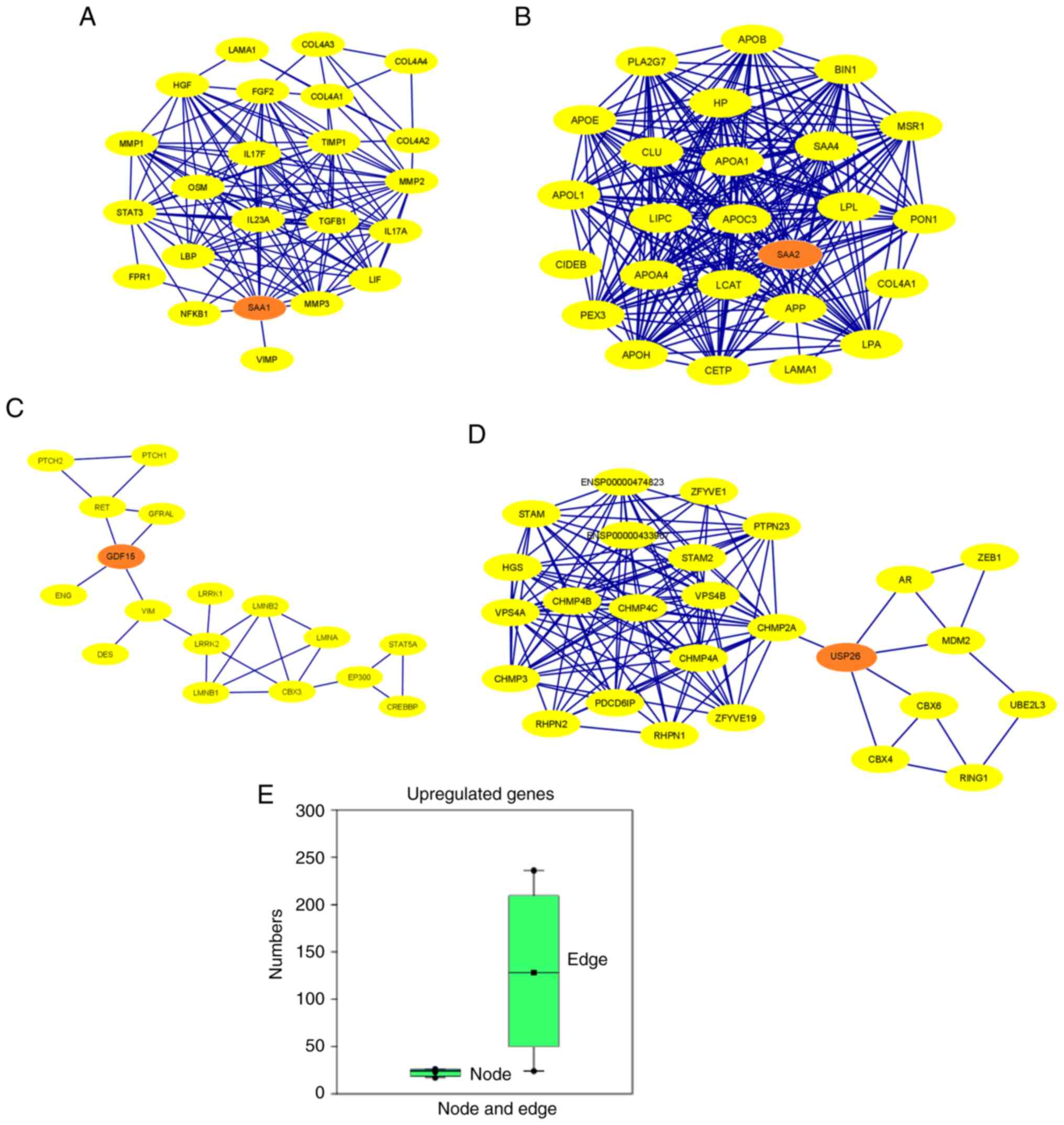
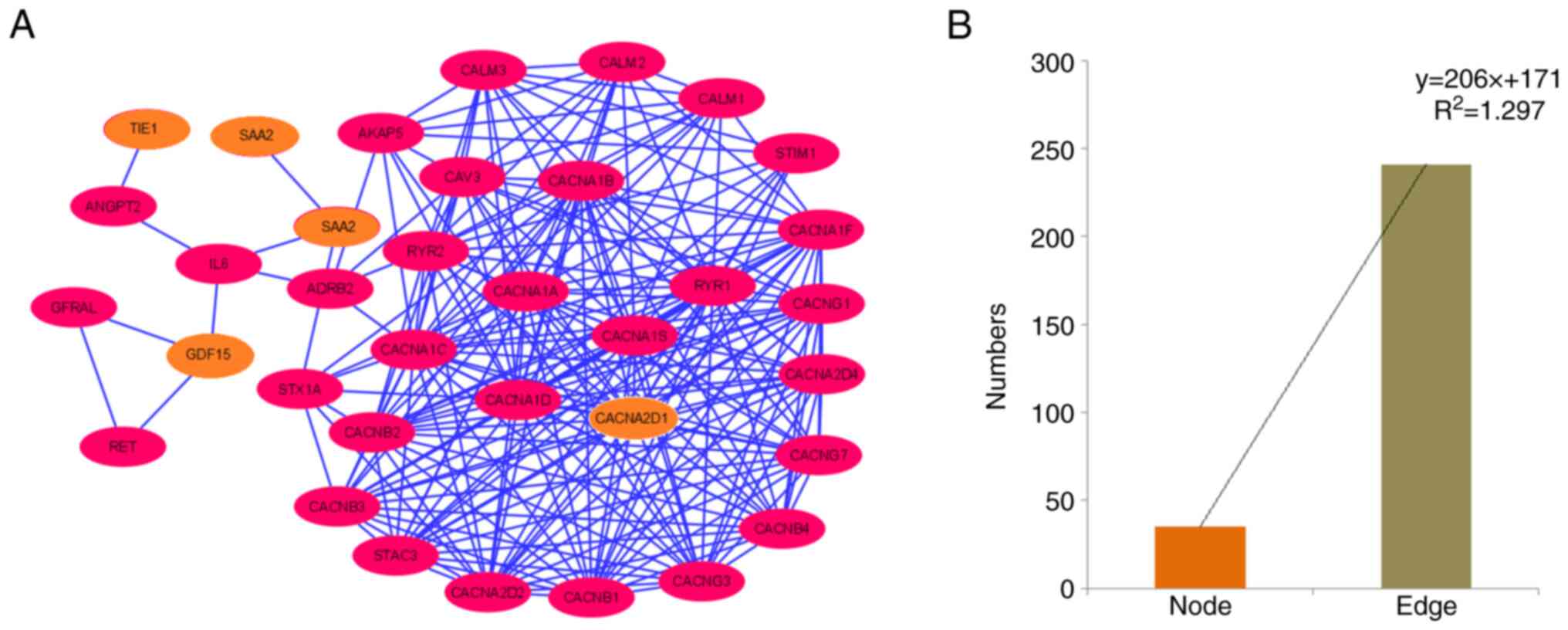
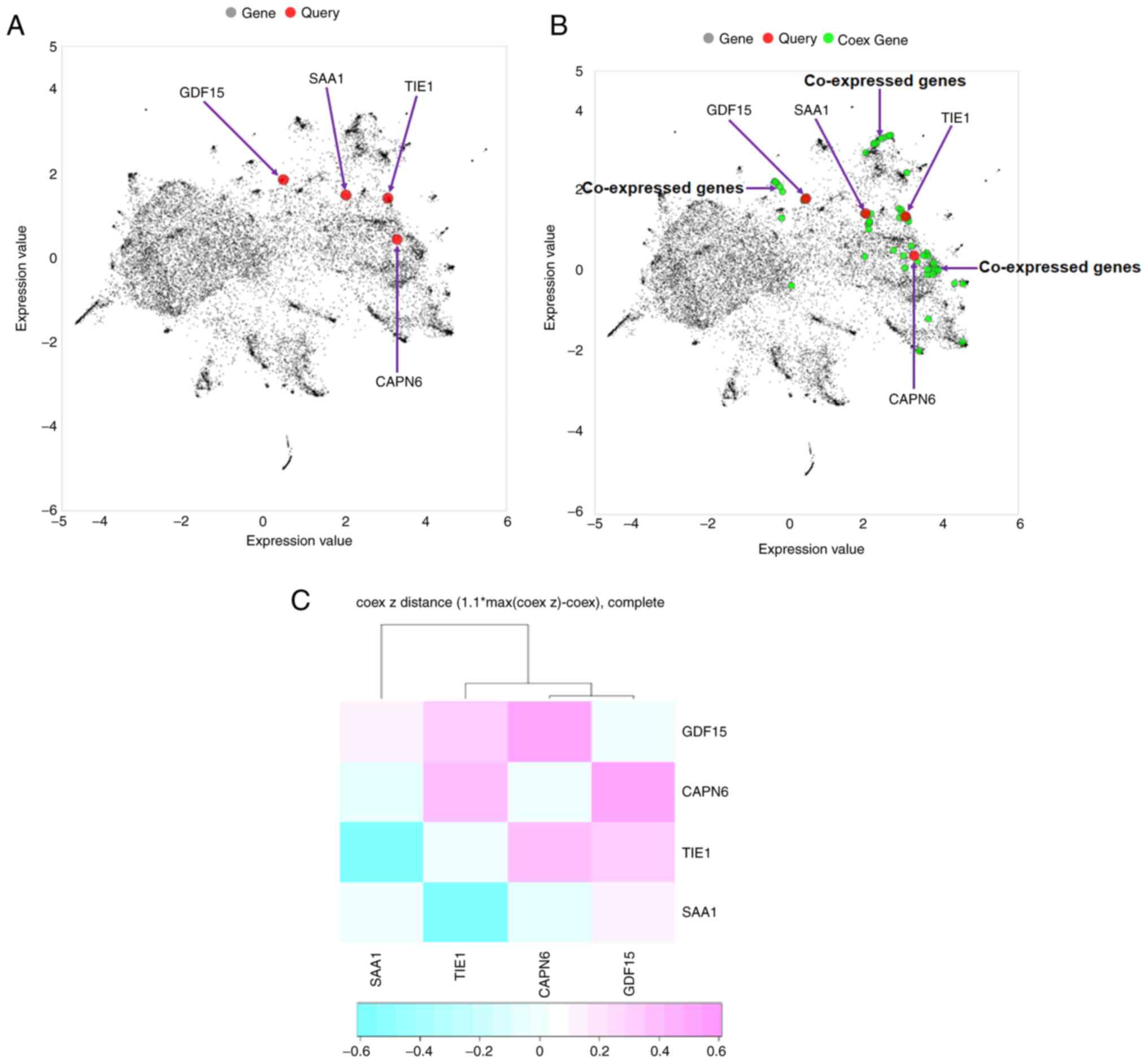
|
1
|
Grech N, Dalli T, Mizzi S, Meilak L,
Calleja N and Zrinzo A: Rising incidence of glioblastoma multiforme
in a well-defined population. Cureus. 12:e81952020.PubMed/NCBI
|
|
2
|
Miller KD, Ostrom QT, Kruchko C, Patil N,
Tihan T, Cioffi G, Fuchs HE, Waite KA, Jemal A, Siegel RL and
Barnholtz-Sloan JS: Brain and other central nervous system tumor
statistics, 2021. CA Cancer J Clin. 71:381–406. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ostrom QT, Cioffi G, Waite K, Kruchko C
and Barnholtz-Sloan JS: CBTRUS statistical report: Primary brain
and other central nervous system tumors diagnosed in the United
States in 2014–2018. Neuro Oncol. 23 (12 Suppl 2):iii1–iii105.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kleihues P and Ohgaki H: Primary and
