Introduction
Colorectal cancer (CRC) is one of the most prevalent
malignant cancers, with the third highest incidence and second
highest mortality rates among all cancers worldwide (1). Surgical resection offers a potentially
high probability of cure, with approximately 90% of the patients
having early-stage primary CRC. However, many patients are
diagnosed at advanced stages, resulting in lower cure rates
(2). According to recent research,
20–34% of patients newly diagnosed with CRC already exhibit
metastatic disease (3), and 50–60%
of individuals initially diagnosed with localized CRC eventually
develop metastases (4). In
particular, the liver is the most prevalent site for CRC
metastasis, and the lungs are the second most prevalent one,
accounting for approximately 10–18% of rectal and 5–6% of colon
cancers (5). Surgical resection and
chemotherapy are currently used to treat metastatic CRC. However,
the clinical outcomes of patients with distant metastases remain
poor, and the survival rate of patients with metastatic CRC remains
below 20% (6). Therefore, the
development of novel treatments based on an understanding of the
molecular mechanisms of CRC progression is required.
In previous studies on metastatic CRC, liver
metastasis is well investigated from the viewpoint of
pathophysiological and molecular aspects owing to its high
probability. During the process, TNF-α and IL-1β upregulate the
expression of E-selectin and other adhesion molecules on liver
sinusoidal endothelial cells and enhance liver metastases; VEGF
mediates the vascularization (7).
Compared to liver metastasis, lung metastasis is
thought to occur through different mechanisms. Lymphatic vessels in
the colon and systemic venous circulation play more significant
roles than those in the liver. Furthermore, the lung
microenvironment is considerably more enriched with several types
of immune cells that reside in the airways and alveoli than in the
liver stroma (8).
SMAD4-deficient CRC cells in a mouse model secrete CCL15,
which recruits CCR1+ tumor-associated neutrophils, resulting in
lung metastasis (9). NDRG1 plays an
important role in MORC2-mediated CRC cell migration and invasion in
vitro and promotes lung metastasis of CRC cells in vivo
(10). Lung metastases share
clonality with primary tumors (including KRAS, TP53, and
APC mutations) through extensive genomic profiling of
clinical samples from three patients with CRC (8). An animal study shows that HDAC1 plays
a key role in lung metastasis by inhibiting EMT (11). Microarray analysis using metastatic
lung tissue from six patients shows that 42 genes are upregulated
in lung metastasis compared to those in the original colon tumor
(12). Although these studies may
lead to novel treatments, there is no consensus on the mechanism of
lung metastasis since the results are inconsistent. We suspect that
the heterogeneity of the samples can lead to this problem because
most previous studies used a small number of tumor tissues derived
from patients with different treatment histories or genetic
backgrounds. Therefore, a more precise analysis of the mechanisms
underlying lung metastasis is required.
We aimed to elucidate the mechanisms underlying lung
metastasis of colorectal cancer. We used in vitro models to
overcome the heterogeneity of the clinical materials. It is
possible to obtain more accurate insights into lung metastasis by
utilizing the experimental animal models and cell lines that we
previously established since the mice we used were genetically
homogeneous and each cell line was a single population.
In this study, we compared the gene expression
profiles of the original cell line and cell lines established from
lung metastasis to reveal the molecular background of lung
metastasis in CRC. We found a unique difference in profilin 2
isoforms between primary and metastatic lung CRC cells.
Materials and methods
Cell culture
The CMT93-PM cell line was established in a previous
study (13). Briefly, CMT93 cells
(CCL-223; ATCC, Manassas, VA, USA) were injected into the tail
veins of six-week-old female C57BL/6 mice. Twenty-five mice were
used for this experiment. Eight weeks after the tail vein
injection, all mice were euthanized by inducing anesthesia with
4.0% isoflurane and maintaining it with 2.0% isoflurane. Once
general anesthesia was achieved, they were quickly sacrificed by
exsanguination. The lungs were extracted and lung metastatic tumors
were visually identified. The tumor tissues were used for primary
tissue culture as previously described (13). The tumor cells were cultured and
maintained in Dulbecco's modified Eagle's medium (Sigma-Aldrich,
St. Louis, MO, USA) supplemented with 10% fetal bovine serum (v/v),
10,000 units/ml penicillin, 10,000 µg/ml streptomycin, and 25 µg/ml
Gibco amphotericin B (Thermo Fisher Scientific, Tokyo, Japan) at
37°C with 5% CO2. Four metastatic tumors were subjected
to primary tissue culture. Tumor cells from one metastatic tumor
grew spontaneously, and the cells underwent more than 20 passages
over a period of three months. The cells were designated as
CMT93-PM and used in this study. Moreover, we assigned CMT93-PM
cell numbers 1, 2, 3, and 4 according to the order in which they
were collected. All animals were housed in a controlled environment
at the Keio University School of Medicine under standard
temperature and light and dark cycles. All animal procedures were
approved by the Laboratory Animal Care and Use Committee of Keio
University School of Medicine (approval no. 15006).
Evaluating metastatic potentials of
CMT93 and CMT93-PM
CMT93-PM and CMT93-PM1 cells were injected into the
eight-week-old male mice via the tail vein, as described above.
Twenty-nine mice were used for this experiment. The number of
metastatic tumors in the lungs was evaluated using computed
tomography (CT) Before euthanasia, pulmonary metastasis was
assessed using an X-ray micro-CT system (R_mCT2; Rigaku, Tokyo,
Japan) under mild inhalation anesthesia using isoflurane. Chest CT
operational parameters were set at 90 kV and 160 µA to utilize a
respiratory and cardiac reconstruction mode according to the
instruction manual. The field of view was 24×24 mm with a pixel
size of 50×50 m. The scanning duration was 4.5 min. The mice were
positioned prone to scanning, and inhalation anesthesia comprising
a mixture of isoflurane (Pfizer Japan, Tokyo, Japan) and oxygen was
administered via a nasal cone. The concentration of isoflurane was
maintained at 2.0% from induction to maintenance. The respiratory
and cardiac reconstruction modes allow X-ray images to be
exclusively captured during the diastolic phase of the heart within
the end-expiratory period. Imaging was performed by simultaneously
tracking the breathing and cardiac movements under radiographic
guidance. Examination was performed twice-at the 8 and 12th week
after injection- to confirm pulmonary metastases by comparing the
metachronous images obtained. All mice were euthanized at 12 weeks
by cervical dislocation under general anesthesia using 4.0%
isoflurane for induction. After euthanasia, the lungs were
extracted and fixed by injecting 4% paraformaldehyde (PFA) (1 ml)
through the trachea. The lung tissues were then immersed in 4% PFA
and stored at room 4°C for 48 h to facilitate fixation.
Subsequently, they were dehydrated in absolute ethanol at 25°C. The
tissues were embedded in paraffin, and the paraffin blocks were cut
into 4-µm thick sections. These sections were stained with Mayer's
hematoxylin solution for 15 min at 25°C, followed by rinsing in tap
water. Finally, staining was conducted with eosin Y ethanol
solution for 1 min at 25°C. Pathological evaluation of pulmonary
metastases was also conducted using Evos FL Auto2 microscope
(Invitrogen, Carlsbad, CA, USA).
RNA extraction and microarrays
Total RNAs were extracted from CMT93 and CMT93-PM1
cells using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Gene
expression profiles were obtained using the Affymetrix GeneChip
Mouse430_2 array following the manufacturer's instructions.
Briefly, the sense-strand cDNA was fragmented and labeled using the
Affymetrix GeneChip WT Terminal Labeling Kit (Santa Clara, CA, USA)
after generating single-stranded cDNA. Following hybridization, the
GeneChip Fluidics Station 450 (Affymetrix) was used to wash the
arrays and scanning was performed using a GeneChip Scanner 3000 7G
(Affymetrix). The raw intensity data derived from the microarray
images were pre-processed using Affymetrix Expression Console
software (Affymetrix). Expression intensities were stored as cell
intensity files and normalized using the robust multi-chip average
method. Subsequently, these datasets were subjected to filtering,
with genes displaying an absolute fold change ≥2 or ≤0.5 identified
as exhibiting differential expression. Subsequently, we conducted
Gene Ontology analysis using the Database for Annotation,
Visualization, and Integrated Discovery (DAVID) tool, and plotted
the enrichment scores for each cluster (14), which served as indicators of the
biological significance of these clusters.
Evaluating PFN2 expression using
real-time-polymerase chain reaction (RT-PCR) and melting curve
analysis
To evaluate the expression pattern of PFN2 in
the cell lines, RT-PCR with high-resolution melting curve analysis
was sequentially performed on an LC480 platform (Roche Applied
Science, Mannheim, Germany) in a reaction mixture containing 5 µl
of 2×THUNDERBIRD™ Next SYBR® qPCR Mix, 1.2 µl of
profilin2-specific primer (forward: 5′-GCCTATACGTTGATGGTGACTG-3′,
and reverse: 5′-ACAAAGACCAAGACTCTCCCG-3′), 1 µl of cDNA and 2.8 µl
of DNase-free water in a total reaction volume of 10 µl per sample.
GAPDH (Hs99999903_m1) purchased from Applied Biosystems
(Foster City, CA, USA) was used as a control. Two micrograms of
total tissue RNA were reverse transcribed to generate cDNA using
the High Capacity RNA-to-cDNA Kit (Applied Biosystems, Foster City,
California, United States). The reaction conditions involved enzyme
activation at 95°Cx30 sec, followed by 50 cycles of denaturation at
95°Cx5 sec, 53°Cx20 sec for annealing, and 60°Cx30 sec for
extension (15). To analyze the
relative changes in the quantitative RT-PCR, we employed the
2(−ΔΔCt) method (16). Following
these reactions, high-resolution melting curve analysis was
conducted by heating the mixture from 60–99°C using a ramping
degree of 4.8°C/sec.
Detecting profilin splice forms by
RT-PCR/Sanger sequencing
Real time PCR was performed and analyzed following
the previously described protocol (15,16).
cDNA was generated as described above. Polymerase chain reaction
amplification was performed using profilin 2-specific
primers. The same 3′ primer (5′-GAGTCAAGGTGGGGAGCCAAC3′) and a
unique internal 5′ primer (PFN2A: 5′-TCAAGTATTTTGCCATTGAGTATGCC3′,
PFN2B: 5′-CCTCTTCAGGTATAAAGCGAGTTC3′) were used. The PCR products
were confirmed by Sanger sequencing. The PCR products were purified
using the ExoSAP-IT® Express PCR Product Clean-up
(Affymetrix, Santa Clara, California, United States) as per the
manufacturer's instructions. Sanger sequencing reactions were
prepared using the BigDye™ Terminator version 3.1 kit (Applied
Biosystems, Foster City, California, United States) as per the
manufacturer's instructions. Briefly, the sequencing reaction
consisted of 1.75 µl Sequencing Reaction Mix (BigDye™ Terminator
v3.1), 1 µl Sequencing Buffer (BigDye™ Terminator v1.1), 4.75 µl
nuclease-free water, 1 µl forward primer, 1 µl reverse primer and 1
µl PCR product. The sequencing reaction was subjected to the
following cycling conditions in a 2720 Thermal Cycler (Applied
Biosystems, Foster City, California, United States): one cycle at
96°C for 1 min, and 25 cycles at 96°C for 13 sec, 50°C for 8 sec,
and 60°C for 4 min. Sequence reactions were cleaned up with the
AxyPrep Mag DyeClean Kit (Axygen, Union City, CA, United States) as
per the manufacturer's instructions. Sequence raw reads were loaded
onto the Seq studio Genetic Analyzer (PE Applied Biosystems) with
Tracking Dye, Hi-Di™ Formamide (PE Applied Biosystems, Fostercity
CA, USA).
Statistical analysis
All results were expressed as means (± standard
deviations) without any notation. All statistical analyses were
performed using Stata software ver.14.2 (Stata Corp., College
Station, TX, USA). Statistical significance was set at P<0.05.
Hierarchical clustering was performed using TAC software (Thermo
Fisher Scientific), which utilizes the Euclidean distance metric
and complete linkage method for clustering. The χ2 test
was performed to compare the original and pulmonary metastatic
cells, and ANOVA followed by Bonferroni as a post hoc test were
used for the other comparisons without any notation.
Results
Evaluating the potential for lung
metastasis of CMT93-PM cells and their original counterpart cells,
CMT93
CMT93 is a commercially available cell line isolated
from the rectum of a mouse with polypoid carcinoma and CMT93-PM1-4
are cell lines that we previously established from lung metastasis
of CMT93 caused by tail vein injection (13). Initially, we confirmed the superior
metastatic function of CMT93-PM1 compared with that of CMT93.
CMT93-PM1 cells demonstrated a much more aggressive metastatic
function than CMT93 cells using the tail vein injection method in
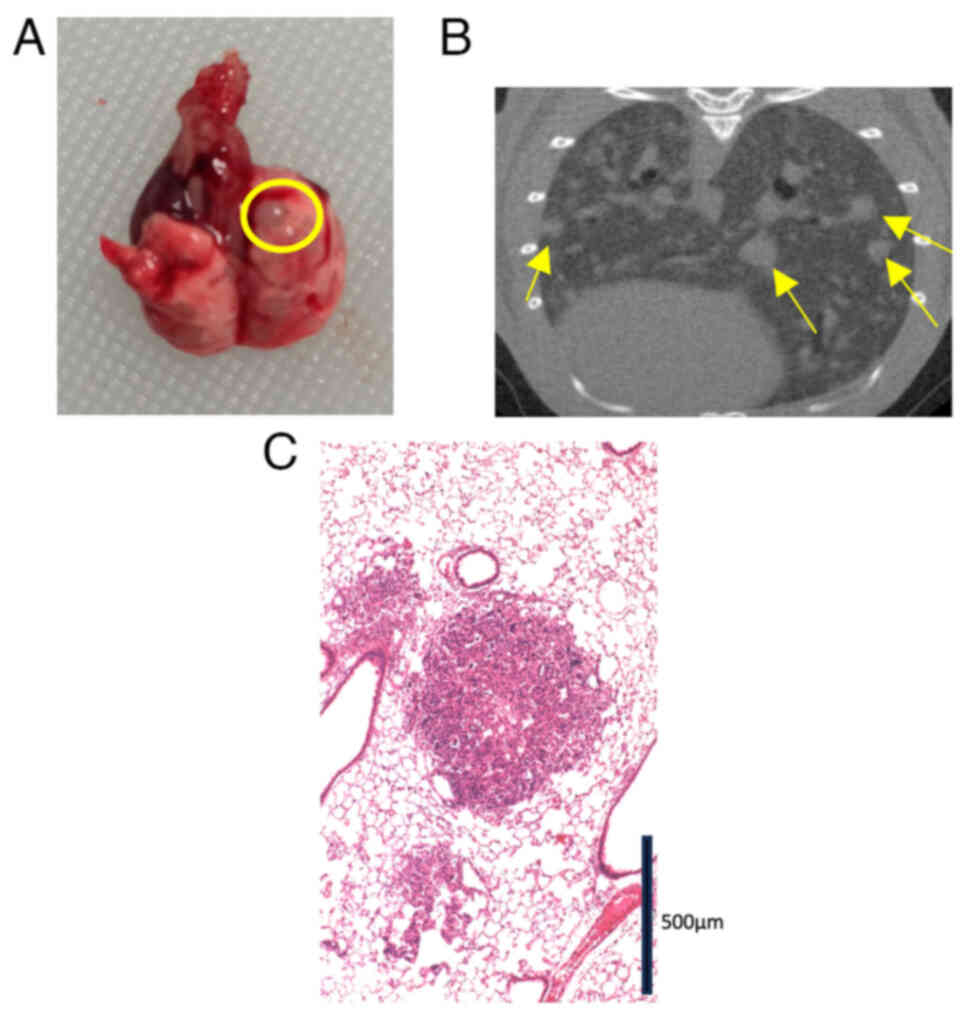
29 mice (Fig. 1). Visual inspection
(Fig. 1A) and CT (Fig. 1B) revealed numerous metastatic
tumors. We microscopically confirmed a metastatic lung lesion
(Fig. 1C). Although the rate of
lung metastasis was only 13.3% (2/15) in the CMT93 group, the
CMT93-PM1 group showed a 100% metastasis rate (14/14) (P<0.001)
(Table I).