secondary glioblastomas: From concept to clinical diagnosis. Neuro
Oncol. 1:44–51. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin D, Wang M, Chen Y, Gong J, Chen L, Shi
X, Lan F, Chen Z, Xiong T, Sun H and Wan S: Trends in Intracranial
glioma incidence and mortality in the United States, 1975–2018.
Front Oncol. 11:7480612021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Upadhyaya SA, Ghazwani Y, Wu S, Broniscer
A, Boop FA, Gajjar A and Qaddoumi I: Mortality in children with
low-grade glioma or glioneuronal tumors: A single-institution
study. Pediatr Blood Cancer. 65:10.1002/pbc.26717. 2018. View Article : Google Scholar
|
|
8
|
Yao M, Li S, Wu X, Diao S, Zhang G, He H,
Bian L and Lu Y: Cellular origin of glioblastoma and its
implication in precision therapy. Cell Mol Immunol. 15:737–739.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Olivier C, Oliver L, Lalier L and Vallette
FM: Drug resistance in glioblastoma: The two faces of oxidative
stress. Front Mol Biosci. 7:6206772021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mansoori B, Mohammadi A, Davudian S,
Shirjang S and Baradaran B: The different mechanisms of cancer drug
resistance: A brief review. Adv Pharm Bull. 7:339–348. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jakoby WB: The glutathione S-transferases:
A group of multifunctional detoxification proteins. Adv Enzymol
Relat Areas Mol Biol. 46:383–414. 1978.PubMed/NCBI
|
|
14
|
Oliver L, Lalier L, Salaud C, Heymann D,
Cartron PF and Vallette FM: Drug resistance in glioblastoma: Are
persisters the key to therapy? Cancer Drug Resist. 3:287–301.
2020.PubMed/NCBI
|
|
15
|
Phi LTH, Sari IN, Yang YG, Lee SH, Jun N,
Kim KS, Lee YK and Kwon HY: Cancer stem cells (CSCs) in drug
resistance and their therapeutic implications in cancer treatment.
Stem Cells Int. 2018:54169232018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu G, Yuan X, Zeng Z, Tunici P, Ng H,
Abdulkadir IR, Lu L, Irvin D, Black KL and Yu JS: Analysis of gene
expression and chemoresistance of CD133+ cancer stem cells in
glioblastoma. Mol Cancer. 5:672006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Behnan J, Finocchiaro G and Hanna G: The
landscape of the mesenchymal signature in brain tumours. Brain.
142:847–866. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu Z, Chen Y, Wang S, Li P, Zhou G and
Yuan Y: Inhibition of NF-κB results in anti-glioma activity and
reduces temozolomide-induced chemoresistance by down-regulating
MGMT gene expression. Cancer Lett. 428:77–89. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yao L, Li J, Zhang X, Zhou L and Hu K:
Downregulated ferroptosis-related gene SQLE facilitates
temozolomide chemoresistance, and invasion and affects immune
regulation in glioblastoma. CNS Neurosci Ther. 28:2104–2115. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cohen AL, Holmen SL and Colman H: IDH1 and
IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 13:3452013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hegi ME, Diserens AC, Gorlia T, Hamou MF,
de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani
L, et al: MGMT gene silencing and benefit from temozolomide in
glioblastoma. N Engl J Med. 352:997–1003. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Perez A and Huse JT: The evolving
classification of diffuse gliomas: World Health Organization
updates for 2021. Curr Neurol Neurosci Rep. 21:672021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun X and Turcan S: From laboratory
studies to clinical trials: Temozolomide use in IDH-mutant gliomas.
Cells. 10:12252021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Han S, Liu Y, Cai SJ, Qian M, Ding J,
Larion M, Gilbert MR and Yang C: IDH mutation in glioma: Molecular
mechanisms and potential therapeutic targets. Br J Cancer.
122:1580–1589. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nobusawa S, Watanabe T, Kleihues P and
Ohgaki H: IDH1 mutations as molecular signature and predictive
factor of secondary glioblastomas. Clin Cancer Res. 15:6002–6007.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Govindarajan V, Shah AH, Di L, Rivas S,
Suter RK, Eichberg DG, Luther E, Lu V, Morell AA, Ivan ME, et al:
Systematic review of epigenetic therapies for treatment of
IDH-mutant glioma. World Neurosurg. 162:47–56. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qi S, Lei Y, Si G, YanQing D, HuiXia H,
XueLin Z, LanXiao W and Fei Y: IDH mutations predict longer
survival and response to temozolomide in secondary glioblastoma.
Cancer Sci. 103:269–273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Munoz JL, Bliss SA, Greco SJ, Ramkissoon
SH, Ligon KL and Rameshwar P: Delivery of functional anti-miR-9 by
mesenchymal stem cell-derived exosomes to glioblastoma multiforme
cells conferred chemosensitivity. Mol Ther Nucleic Acids.
2:e1262013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and Ballgown. Nat Protoc.
11:1650–1667. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gentles AJ, Newman AM, Liu CL, Bratman SV,
Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al: The
prognostic landscape of genes and infiltrating immune cells across
human cancers. Nat Med. 21:938–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dudley WN, Wickham R and Coombs N: An
introduction to survival statistics: Kaplan-Meier analysis. J Adv
Pract Oncol. 7:91–100. 2016.PubMed/NCBI
|
|
35
|
Lubbock ALR, Katz E, Harrison DJ and
Overton IM: TMA navigator: Network inference, patient
stratification and survival analysis with tissue microarray data.