 | Table I.Comparison of lung metastasis rates
between CMT93 and CMT93-PM1. |
Table I.
Comparison of lung metastasis rates
between CMT93 and CMT93-PM1.
| Cell line | Total mice, n | Mice with lung
metastasis, n | Rate of lung
metastasis, % | P-value |
|---|
| CMT93 | 15 | 2 | 13.3 | P<0.001 |
| CMT93-PM1 | 14 | 14 | 100.0 |
|
Microarray, cluster analysis, and
enrichment analysis by DAVID
We analyzed the gene expression profiles using
microarray analysis to investigate the mechanisms underlying lung
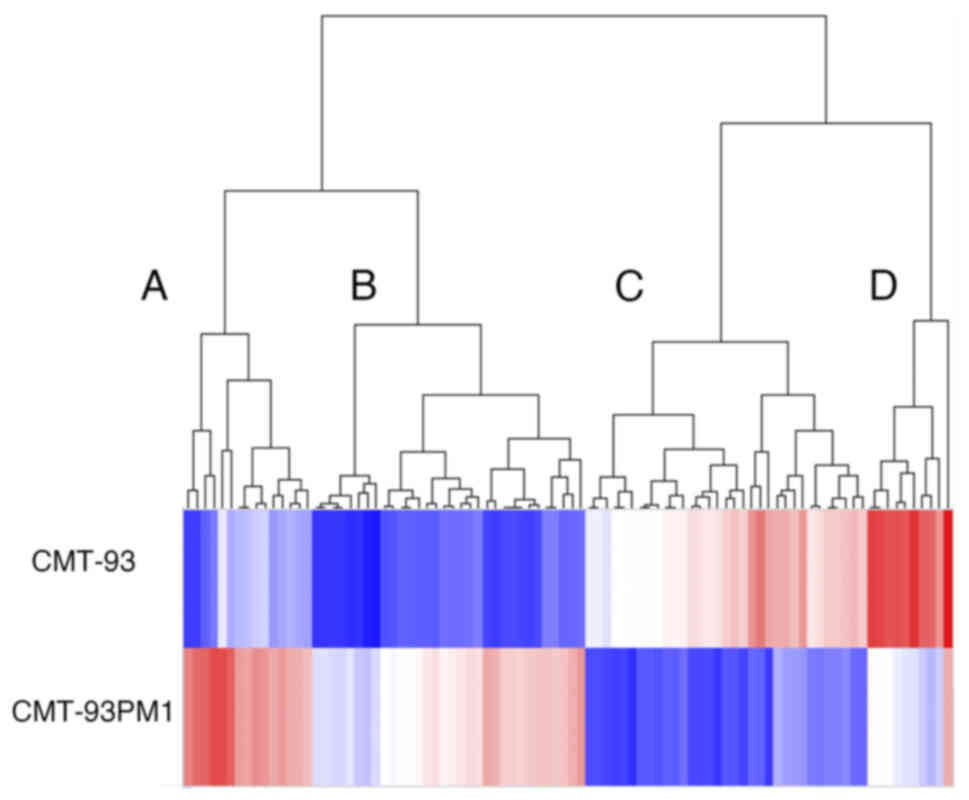
metastasis. We observed that 2241 and 88 genes showed differential
expression CMT93 and CMT93-PM1 with a fold change of more than 2.0
and 10, respectively (Fig. 2).
Among these 88 genes, 46 were upregulated and 42 were
downregulated. The 88 genes were further divided into two groups
using clustering analysis: genes that were upregulated or
downregulated in CMT93-PM1 cells. The genes in these two groups
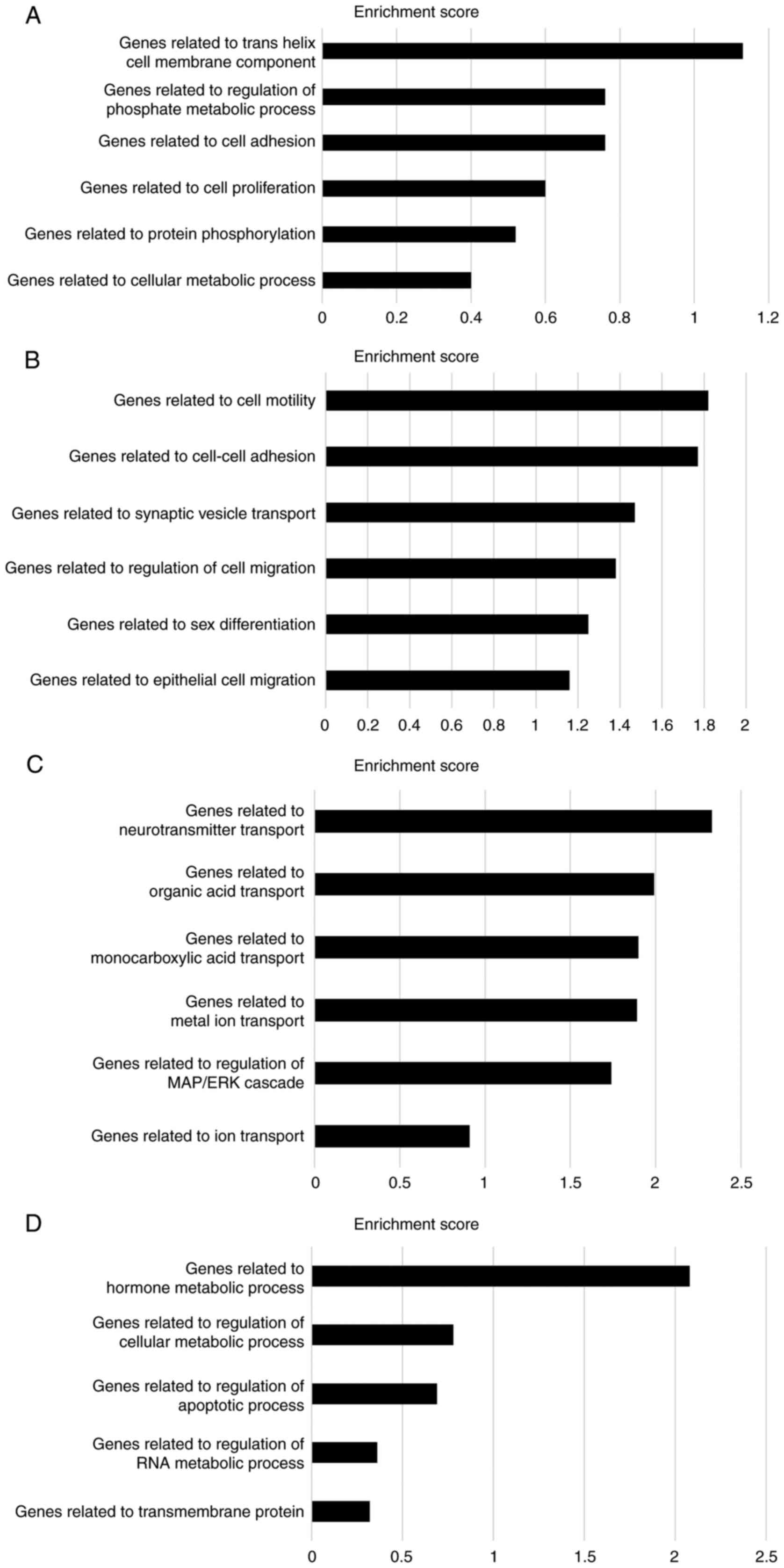
were further subdivided into two groups. Cluster A (the most
upregulated cluster in CMT93-PM1 cells compared to that in CMT93
cells) demonstrated phosphate metabolic processes, cell adhesion,
cell proliferation, protein phosphorylation, and cellular metabolic
processes. Cluster B (which included profilin 2) demonstrated
upregulation of genes related to cell motility, adhesion, and
migration. Clusters C and D consisted of genes that were
downregulated in CMT93-PM1 cells compared to those in CMT93 cells.
Cluster C contains several genes related to molecular transport.
Cluster D contains genes related to hormone metabolism and
regulation of cellular metabolic processes (Fig. 3A-D).
Evaluating PFN2 expression in CMT93-PM
using RT-PCR and melting curve analysis
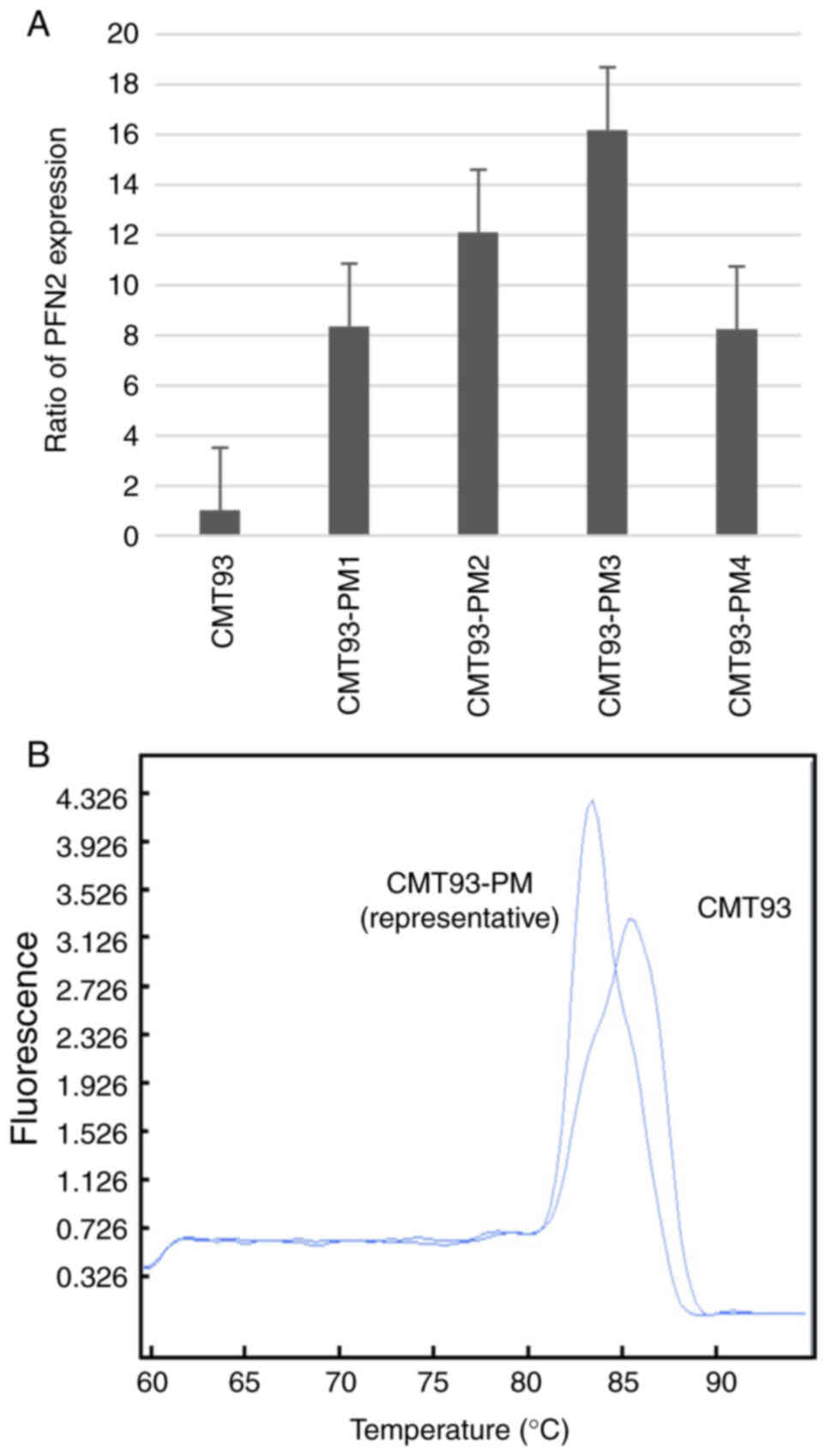
We focused on PFN2 because it showed the highest
level of overexpression in CMT93-PM1 cells (Table SI). We confirmed the differential
expression of PFN2 in the four cell lines of the CMT93-PM series
using RT-PCR. The PFN2 expression level was higher in
CMT93-PM1-4 cells than in CMT93 cells, however we could not confirm
a significant difference (Fig. 4A).