Nucleic Acids Res. 41((Web Server Issue)): W562–W568. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Stelzer G, Dalah I, Stein TI, Satanower Y,
Rosen N, Nativ N, Oz-Levi D, Olender T, Belinky F, Bahir I, et al:
In-silico human genomics with GeneCards. Hum Genomics. 5:709–717.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stelzer G, Rosen N, Plaschkes I, Zimmerman
S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et
al: The GeneCards suite: From gene data mining to disease genome
sequence analyses. Curr Protoc Bioinformatics. 54:1.30.1–1.30.33.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43((Database Issue)): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Maglott D, Ostell J, Pruitt KD and
Tatusova T: Entrez gene: Gene-centered information at NCBI. Nucleic
Acids Res. 33((Database Issue)): D54–D58. 2005.PubMed/NCBI
|
|
40
|
Crowe AR and Yue W: Semi-quantitative
determination of protein expression using immunohistochemistry
staining and analysis: An Integrated Protocol. Bio Protoc.
9:e34652019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yuan JX and Munson JM: Quantitative
immunohistochemistry of the cellular microenvironment in patient
glioblastoma resections. J Vis Exp. 560252017.PubMed/NCBI
|
|
42
|
Burley SK, Berman HM, Kleywegt GJ, Markley
JL, Nakamura H and Velankar S: Protein data bank (PDB): The single
global macromolecular structure archive. Methods Mol Biol.
1607:627–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yuan S, Chan HCS, Filipek S and Vogel H:
PyMOL and inkscape bridge the data and the data visualization.
Structure. 24:2041–2042. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jumper J, Evans R, Pritzel A, Green T,
Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A,
Potapenko A, et al: Highly accurate protein structure prediction
with AlphaFold. Nature. 596:583–589. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
de Beer TAP, Berka K, Thornton JM and
Laskowski RA: PDBsum additions. Nucleic Acids Res. 42((Database
Issue)): D292–D296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hammer Ø, Harper DAT and Ryan PD: PAST:
Paleontological statistics software package for education and data
analysis. Palaeontol Electron. 4:92001.
|
|
47
|
MATLAB, . High performance numeric
computation and visualization software: User's guide: For UNIX
workstations. Mathworks Incorporated. 2003.
|
|
48
|
Singh N, Miner A, Hennis L and Mittal S:
Mechanisms of temozolomide resistance in glioblastoma-a
comprehensive review. Cancer Drug Resist. 4:17–43. 2021.PubMed/NCBI
|
|
49
|
Chen X, Zhang M, Gan H, Wang H, Lee JH,
Fang D, Kitange GJ, He L, Hu Z, Parney IF, et al: A novel enhancer
regulates MGMT expression and promotes temozolomide resistance in
glioblastoma. Nat Commun. 9:29492018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bukowski K, Kciuk M and Kontek R:
Mechanisms of multidrug resistance in cancer chemotherapy. Int J
Mol Sci. 21:32332020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rapin N, Bagger FO, Jendholm J,
Mora-Jensen H, Krogh A, Kohlmann A, Thiede C, Borregaard N,
Bullinger L, Winther O, et al: Comparing cancer vs normal gene
expression profiles identifies new disease entities and common
transcriptional programs in AML patients. Blood. 123:894–904. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cheng SY, Chen NF, Wen ZH, Yao ZK, Tsui
KH, Kuo HM and Chen WF: Glutathione S-transferase M3 is associated
with glycolysis in intrinsic temozolomide-resistant glioblastoma
multiforme cells. Int J Mol Sci. 22:70802021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gordinier ME, Schau GF, Pollock SB,
Shields LBE and Talwalkar S: Genomic characterization of vulvar
squamous cell carcinoma reveals differential gene expression based
on clinical outcome. Gynecol Oncol. 180:111–117. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hu G, Wei B, Wang L, Wang L, Kong D, Jin Y
and Sun Z: Analysis of gene expression profiles associated with
glioma progression. Mol Med Rep. 12:1884–1890. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kothari C, Osseni MA, Agbo L, Ouellette G,
Déraspe M, Laviolette F, Corbeil J, Lambert JP, Diorio C and
Durocher F: Machine learning analysis identifies genes
differentiating triple negative breast cancers. Sci Rep.