These two PFN2 isoforms have distinct functions in malignancy
(17). Two different melting
temperatures suggested the presence of two different types of
PFN2 following melting curve analysis (Fig. 4B). PFN2A and PFN2B isoforms are
splicing variants of PFN2 with different distributions
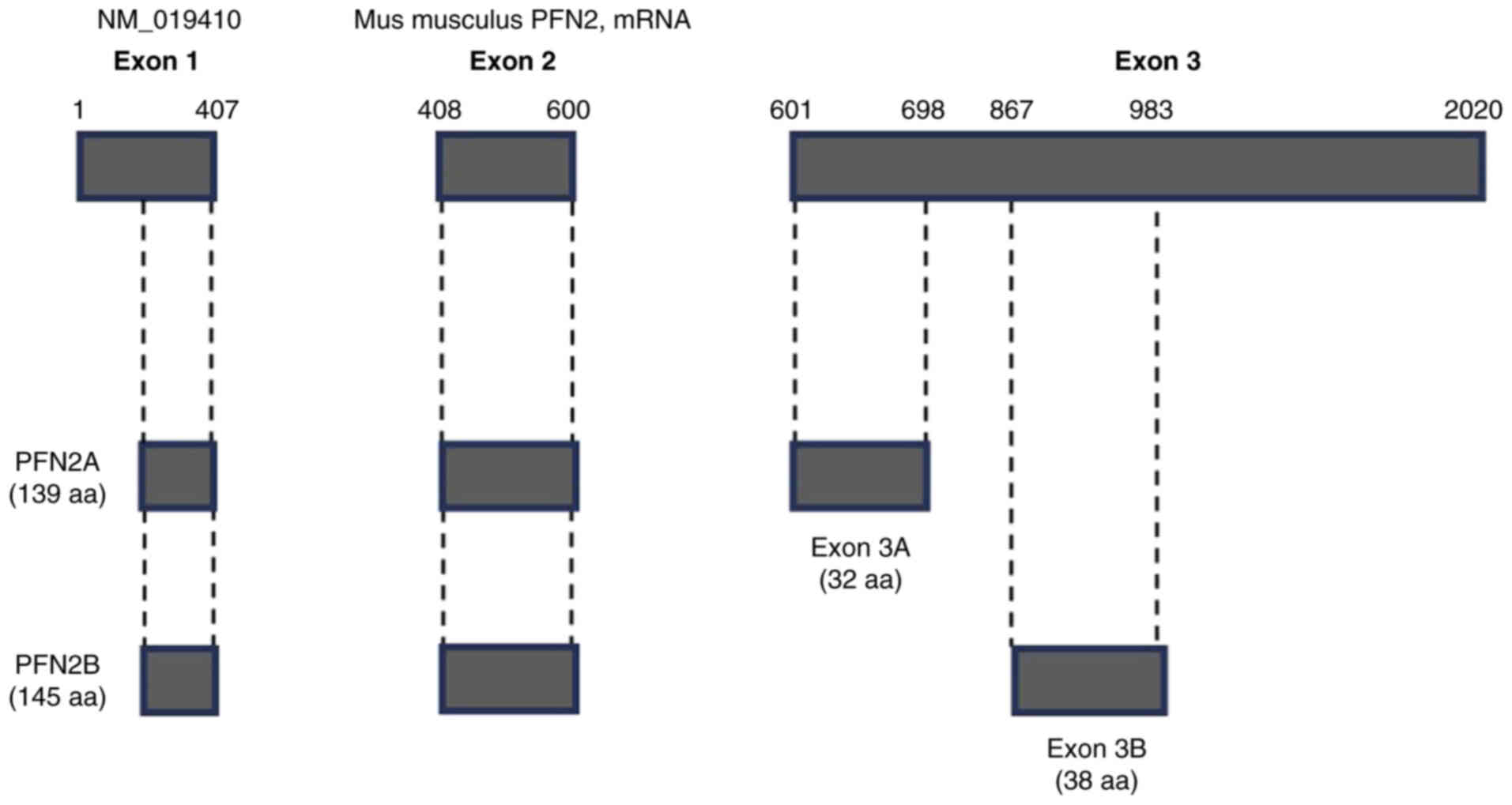
(18). The overall structure of
PFN2 is illustrated in Fig. 5
(18,19). We confirmed that PFN2A showed
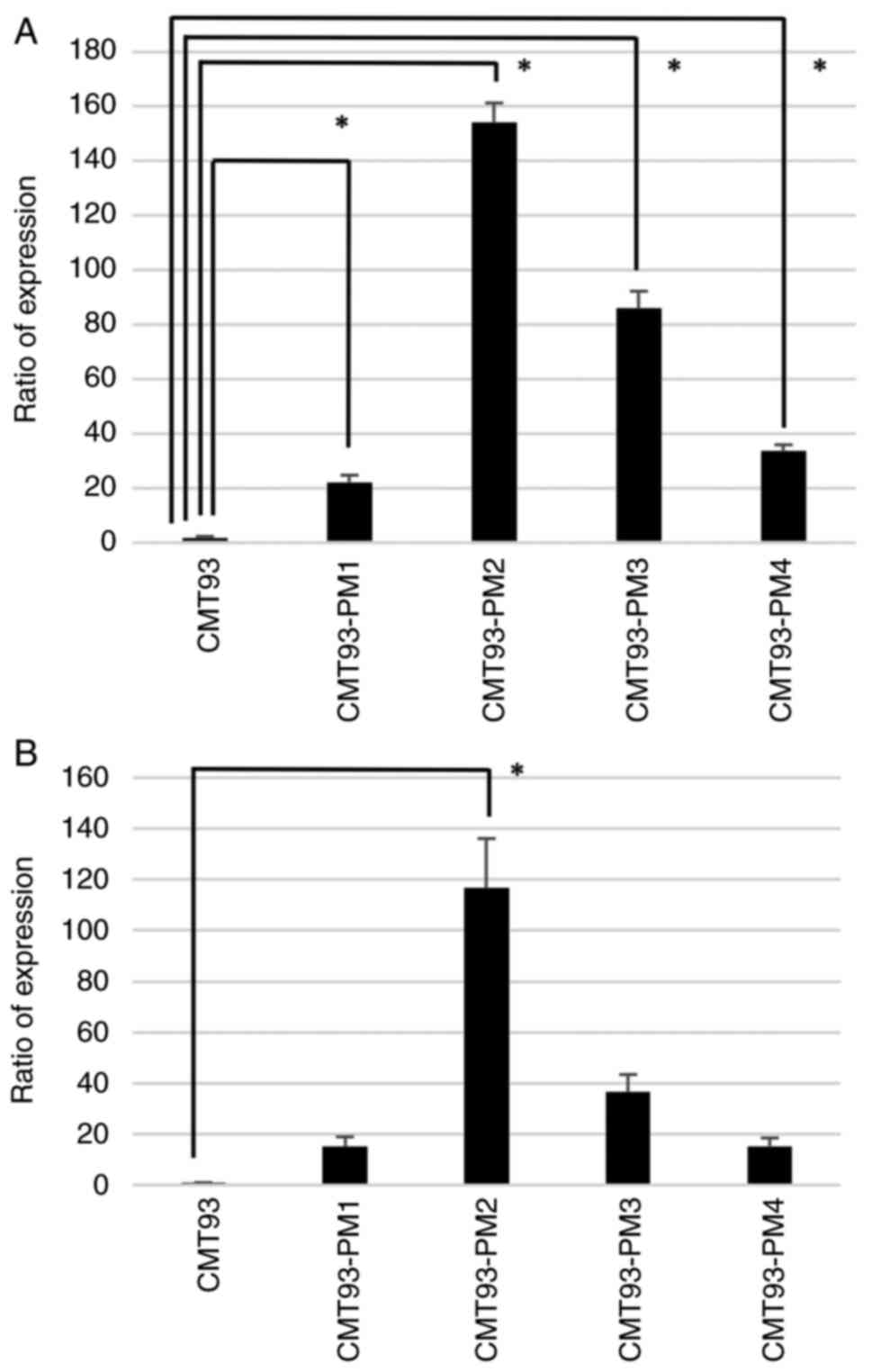
significantly higher expression in CMT93-PM1-4 cell lines than in
CMT93 cells (P<0.01) (Fig. 6A).
As for PFN2B, we observed overexpression in CMT93-PM1-4 cell lines
and a significant difference in CMT93-PM2 cells (P<0.01).
Discussion
The molecular mechanisms underlying lung metastasis
in CRC have been extensively studied (8,9,12,13,20–23).
However, the number of patients included in these studies was
insufficient to obtain conclusive results owing to the
heterogeneity of metastatic tumors (8,12,21).
We can avoid the effects of tumor heterogeneity owing to varying
patient backgrounds (such as a history of smoking, obesity,
familial genomic problems, or other diseases, and previous
treatments, such as chemotherapy and radiotherapy) using
experimental animal models and cell lines. In a previous study, we
injected mouse rectal cancer cells into the tail vein and observed
lung metastases (13). We retrieved
metastatic tumors and established their cell lines. To the best of
our knowledge, this is the first cell line reported for the
treatment of metastatic lung colorectal cancer. While three studies
report the establishment or utilization of cell lines for liver
metastases (24–26), none have focused on the
establishment of cell lines for lung metastases. We found that the
lung surfactant SP-D suppressed lung metastasis by inhibiting the
epidermal growth factor receptor signaling using established cell
lines. Our previous study suggests that environmental factors might
affect lung metastasis in CRC (13). In this study, we conducted a global
gene expression analysis to reveal the background of lung
metastasis.
Initially, we confirmed the metastatic ability of
CMT93-PM1 cells. Metastatic sites were formed by numerous
homogeneous lesions in all the lungs. This form of lung metastasis
differs from that observed in clinical situations because most
human lung metastatic lesions form distinct nodules (27). However, this may be owing to the
high clonality of the tumor cells, immune evasion specialized in
the lung immune system, and the tail vein injection model. Multiple
mechanisms can influence this phenomenon, including the
upregulation of cell motility, cell adhesion, and invasion as well
as the downregulation of tumor suppressor genes and the immune
system. Therefore, we believe that employing technologies such as
microarrays and investigating changes in a wide range of genomes
are essential. This study provides new insights from this
perspective.
Microarray analysis revealed that 88 genes were
expressed with a fold change exceeding 10 times in CMT93-PM cells
compared to that in CMT93 cells. Cluster analysis revealed four
distinct clusters and subsequent enrichment analysis indicated that
these clusters exhibited distinctive features. Clusters A and B
consisting of genes upregulated in CMT93-PM1 cells were primarily
associated with cell adhesion, proliferation, and motility. These
functions are crucial for the formation of distant metastases,
consistent with the results of previous studies (28–30).
Notably, N-cadherin and VE-cadherin found within this cluster play
significant roles in cell adhesion and migration and exhibit strong
interactions with the wnt/β-catenin signaling pathway (31). The upregulation of N-cadherin
expression in CRC tissues is negatively correlated with E-cadherin
expression; furthermore, high N-cadherin expression promotes
proliferation and migration of CRC cells by inducing EMT (32). Six4 is another upregulated
gene in cluster A and B that activates Akt and triggers the
PI3K/Akt pathway (33). The
PI3K/AKT/mammalian target of rapamycin signaling pathway is one of
the most pivotal intracellular pathways, serving as a master
regulator of cancer and enhancing cell growth and proliferation
(34). Additionally, VCAM1 in this
cluster promotes CRC invasion and metastasis by activating the EMT
(35).
In contrast, clusters C and D comprised
downregulated genes in CMT93-PM, and the roles of the genes in
these clusters were markedly different from those in clusters A and
B. Notably, a tumor suppressor gene [tissue inhibitor of
metalloproteinase-3 (Timp3)] was found in these
clusters. TIMP3 plays various roles, one of which is the induction
of apoptosis through caspase-dependent and-independent pathways
(36). A decrease in Timp3
expression correlates with the malignant behavior of colorectal
cancer and TIMP3 overexpression results in reduced adhesion,
migration, and invasion of human CRC cells in vitro
(37). PAX6 was detected in
clusters C and D; it serves as a master control gene for the
development of the eyes, other sensory organs, certain neural and
epidermal tissues, and other homologous structures typically
derived from ectodermal tissues. PAX6 is a potential tumor
suppressor (38). Further
investigations are required to assess the relationship between PAX6
and colorectal cancer and the several differences between CMT93 and
CMT93-PM cells shown in this study.
We observed an overexpression of PFN2 in the
CMT93-PM series compared to that in the original CMT93 cells.
Furthermore, PFN2a and PFN2b splicing variants were
upregulated.
Profilins are actin-binding proteins that regulate
cell structure by controlling signal-dependent actin polymerization
(39). The profilin gene
family comprises two major isoforms: PFN1 and PFN2
(40). Mouse profilin I is
ubiquitously expressed in all tissues except skeletal muscle,
whereas profilin IIA is mainly expressed in neuronal tissues
(41). Thus, profilin I
appears to be a housekeeping gene that is important for vital
functions, whereas profilin II was later developed to serve
specific functions, mainly in the brain. PFN2 is alternately
spliced into PFN2a and 2b. Among these, PFN2a is the predominant
splice form (18,19) that is conserved across vertebrates,
including humans, mice, chickens, and cattle. Research on the
functions of PFN2a has primarily focused on cell migration and the
mammalian nervous system, including synaptic vesicle exocytosis and
neuronal excitability (42). The
relationship between PFN2 and lung, breast, and prostate cancers
was previously investigated (17,39,43,44).
PFN2 regulates growth by inhibiting nuclear localization of histone
deacetylase 1 (HDAC1) in lung cancer (43). It promotes the progression of small
cell lung cancer in vitro and in vivo and regulates
angiogenesis in the tumor microenvironment through cancer-derived
exosomes (45).
Only two reports have described the role of PFN2 in
colorectal cancer and no previous study has described the
relationship between PFN2 isoforms and colorectal cancer. PFN2
promotes the metastatic potential and stemness of colorectal CSCs
by regulating EMT- and stemness-related proteins (45). These findings were consistent with
our results. In contrast, the loss of PFN2 contributes to
enhanced epithelial-mesenchymal transition and metastasis (46). Their observations are inconsistent
with our results, since they concluded that the loss of PFN2
exacerbates metastasis, leading them to insist that PFN2 inhibits
tumor metastasis. However, their study has several limitations.
First, although they concluded that PFN2 downregulation enhanced
cell migration, they compared two different cell lines: SW620 and
HCT116. Different cell lines have distinct genetic backgrounds in
addition to PFN2 expression. Therefore, comparisons between
different cell lines are not particularly informative. Second, a
retrovirus was used to transfect PFN2 into SW620 cells.
However, the retroviral vector method does not offer precise
control over the location of transfection and is often associated
with the activation of tumor-related genes. Third, in vitro
experiments were conducted, and the results were validated in
vivo. These factors could have contributed to the potential
differences between the results of their study and ours.
Our study has some limitations. First, we used a
single cell line (CMT93) as a model. Therefore, it is essential to
examine multiple cell lines to validate our results. Secondly, we
used the tail vein injection method with CMT93-PM and confirmed a
significant difference in metastatic function between CMT93-PM and
CMT93. However, the tail vein injection method has positive and
negative aspects for evaluating lung metastasis. In this method,
metastasis is initiated via tail vein injection, which mimics the
natural progression of colorectal cancer. After injection, tumor
cells were disseminated swiftly through systemic venous perfusion.