10:104642020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kumar SU, Kumar DT, Siva R, Doss CGP and
Zayed H: Integrative bioinformatics approaches to map potential
novel genes and pathways involved in ovarian cancer. Front Bioeng
Biotechnol. 7:3912019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chakraborty C, Sharma AR, Bhattacharya M,
Zayed H and Lee SS: Understanding gene expression and transcriptome
profiling of COVID-19: An initiative towards the mapping of
protective immunity genes against SARS-CoV-2 infection. Front
Immunol. 12:7249362021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chakraborty C, Bhattacharya M, Dhama K and
Lee SS: Evaluation of differentially expressed genes during
replication using gene expression landscape of monkeypox-infected
MK2 cells: A bioinformatics and systems biology approach to
understanding the genomic pattern of viral replication. J Infect
Public Health. 16:399–409. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dunn SL, Soul J, Anand S, Schwartz JM,
Boot-Handford RP and Hardingham TE: Gene expression changes in
damaged osteoarthritic cartilage identify a signature of
non-chondrogenic and mechanical responses. Osteoarthritis
Cartilage. 24:1431–1440. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen SJ, Liao DL, Chen CH, Wang TY and
Chen KC: Construction and analysis of protein-protein interaction
network of heroin use disorder. Sci Rep. 9:49802019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Raman K: Construction and analysis of
protein-protein interaction networks. Autom Exp. 2:22010.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Silverbush D and Sharan R: A systematic
approach to orient the human protein-protein interaction network.
Nat Commun. 10:30152019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lee SY: Temozolomide resistance in
glioblastoma multiforme. Genes Dis. 3:198–210. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Mrugala MM and Chamberlain MC: Mechanisms
of disease: Temozolomide and glioblastoma-look to the future. Nat
Clin Pract Oncol. 5:476–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Woo PYM, Li Y, Chan AHY, Ng SCP, Loong
HHF, Chan DTM, Wong GKC and Poon WS: A multifaceted review of
temozolomide resistance mechanisms in glioblastoma beyond
O-6-methylguanine-DNA methyltransferase. Glioma. 2:68–82. 2019.
View Article : Google Scholar
|
|
66
|
Kitange GJ, Carlson BL, Schroeder MA,
Grogan PT, Lamont JD, Decker PA, Wu W, James CD and Sarkaria JN:
Induction of MGMT expression is associated with temozolomide
resistance in glioblastoma xenografts. Neuro Oncol. 11:281–291.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Barthel FP, Johnson KC, Varn FS, Moskalik
AD, Tanner G, Kocakavuk E, Anderson KJ, Abiola O, Aldape K, Alfaro
KD, et al: Longitudinal molecular trajectories of diffuse glioma in
adults. Nature. 576:112–120. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Jonsson P, Lin AL, Young RJ, DiStefano NM,
Hyman DM, Li BT, Berger MF, Zehir A, Ladanyi M, Solit DB, et al:
Genomic correlates of disease progression and treatment response in
prospectively characterized gliomas. Clin Cancer Res. 25:5537–5547.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ohba S, Mukherjee J, See WL and Pieper RO:
Mutant IDH1-driven cellular transformation increases RAD51-mediated
homologous recombination and temozolomide resistance. Cancer Res.
74:4836–4844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chen T, Wang J, Xue B, Kong Q, Liu Z and
Yu B: Identification and characterization of a novel porcine
endothelial cell-specific Tie1 promoter. Xenotransplantation.
20:438–448. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Rodewald HR and Sato TN: Tie1, a receptor
tyrosine kinase essential for vascular endothelial cell integrity,
is not critical for the development of hematopoietic cells.
Oncogene. 12:397–404. 1996.PubMed/NCBI
|
|
72
|
Woo KV and Baldwin HS: Role of Tie1 in
shear stress and atherosclerosis. Trends Cardiovasc Med.
21:118–123. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Meltzer M, Eliash N, Azoulay Z, Hadad U
and Papo N: In vitro inhibition of cancer angiogenesis and
migration by a nanobody that targets the orphan receptor Tie1. Cell
Mol Life Sci. 79:3122022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Dolphin AC: Voltage-gated calcium channel
α 2δ subunits: an assessment of proposed novel roles.
F1000Res. 7:F1000 Faculty Rev. –1830. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Panebianco M, Al-Bachari S, Hutton JL and
Marson AG: Gabapentin add-on treatment for drug-resistant focal
epilepsy. Cochrane Database Syst Rev. 1:CD0014152021.PubMed/NCBI
|
|
76
|
Derry S, Bell RF, Straube S, Wiffen PJ,
Aldington D and Moore RA: Pregabalin for neuropathic pain in
adults. Cochrane Database Syst Rev. 1:CD0070762019.PubMed/NCBI
|
|
77
|
Alles SRA, Cain SM and Snutch TP:
Pregabalin as a pain therapeutic: Beyond calcium channels. Front
Cell Neurosci. 14:832020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Verma V, Singh N and Singh Jaggi A:
Pregabalin in neuropathic pain: Evidences and possible mechanisms.