However, our experiment did not recapitulate metastasis in these
patients because the tumor cells were dispersed by proteinase
before injection. Thirdly, we used the cells derived from the
metastatic tumors in lungs, and the similarity of the established
cell lines with the original tumors, and the homogeneity of the
cells from the metastatic lesions were not be verified. Fourth, our
experiments are solely based on RNA and cDNA, lacking evidence of
protein expression levels for the two different types of PFN2. This
was due to the unavailability of the corresponding antibodies
commercially. Finally, we did not study the effects of PFN2
overexpression on cell behavior. Further investigations into the
function of PFN2 and the relationship between lung metastasis and
PFN2 are necessary to validate the clinical utility of our
findings. This validation may be achieved using pathological
materials and publicly available datasets. Additionally, the role
of each variant remains unclear. Studies on PFN2a and PFN2b will
provide deeper insights into the molecular mechanisms of lung
metastasis in CRC.
We established an in vitro lung metastasis
model using colorectal cancer cells and mice. Differential gene
expression analysis was used to identify genes associated with lung
metastasis. Thus, our model provides a unique platform to study
lung metastasis. The functional significance and clinical utility
of these two PFN2 isoforms should be considered in future
studies.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms. Kaoru Hattori
(Department of Surgery, Keio University School of Medicine, Tokyo,
Japan) for their technical support and handling of the mice, Dr.
Shingo Akimoto (Department of Surgery, Keio University School of
Medicine, Tokyo, Japan) for their support during this study, and
Dr. Shinya Saito (Oncogene Research Unit/Cancer Prevention Unit,
Tochigi Cancer Center Research Institute, Utsunomiya, Japan) for
providing insightful instructions and support during the
experiments.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated and analyzed in the current
study are available from the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE252603).
GEO accession number is GSE252603. The rest of the data generated
in the present study may be requested from the corresponding
author.
Authors' contributions
NT and MT participated in the study design,
coordination, and drafting of manuscript. YT and YY performed the
experiments and acquired data. KS and KO participated in the study
design and performed statistical analyses. MT and TK confirm the
authenticity of all the raw data. HH, SF, TK, IO, and YK conceived
the study, participated in its design and coordination, and helped
draft the manuscript. All authors have read and approved the final
manuscript and agree to be accountable for all aspects of the
study.
Ethics approval and consent to
participate
All procedures were performed with the approval of
the Laboratory Animal Care and Use Committee at the Keio University
School of Medicine (approval no. 15006).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
CT
|
computed tomography
|
|
DAVID
|
Database for Annotation, Visualization
and Integrated Discovery
|
|
HDAC1
|
histone deacetylase 1
|
|
PFN2
|
Profilin 2
|
|
RT-PCR
|
reverse transcription PCR
|
|
TIMP3
|
tissue inhibitor of
metalloproteinase-3
|
References
|
1
|
Keum N and Giovannucci E: Global burden of
colorectal cancer: Emerging trends, risk factors and prevention
strategies. Nat Rev Gastroenterol Hepatol. 16:713–732. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Van Cutsem E, Cervantes A, Adam R, Sobrero
A, Van Krieken JH, Aderka D, Aguilar EA, Bardelli A, Benson A,
Bodoky G, et al: ESMO consensus guidelines for the management of
patients with metastatic colorectal cancer. Ann Oncol.
27:1386–1422. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Muratore A, Zorzi D, Bouzari H, Amisano M,
Massucco P, Sperti E and Capussotti L: Asymptomatic colorectal
cancer with un-resectable liver metastases: Immediate colorectal
resection or up-front systemic chemotherapy? Ann Surg Oncol.
14:766–770. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Van Cutsem E, Nordlinger B, Adam R, Köhne
CH, Pozzo C, Poston G, Ychou M and Rougier P; European Colorectal
Metastases Treatment Group, : Towards a pan-European consensus on
the treatment of patients with colorectal liver metastases. Eur J
Cancer. 42:2212–2221. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Parnaby CN, Bailey W, Balasingam A,
Beckert L, Eglinton T, Fife J, Frizelle FA, Jeffery M and Watson
AJ: Pulmonary staging in colorectal cancer: A review. Colorectal
Dis. 14:660–670. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Watanabe T, Muro K, Ajioka Y, Hashiguchi
Y, Ito Y, Saito Y, Hamaguchi T, Ishida H, Ishiguro M, Ishihara S,
et al: Japanese society for cancer of the colon and rectum (JSCCR)
guidelines 2016 for the treatment of colorectal cancer. Int J Clin
Oncol. 23:1–34. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chandra R, Karalis JD, Liu C, Murimwa GZ,
Voth Park J, Heid CA, Reznik SI, Huang E, Minna JD and Brekken RA:
The colorectal cancer tumor microenvironment and its impact on
liver and lung metastasis. Cancers (Basel). 13:62062021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang N, Di J, Wang Z, Gao P, Jiang B and
Su X: Genomic profiling of colorectal cancer with isolated lung
metastasis. Cancer Cell Int. 20:2812020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamamoto T, Kawada K, Itatani Y, Inamoto
S, Okamura R, Iwamoto M, Miyamoto E, Chen-Yoshikawa TF, Hirai H,
Hasegawa S, et al: Loss of SMAD4 promotes lung metastasis of
colorectal cancer by accumulation of CCR1+ tumor-associated
neutrophils through CCL15-CCR1 axis. Clin Cancer Res. 23:833–844.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu J, Shao Y, He Y, Ning K, Cui X, Liu F,
Wang Z and Li F: MORC2 promotes development of an aggressive
colorectal cancer phenotype through inhibition of NDRG1. Cancer
Sci. 110:135–146. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu XT, Xing W, Zhao RS, Tan Y, Wu XF, Ao
LQ, Li Z, Yao MW, Yuan M, Guo W, et al: HDAC2 inhibits EMT-mediated
cancer metastasis by downregulating the long noncoding RNA H19 in
colorectal cancer. J Exp Clin Cancer Res. 39:2702020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim SH, Choi SJ, Cho YB, Kang MW, Lee J,
Lee WY, Chun HK, Choi YS, Kim HK, Han J and Kim J: Differential
gene expression during colon-to-lung metastasis. Oncol Rep.
25:629–636. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tajima Y, Tsuruta M, Hasegawa H,
Okabayashi K, Ishida T, Yahagi M, Makino A, Koishikawa K, Akimoto
S, Sin DD and Kitagawa Y: Association of surfactant protein D with
pulmonary metastases from colon cancer. Oncol Lett. 20:3222020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
David bioinformatics resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35:W169–W175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang R, Yuan DJ, Li S, Liang XS, Gao Y,
Lan XY, Qin HM, Ma YF, Xu GY, Schachner M, et al: NCAM regulates
temporal specification of neural progenitor cells via profilin2
during corticogenesis. J Cell Biol. 219:e2019021642020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mouneimne G, Hansen SD, Selfors LM, Petrak
L, Hickey MM, Gallegos LL, Simpson KJ, Lim J, Gertler FB, Hartwig
JH, et al: Differential remodeling of actin cytoskeleton
architecture by profilin isoforms leads to distinct effects on cell
migration and invasion. Cancer Cell. 22:615–630. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lambrechts A, Braun A, Jonckheere V,
Aszodi A, Lanier LM, Robbens J, Van Colen I, Vandekerckhove J,
Fässler R and Ampe C: Profilin II is alternatively spliced,
resulting in profilin isoforms that are differentially expressed
and have distinct biochemical properties. Mol Cell Biol.
20:8209–8219. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Di Nardo A, Gareus R, Kwiatkowski D and
Witke W: Alternative splicing of the mouse profilin II gene
generates functionally different profilin isoforms. J Cell Sci.
113:3795–3803. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang H, Fu W, Im JH, Zhou Z, Santoro SA,
Iyer V, DiPersio CM, Yu QC, Quaranta V, Al-Mehdi A and Muschel RJ:
Tumor cell alpha3beta1 integrin and vascular laminin-5 mediate
pulmonary arrest and metastasis. J Cell Biol. 164:935–941. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shu R, Xu Y, Tian Y, Zeng Y, Sun L, Gong
F, Lei Y, Wang K and Luo H: Differential expression profiles of
long noncoding RNA and mRNA in colorectal cancer tissues from
patients with lung metastasis. Mol Med Rep. 17:5666–5675.
2018.PubMed/NCBI
|
|
22
|
Tang L, Lei YY, Liu YJ, Tang B and Yang
SM: The expression of seven key genes can predict distant
metastasis of colorectal cancer to the liver or lung. J Dig Dis.
21:639–649. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dai W, Guo C, Wang Y, Li Y, Xie R, Wu J,
Yao B, Xie D, He L, Li Y, et al: Identification of hub genes and
pathways in lung metastatic colorectal cancer. BMC Cancer.
23:3232023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu M, Zessin AS, Glover W and Hsu DS:
Activation of the mTOR pathway by oxaliplatin in the treatment of
colorectal cancer liver metastasis. PLoS One. 12:e01694392017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li LN, Zhang HD, Yuan SJ, Tian ZY and Sun
ZX: Establishment and characterization of a novel human colorectal
cancer cell line (CLY) metastasizing spontaneously to the liver in
nude mice. Oncol Rep. 17:835–840. 2007.PubMed/NCBI
|
|
26
|
Govaert KM, Emmink BL, Nijkamp MW, Cheung
ZJ, Steller EJ, Fatrai S, de Bruijn MT, Kranenburg O and Rinkes IH:
Hypoxia after liver surgery imposes an aggressive cancer stem cell
phenotype on residual tumor cells. Ann Surg. 259:750–759. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Salah S, Ardissone F, Gonzalez M, Gervaz
P, Riquet M, Watanabe K, Zabaleta J, Al-Rimawi D, Toubasi S, Massad
E, et al: Pulmonary metastasectomy in colorectal cancer patients
with previously resected liver metastasis: Pooled analysis. Ann
Surg Oncol. 22:1844–1850. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang JY, Sun J, Huang MY, Wang YS, Hou MF,
Sun Y, He H, Krishna N, Chiu SJ, Lin S, et al: STIM1 overexpression
promotes colorectal cancer progression, cell motility and COX-2
expression. Oncogene. 34:4358–4367. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jackstadt R, Röh S, Neumann J, Jung P,
Hoffmann R, Horst D, Berens C, Bornkamm GW, Kirchner T, Menssen A
and Hermeking H: AP4 is a mediator of epithelial-mesenchymal
transition and metastasis in colorectal cancer. J Exp Med.
210:1331–1350. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eckey M, Kuphal S, Straub T, Rümmele P,
Kremmer E, Bosserhoff AK and Becker PB: Nucleosome remodeler SNF2L
suppresses cell proliferation and migration and attenuates Wnt
signaling. Mol Cell Biol. 32:2359–2371. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Katoh M: Multi-layered prevention and
treatment of chronic inflammation, organ fibrosis and cancer
associated with canonical WNT/β-catenin signaling activation
(Review). Int J Mol Med. 42:713–725. 2018.PubMed/NCBI
|
|
32
|
Yan X, Yan L, Liu S, Shan Z, Tian Y and
Jin Z: N-cadherin, a novel prognostic biomarker, drives malignant
progression of colorectal cancer. Mol Med Rep. 12:2999–3006. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun X, Hu F, Hou Z, Chen Q, Lan J, Luo X,
Wang G, Hu J and Cao Z: SIX4 activates Akt and promotes tumor
angiogenesis. Exp Cell Res. 383:1114952019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang J, Nie J, Ma X, Wei Y, Peng Y and Wei
X: Targeting PI3K in cancer: Mechanisms and advances in clinical
trials. Mol Cancer. 18:262019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang D, Bi J, Liang Q, Wang S, Zhang L,
Han F, Li S, Qiu B, Fan X, Chen W, et al: VCAM1 promotes tumor cell
invasion and metastasis by inducing EMT and transendothelial
migration in colorectal cancer. Front Oncol. 10:10662020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kallio JP, Hopkins-Donaldson S, Baker AH
and Kähäri VM: TIMP-3 promotes apoptosis in nonadherent small cell
lung carcinoma cells lacking functional death receptor pathway. Int
J Cancer. 128:991–996. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin H, Zhang Y, Wang H, Xu D, Meng X, Shao
Y, Lin C, Ye Y, Qian H and Wang S: Tissue inhibitor of
metalloproteinases-3 transfer suppresses malignant behaviors of
colorectal cancer cells. Cancer Gene Ther. 19:845–851. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kiselev Y, Andersen S, Johannessen C,
Fjukstad B, Olsen KS, Stenvold H, Al-Saad S, Donnem T, Richardsen
E, Bremnes RM and Busund LT: Transcription factor PAX6 as a novel
prognostic factor and putative tumour suppressor in non-small cell
lung cancer. Sci Rep. 8:50592018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ma CY, Zhang CP, Zhong LP, Pan HY, Chen
WT, Wang LZ, Andrew OW, Ji T and Han W: Decreased expression of
profilin 2 in oral squamous cell carcinoma and its
clinicopathological implications. Oncol Rep. 26:813–823.
2011.PubMed/NCBI
|
|
40
|
Jeong DH, Choi YN, Seo TW, Lee JS and Yoo
SJ: Ubiquitin-proteasome dependent regulation of Profilin2 (Pfn2)
by a cellular inhibitor of apoptotic protein 1 (cIAP1). Biochem
Biophys Res Commun. 506:423–428. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Witke W, Podtelejnikov AV, Di Nardo A,
Sutherland JD, Gurniak CB, Dotti C and Mann M: In mouse brain
profilin I and profilin II associate with regulators of the
endocytic pathway and actin assembly. EMBO J. 17:967–976. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Boyl PP, Di Nardo A, Mulle C,
Sassoè-Pognetto M, Panzanelli P, Mele A, Kneussel M, Costantini V,
Perlas E, Massimi M, et al: Profilin2 contributes to synaptic
vesicle exocytosis, neuronal excitability, and novelty-seeking
behavior. EMBO J. 26:2991–3002. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tang YN, Ding WQ, Guo XJ, Yuan XW, Wang DM
and Song JG: Epigenetic regulation of Smad2 and Smad3 by profilin-2
promotes lung cancer growth and metastasis. Nat Commun. 6:82302015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Obinata D, Funakoshi D, Takayama K, Hara
M, Niranjan B, Teng L, Lawrence MG, Taylor RA, Risbridger GP,
Suzuki Y, et al: OCT1-target neural gene PFN2 promotes tumor growth
in androgen receptor-negative prostate cancer. Sci Rep.
12:60942022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cao Q, Liu Y, Wu Y, Hu C, Sun L, Wang J,
Li C, Guo M, Liu X, Lv J, et al: Profilin 2 promotes growth,
metastasis, and angiogenesis of small cell lung cancer through
cancer-derived exosomes. Aging (Albany NY). 12:25981–25999. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang H, Yang W, Yan J, Zhou K, Wan B, Shi
P, Chen Y, He S and Li D: Loss of profilin 2 contributes to
enhanced epithelial-mesenchymal transition and metastasis of
colorectal cancer. Int J Oncol. 53:1118–1128. 2018.PubMed/NCBI
|