Curr Neuropharmacol. 12:44–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Tonami K, Kurihara Y, Aburatani H,
Uchijima Y, Asano T and Kurihara H: Calpain 6 is involved in
microtubule stabilization and cytoskeletal organization. Mol Cell
Biol. 27:2548–2561. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ono Y and Sorimachi H: Calpains: An
elaborate proteolytic system. Biochim Biophys Acta. 1824:224–236.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Chen L, Xiao D, Tang F, Gao H and Li X:
CAPN6 in disease: An emerging therapeutic target (review). Int J
Mol Med. 46:1644–1652. 2020.PubMed/NCBI
|
|
82
|
Kelwick R, Desanlis I, Wheeler GN and
Edwards DR: The ADAMTS (a disintegrin and metalloproteinase with
thrombospondin motifs) family. Genome Biol. 16:1132015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Xie Y, Gou Q, Xie K, Wang Z, Wang Y and
Zheng H: ADAMTS6 suppresses tumor progression via the ERK signaling
pathway and serves as a prognostic marker in human breast cancer.
Oncotarget. 7:61273–61283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Mori M, Tian G, Ishikawa A and Higuchi K:
Diversity and complexity of the mouse Saa1 and Saa2 genes. Exp
Anim. 63:99–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Jumeau C, Awad F, Assrawi E, Cobret L,
Duquesnoy P, Giurgea I, Valeyre D, Grateau G, Amselem S, Bernaudin
JF and Karabina SA: Expression of SAA1, SAA2 and SAA4 genes in
human primary monocytes and monocyte-derived macrophages. PLoS One.
14:e02170052019. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Li W, Wang W, Zuo R, Liu C, Shu Q, Ying H
and Sun K: Induction of pro-inflammatory genes by serum amyloid A1
in human amnion fibroblasts. Sci Rep. 7:6932017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Abouelasrar Salama S, De Bondt M, De Buck
M, Berghmans N, Proost P, Oliveira VLS, Amaral FA, Gouwy M, Van
Damme J and Struyf S: Serum amyloid A1 (SAA1) revisited: Restricted
leukocyte-activating properties of homogeneous SAA1. Front Immunol.
11:8432020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Li S, Cheng Y, Cheng G, Xu T, Ye Y, Miu Q,
Cao Q, Yang X, Ruan H and Zhang X: High SAA1 expression predicts
advanced tumors in renal cancer. Front Oncol. 11:6497612021.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Sack GH Jr: Serum amyloid A-a review. Mol
Med. 24:462018. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Kim YJ, Gallien S, El-Khoury V, Goswami P,
Sertamo K, Schlesser M, Berchem G and Domon B: Quantification of
SAA1 and SAA2 in lung cancer plasma using the isotype-specific PRM
assays. Proteomics. 15:3116–3125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wischhusen J, Melero I and Fridman WH:
Growth/differentiation factor-15 (GDF-15): From biomarker to novel
targetable immune checkpoint. Front Immunol. 11:9512020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Arinaga-Hino T, Ide T, Akiba J, Suzuki H,
Kuwahara R, Amano K, Kawaguchi T, Sano T, Inoue E, Koga H, et al:
Growth differentiation factor 15 as a novel diagnostic and
therapeutic marker for autoimmune hepatitis. Sci Rep. 12:87592022.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Gkretsi V, Louca M, Stylianou A, Minadakis
G, Spyrou GM and Stylianopoulos T: Inhibition of breast cancer cell
invasion by ras suppressor-1 (RSU-1) silencing is reversed by
growth differentiation factor-15 (GDF-15). Int J Mol Sci.
20:1632019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Sakai K, Ito C, Wakabayashi M, Kanzaki S,
Ito T, Takada S, Toshimori K, Sekita Y and Kimura T: Usp26 mutation
in mice leads to defective spermatogenesis depending on genetic
background. Sci Rep. 9:137572019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Guo L, Chen Y, Hu S, Gao L, Tang N, Liu R,
Qin Y, Ren C and Du S: GDF15 expression in glioma is associated
with malignant progression, immune microenvironment, and serves as
a prognostic factor. CNS Neurosci Ther. 28:158–171. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zhang H, Xu Y, Deng G, Yuan F, Tan Y, Gao
L, Sun Q, Qi Y, Yang K, Geng R and Jiang H: SAA1 knockdown promotes
the apoptosis of glioblastoma cells via downregulation of AKT
signaling. J Cancer. 12:2756–2767. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Tommasini-Ghelfi S, Murnan K, Kouri FM,
Mahajan AS, May JL and Stegh AH: Cancer-associated mutation and
beyond: The emerging biology of isocitrate dehydrogenases in human
disease. Sci Adv. 5:eaaw45432019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Immanuel SRC, Ghanate AD, Parmar DS, Yadav
R, Uthup R, Panchagnula V and Raghunathan A: Integrated genetic and
metabolic landscapes predict vulnerabilities of temozolomide
resistant glioblastoma cells. NPJ Syst Biol Appl. 7:22021.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lee JY, Hall JA, Kroehling L, Wu L, Najar
T, Nguyen HH, Lin WY, Yeung ST, Silva HM, Li D, et al: Serum
amyloid A proteins induce pathogenic Th17 cells and promote
inflammatory disease. Cell. 180:79–91.e16. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Takehara M, Sato Y, Kimura T, Noda K,
Miyamoto H, Fujino Y, Miyoshi J, Nakamura F, Wada H, Bando Y, et
al: Cancer-associated adipocytes promote pancreatic cancer
progression through SAA1 expression. Cancer Sci. 111:2883–2894.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Kang YE, Kim JM, Lim MA, Lee SE, Yi S, Kim
JT, Oh C, Liu L, Jin Y, Jung SN, et al: Growth differentiation
factor 15 is a cancer cell-induced mitokine that primes thyroid
cancer cells for invasiveness. Thyroid. 31:772–786. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Wosnitzer MS, Mielnik A, Dabaja A,
Robinson B, Schlegel PN and Paduch DA: Ubiquitin specific protease
26 (USP26) expression analysis in human testicular and extragonadal
tissues indicates diverse action of USP26 in cell differentiation
and tumorigenesis. PLoS One. 9:e986382014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Parker SJ and Metallo CM: Metabolic
consequences of oncogenic IDH mutations. Pharmacol Ther. 152:54–62.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Dang L, White DW, Gross S, Bennett BD,
Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et
al: Cancer-associated IDH1 mutations produce 2-hydroxyglutarate.
Nature. 462:739–744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Turkalp Z, Karamchandani J and Das S: IDH
mutation in glioma: New insights and promises for the future. JAMA
Neurol. 71:1319–1325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Peng H, Li Z, Fu J and Zhou R: Growth and
differentiation factor 15 regulates PD-L1 expression in
glioblastoma. Cancer Manag Res. 11:2653–2661. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Wang S, Yao F, Lu X, Li Q, Su Z, Lee JH,
Wang C and Du L: Temozolomide promotes immune escape of GBM cells
via upregulating PD-L1. Am J Cancer Res. 9:1161–1171.
2019.PubMed/NCBI
|
|
108
|
Jin Y, Cui D, Ren J, Wang K, Zeng T and
Gao L: CACNA2D3 is downregulated in gliomas and functions as a
tumor suppressor. Mol Carcinog. 56:945–959. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Nie C, Qin X, Li X, Tian B, Zhao Y, Jin Y,
Li Y, Wang Q, Zeng D, Hong A and Chen X: CACNA2D3 enhances the
chemosensitivity of esophageal squamous cell carcinoma to cisplatin
via inducing Ca2+-mediated apoptosis and suppressing
PI3K/Akt pathways. Front Oncol. 9:1852019. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Dai X, Tian X, Gu S, Yang Y, Li H, Gao P,
Lan Q and Cheng H: Hybrid biofabrication of neurosecretory
structures as a model for neurosecretion. Int J Bioprint.
9:6592022. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Dai X, Shao Y, Tian X, Cao X, Ye L, Gao P,
Cheng H and Wang X: Fusion between glioma stem cells and
mesenchymal stem cells promotes malignant progression in
3D-bioprinted models. ACS Appl Mater Interfaces. 14:35344–35356.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Dai X, Ye L, Li H, Dong X, Tian H, Gao P,
Dong J and Cheng H: Crosstalk between microglia and neural stem
cells influences the relapse of glioblastoma in GBM immunological
microenvironment. Clin Immunol. 251:1093332023. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Liu D, Dai X, Zhang W, Zhu X, Zha Z, Qian
H, Cheng L and Wang X: Liquid exfoliation of ultrasmall zirconium
carbide nanodots as a noninflammatory photothermal agent in the
treatment of glioma. Biomaterials. 292:1219172023. View Article : Google Scholar : PubMed/NCBI
|